Abstract
Staphylococcus aureus regulates the production of extracellular virulence factors using the agr quorum-sensing system. This regulatory system responds to a secreted peptide thiolactone signal called an autoinducing peptide (AIP). The biosynthesis of AIP requires AgrD, the peptide precursor of AIP, and the integral membrane endopeptidase AgrB. In this study, we performed a molecular analysis of AgrD to identify peptide regions important for processing and AIP secretion. As a lead-in to this study, we discovered that AIP type I could be generated in Escherichia coli through the heterologous expression of the agrBD genes, allowing the use of E. coli as an expression host for investigating the biosynthetic pathway. One of the most conserved regions of AgrD is the charged C-terminal tail, and through truncation analysis, the first nine residues were found to be essential for AIP production and AgrB endopeptidase activity. Within this essential region, mutation of residues glutamate 34 or leucine 41 inhibited AIP production and AgrB activity. Following cleavage, AgrB is hypothesized to form an enzyme-bound intermediate with the AgrD N-terminal region, but clear evidence of this intermediate has never been presented. By inactivating the AgrD cysteine 28 residue, an AgrD-AgrB structure was stabilized and detected in immunoblots using N-terminal His6-tagged AgrD. Formation of the structure could not be detected using the AgrB C84S mutation, indicating the cysteine residue is essential for its formation. These studies provide new insights on the requirements and mechanism of S. aureus AIP biosynthesis.
Staphylococcus aureus is a commensal of humans and other mammals and an opportunistic pathogen capable of causing a wide range of infections (1). S. aureus secretes a diverse arsenal of invasive virulence factors, including hemolysins, superantigens, and tissue-degrading enzymes, which all contribute to pathogenesis. The temporal regulation of these virulence factors is controlled in large part by the accessory gene regulator (agr) quorum-sensing system (2). The agr system responds to the extracellular concentration of a peptide signal, called an autoinducing peptide (AIP).2 Once AIP accumulates to a critical concentration, the agr system activates and up-regulates secretion of virulence factors while down-regulating production of surface adhesins (3). The importance of the agr locus has been tested in various animal infection models, which have demonstrated that inhibiting agr reduces acute virulence (4–7). Recently, the agr system has also been found to be a lifestyle switch that can control the S. aureus decision to attach and develop into a biofilm or remain in a planktonic, invasive state (8).
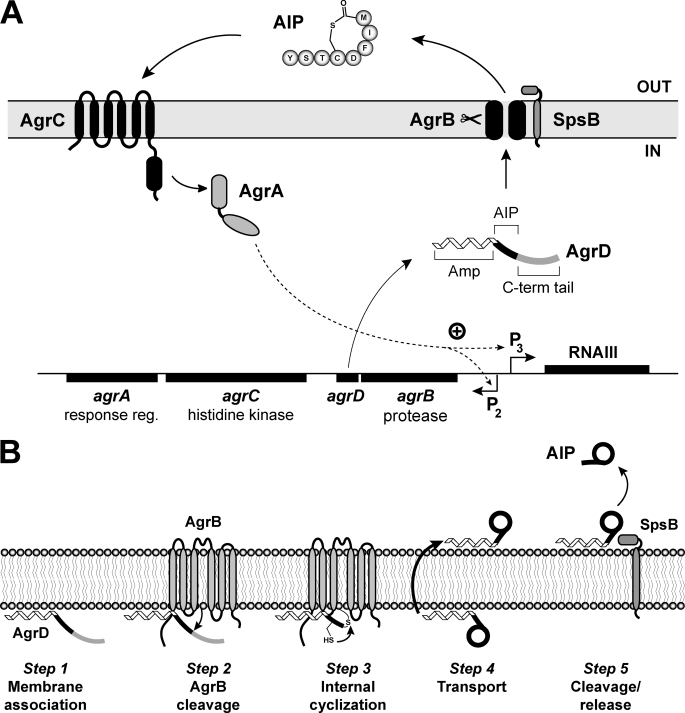
The agr locus has been the focus of extensive characterization and is composed of two divergently transcribed RNA molecules (Fig. 1A). The agr P2 promoter produces the RNAII transcript that contains the agrBDCA operon, encoding the majority of factors necessary for a functional agr system. In order of genetic encoding, AgrB is an integral membrane endopeptidase essential for AIP production, and AgrD is the propeptide precursor of AIP. AgrC and AgrA work together as a two-component regulatory pair, and these two proteins function as a histidine kinase and response regulator, respectively. Once AIP has accumulated to a critical concentration, the signal will bind to an extracellular receptor on AgrC, activating the kinase. Then AgrC will phosphorylate AgrA, and in turn, activated AgrA will bind and induce transcription from the agr P2 and P3 promoters (9). The primary effector of the agr cascade is the RNAIII transcript, which is a large regulatory RNA molecule produced from the divergent agr P3 promoter. RNAIII production then leads to altered expression of virulence factors (10).
FIGURE 1.
Schematic of the S. aureus agr system and a model for AIP biosynthesis. A, chromosomal agr locus is composed of two divergently expressed transcripts. Promoter P2 drives expression of the agrBDCA operon. AgrD is the peptide precursor of AIP (type I is shown) that is composed of three parts, including an N-terminal amphipathic helix (Amp), a middle region that becomes AIP, and a C-terminal (C-term) charged tail. AgrB is an integral membrane endopeptidase essential for AIP production. AgrC is a membrane histidine kinase that has an extracellular receptor for AIP binding. AgrA is a response regulator and part of a two-component pair with AgrC. Activated AgrA induces transcription from promoters P2 and P3, and the P3 promoter drives expression of RNAIII, the primary effector of the agr system. SpsB is the housekeeping type I signal peptidase that completes the AIP biosynthetic pathway. B, AIP biosynthetic pathway model. Step 1, the N-terminal amphipathic helix associates with the cytoplasmic membrane. Step 2, AgrB removes the C-terminal tail of AgrD. Step 3, the remaining AgrD peptide fragment (N-terminal and AIP portion) is bound to AgrB as a thioester at AgrB residue Cys-84. The AgrD cysteine residue can attack the thioester bond displacing the peptide and forming the thiolactone ring. Step 4, the AIP precursor (N-terminal helix and AIP thiolactone) is transported to the outer face of the membrane. Step 5, SpsB removes the amphipathic helix, releasing AIP from the membrane.
Although many studies have examined the downstream effects of agr activation, the mechanistic steps that lead to AIP secretion remain to be fully clarified. Summarizing published reports (11–14), a consensus model for the AIP biosynthetic pathway is shown in Fig. 1B. In step 1, AgrD propeptide localizes to the cytoplasmic membrane using the N-terminal amphipathic leader. In step 2, the AgrB endopeptidase activity removes the AgrD C-terminal tail. In step 3, following cleavage, an AgrD-AgrB acyl-enzyme intermediate forms that is composed of the AgrD N terminus (leader and AIP regions) bound through a thioester to cysteine 84 of AgrB. In the intermediate structure, AgrD cysteine residue 28 catalyzes thioester exchange, allowing thiolactone ring formation and release from AgrB. In step 4, through an undetermined mechanism, the AIP precursor (N-terminal leader fused to AIP) is transported to the outer face of the cell membrane, either by AgrB function or an unknown protein. Finally, in step 5, type I signal peptidase SpsB finishes the pathway and removes the N-terminal amphipathic helix (14). Although this model provides a potential pathway through which the AIP molecule could be synthesized, many of the mechanistic details have yet to be demonstrated experimentally.
In this study, we have focused on steps 2 and 3 of this model and investigated the AgrD features required for AgrB cleavage and AIP production. Aiding these studies was our discovery that AIP can be produced through heterologous expression in E. coli. Through the construction and characterization of AgrD mutants, we have determined that the C-terminal tail plays an essential role in AgrB cleavage and AIP production. By interrupting the biosynthetic pathway, we have detected a structure with AgrD bound to AgrB. The results of the studies provide new insights on the mechanism of AIP biosynthesis.
EXPERIMENTAL PROCEDURES
Strains, Media, and Growth Conditions
Escherichia coli cultures were maintained in Luria broth (LB) at 37 °C (Table 1). Antibiotic concentrations for E. coli plasmids were 100 μg/ml for ampicillin and 50 μg/ml for kanamycin. S. aureus cultures were maintained in tryptic soy broth (TSB) at 37 °C. Antibiotic concentrations for S. aureus plasmids were chloramphenicol (Cam) 10 μg/ml.
TABLE 1.
Strain and plasmid list
The abbreviations used are as follows: Amp, ampicillin; Kan, kanamycin.
| Strain or plasmid | Genotype | Resistance | Source or Ref. |
|---|---|---|---|
| Escherichia coli | |||
| DH5α-E | Cloning strain | None | Invitrogen |
| BW25141 | Cloning strain | None | 15 |
| ER2566 | Protein expression strain | None | New England Biolabs |
| Staphylococcus aureus | |||
| RN4220 | Restriction modification-deficient | None | 32 |
| ROJ143 | Δagr ::Erm pAgr-lux/pAgrC1agrA | Cam | 24 |
| SH1000 | sigB+ derivative of NCTC8325-4 | None | 33 |
| SH1001 | SH1000/Δagr ::tetM | Tet | 33 |
| AH430 | SA502A/pDB59 | Cam | 23 |
| Plasmids | |||
| pAgrD1 | pEPSA5/agrBD | Amp, Cam | This work |
| pAgrD7a | pEPSA5/agrB | Amp, Cam | This work |
| pAIP1 | pBAD18/agrBD | Amp | This work |
| pBAD18 | Arabinose-inducible expression vector | Amp | 17 |
| pCOLADuet-1b | Expression vector | Kan | Novagen |
| pEPSA5 | Expression vector | Amp, Cam | 18 |
a The AgrD C-terminal truncation and point mutants built on pEPSA5 are not listed.
b The AgrD C-terminal truncation and point mutants built on pCOLADuet-1 are not listed.
Recombinant DNA and Genetic Techniques
Restriction and modification enzymes were purchased from New England Biolabs (Beverly, MA) and were used according to the manufacturer's instructions. All DNA manipulations were performed in E. coli strain BW25141 (15). Plasmids were transformed into S. aureus by electroporation as described previously (16). Plasmid constructions were confirmed by DNA sequencing. Nonradioactive sequencing was performed at the DNA sequencing facility at the University of Iowa. Oligonucleotides were synthesized at Integrated DNA Technologies (Coralville, IA). S. aureus chromosomal DNA was prepared using a Puregene DNA purification kit (Gentra Systems).
Construction of Plasmids
pAIP1
The agrBD genes were amplified from SH1000 genomic DNA using oligonucleotides ARH60 (5′-GTTGTTGAATTCGACAGTGAGGAGAGTGGTGTAAAATTGAAT-3′) and ARH61 (5′-GTTGTTTCTAGACTATTTAAATTATTCGTGTAATTGTG-3′). The PCR product was purified, digested with EcoRI and XbaI, and cloned into the same sites in E. coli expression plasmid pBAD18 (17).
pAgrD1, pAgrD7, and AgrD Truncation Plasmids
For construction of plasmid pAgrD1, the agrBD genes were amplified by PCR from SH1000 genomic DNA using oligonucleotides ARH60 and ARH61. The PCR product was purified, digested with EcoRI and XbaI enzymes, and cloned into the same sites in S. aureus expression plasmid pEPSA5 (18). Plasmid pAgrD7 was constructed in the same manner using oligonucleotides ARH60 and ARH80 (5′-GTTGTTTCTAGATCATTTTAAGTCCTCCTTAATAAAGAA-3′) to amplify only the agrB gene. For investigating the AgrD C terminus, a series of agrD truncations were constructed through PCR amplification of the agrBD genes using oligonucleotide ARH60 paired with oligonucleotides ARH111–ARH115 and MJT001–MJT002 (supplemental Table 1). Each PCR product was purified and cloned into pEPSA5 in the same manner as outlined above.
AgrD Point Mutations
Point mutations in AgrD were created using overlap extension PCR (19). The 5′ PCR fragment was generated using MJT061 and agrD reverse oligonucleotides listed in supplemental Table S1. The 3′ PCR fragment was generated with EPSArev (5′-GGCAAATTCTGTTTTATCAGACCG-3′) and agrD forward oligonucleotides (supplemental Table 1). In each case, plasmid pAgrD1 was used as the PCR template, and AgrD residues were mutated to alanine. The PCR products were joined together and cloned into the EcoRI and XbaI sites of pEPSA5 to generate the AgrD point mutant constructs.
pCOLADuet Constructions
To produce AgrB in E. coli, the agrB gene was amplified from SH1000 genomic DNA with oligonucleotides ARH128 (5′-GTTGTTCATATGAATTATTTTGATAATAAAATTGACCAG-3′) and MJT046 (5′-GTTGTTCTCGAGTCATTTTAAGTCCTCCTTAA-3′). The PCR product was purified and cloned into the NdeI and XhoI sites of pCOLADuet (Novagen). To create His-tagged AgrD, agrD was amplified using oligonucleotides MJT061 (5′-GGTGTTGAATTCAATGAATACATTATTTAACTTATTT-3′) and EPSArev. The PCR product was cloned into pCOLADuet in-frame with the His6 tag using the EcoRI and HindIII restriction sites. Each of the AgrD truncations and point mutations was also cloned into pCOLADuet by amplifying the various constructs from the pEPSA5-based plasmids using oligonucleotides MJT061 and EPSArev. In parallel, the agrD wild type, point mutant, and truncation constructs was also cloned upstream of agrB in pCOLADuet.
AgrB C84S Mutant and T7 Tag
The AgrB C84S point mutation was constructed using overlap extension PCR. The 5′ PCR fragment was generated with oligonucleotide pair ARH159 and ARH153, and the 3′ fragment was generated with ARH154 and MJT046. The resulting PCR products were joined and cloned into the NdeI and XhoI sites of pCOLADuet. T7-tagged AgrB was constructed in a two-step process. First, the agrB gene was amplified using oligonucleotides ARH159 (5′-GTTGTTGGATCCAATTATTTTGATAATAAAATTGACCAG-3′) and ARH129 (5′-GTTGTTAAGCTTTCATTTTAAGTCCTCCTTAATAAAGAA-3′) and cloned into the BamHI and HindIII sites of pET23a (Novagen). In the pET23a-agrB plasmid, the agrB gene is in-frame with the T7 N-terminal tag, allowing removal of T7-agrB through PCR amplification with T7prom (5′-TAATACGACTCACTATAGGG-3′) and MJT046 oligonucleotides. Second, the resulting product was cloned into the NdeI and XhoI sites of pCOLADuet with and without various agrD constructs.
Production of AIP Containing Supernatants
Overnight cultures of S. aureus containing plasmids encoding agrBD were diluted 1:100 in 5 ml of TSB containing antibiotics. Cells were grown by shaking at 37 °C to an optical density of 2 at 600 nm. Supernatants were then prepared by passing through 0.22-μm Spin-X microcentrifuge filters. Samples from E. coli were prepared by diluting overnight cultures 1:100 in LB containing antibiotics. Cells were grown by shaking at 37 °C for 2.5 h and then induced with 1 mm isopropyl β-d-thiogalactopyranoside or arabinose for 1 h. Supernatants were prepared by passing cultures through 0.22-μm filters.
AIP Inhibition Bioassay
A bioassay for testing AIP content in supernatants was performed as described previously (14). Briefly, overnight cultures of reporter strains containing the agr P3 promoter driving green fluorescent protein (pDB59) were diluted 1:500 into 25 ml of TSB containing Cam and grown to an OD at 600 nm of 0.05. At this point, 180-μl aliquots of reporter culture were placed in 96-well microtiter plates. Culture supernatants to be tested were prepared as described above, and 20-μl aliquots were mixed with the reporter cells in triplicate. Microtiter plates were incubated with shaking at 37 °C. Cell growth and fluorescence were monitored periodically using a Tecan GENios microplate reader by measuring OD at 595 nm and fluorescence at 485 nm excitation and 535 nm emission. AIP activity was assessed by measuring the inhibition of agr activity in reporter strains.
AIP Activation Bioassay
Culture supernatants were prepared as described above. Overnight cultures of reporter strain ROJ143 were diluted 1:50 into TSB containing Cam and grown to an OD at 600 nm of 1.0. For AIP testing, 600 μl of supernatant was mixed with 2.4 μl of ROJ143 reporter culture in triplicate. After shaking for 1 h at 37 °C, 200 μl of the reaction was transferred to a 96-well plate, and luminescence was measured using a Tecan Infinite M200 microplate reader.
Immunoblotting
Cell pellets from 1 ml of culture were resuspended in 250 μl of Tris/Tricine sample buffer with 1% β-mercaptoethanol (Bio-Rad) and boiled for 5 min. 10 μl of cell lysates were electrophoresed using Tricine/SDS-PAGE containing 18% acrylamide and 6 m urea as described previously (20). Proteins were transferred to Immobilon-PSQ polyvinylidene difluoride (Millipore) membranes. His6-tagged proteins were detected using Qiagen Penta-His monoclonal antibody conjugated to horseradish peroxidase. His blots were blocked overnight with 5% bovine serum albumin in Tris-buffered saline containing 0.1% Tween 20 (TBST). The membranes were washed three times for 20 min in TBST followed by 1 h of incubation with His-horseradish peroxidase antibody diluted 1:3000 in blocking buffer. Membranes were washed three times for 20 min with TBST and detected using the SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) followed by exposure to Kodak BioMax XAR film. T7-tagged proteins were detected using an anti-T7 antibody conjugated to horseradish peroxidase (Novagen) according to the manufacturer's instructions. All steps were carried out at room temperature.
Membrane Fraction Purification
E. coli membranes were purified as described previously with modifications (21). One-liter cultures of cells were grown, and expression was induced as in previous experiments. After centrifugation, cell pellets were resuspended in 25 ml of modified AH buffer (5 mm MgSO4, 50 mm MES, pH 6) containing protease inhibitors. Cells were lysed by four passages in a French press. Cell lysates were centrifuged at 10,000 × g for 30 min, after which the supernatant was removed and layered over 12 ml of AH buffer containing 20% sucrose in a new centrifuge tube. After spinning at 48,400 × g for 2 h, the supernatant was removed, and the pelleted membranes were removed and resuspended in 1 ml of AH buffer using a Dounce homogenizer. Samples were kept stored at −80 °C. Protein content in membrane samples was quantified using the Bio-Rad protein assay. For SDS-PAGE, membrane fractions containing a total of 0.25 μg of protein were loaded without β-mercaptoethanol onto the gel.
RESULTS
AgrB and AgrD Are Sufficient for AIP Production in E. coli
For our investigation into S. aureus AIP biosynthesis, we have chosen to focus on agr type I proteins unless otherwise noted. To facilitate this study, it would be advantageous to have an expression host with improved molecular and biochemical tools for carrying out detailed studies. Additionally, successful heterologous production would indicate that AgrB and AgrD are the only proteins uniquely required for the AIP biosynthetic pathway. E. coli possesses an abundant methodology toolbox for molecular and genetic manipulations, making it ideal as a potential expression host. Also, the absence of signal peptidase SpsB should not hinder production because the S. aureus and E. coli enzymes have been demonstrated to be functionally interchangeable (22). However, it has been reported that AIP cannot be generated in E. coli by expressing the agrB and agrD genes (13). Unfortunately, the details are unavailable for this experiment, and thus it is not evident why a negative result was obtained.
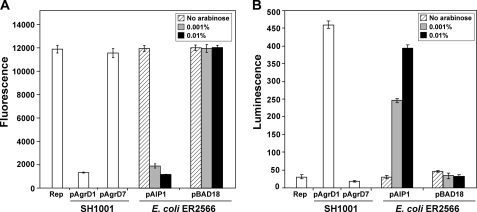
To evaluate E. coli as a heterologous host, we adopted a strategy of expressing the agrB and agrD genes and testing whether AIP was produced. The type I agrBD genes were cloned into the arabinose-inducible pBAD18 expression vector (17), generating plasmid pAIP1, and AIP production was tested using the E. coli protein expression strain ER2566 (New England Biolabs). AIP levels were detected using an inhibition assay that relies on the ability of the signal to cross-inhibit agr function of the type II system (23). In this assay, an agr type II fluorescent reporter strain AH430 displays high levels of fluorescence in the absence of inhibitors, which we confirmed in a control experiment (Fig. 2A). As an additional control, the type I agrBD genes were expressed in an Δagr deletion mutant (SH1001 with pAgrD1), and cell-free supernatant generated from this strain inhibited the fluorescence produced in strain AH430 (Fig. 2A). When only the agrB gene was expressed in the deletion mutant (SH1001 with pAgrD7), no fluorescence inhibition was observed. To test AIP production in E. coli, strain ER2566 with plasmid pAIP1 was grown with a range of l-arabinose (0 to 0.01%), and cell-free supernatants were tested for AIP in the inhibition assay. Without induction, fluorescence inhibition was not observed, but at 0.001% arabinose, significant inhibition became apparent (Fig. 2A). At 0.01% arabinose, full inhibition of the type II agr system was observed, and this inhibition level remained constant even at levels of arabinose up to 0.1% (data not shown). In an ER2566 control strain with pBAD18 alone, no agr inhibition was observed at any level of arabinose (Fig. 2A). This result provided initial evidence that E. coli can generate AIP signal when the type I agrBD genes are provided. The result also confirmed that the housekeeping level of E. coli LepB signal peptidase is sufficient to carry out AIP biosynthesis.
FIGURE 2.
Heterologous production of AIP in E. coli. E. coli strain ER2566 was grown with either plasmid pAIP1 or pBAD18 at l-arabinose concentrations of 0, 0.001, or 0.01% as indicated. As controls, cultures of SH1001 (Δagr::Tet) with either plasmid pAgrD1 (agrBD+) or pAgrD7 (agrB+) were also grown. Cell-free supernatants were prepared and tested in both agr inhibition (A) and agr activation bioassays (B). In the inhibition bioassay (A), Rep refers to the S. aureus type II reporter strain AH430 without anything added. A low fluorescence reading is indicative of AIP production and failure to induce the agr P3-GFP reporter. In the activation bioassay (B), Rep refers to the S. aureus ROJ143 reporter strain without anything added. A high luminescence reading is indicative of AIP production and induction of agr P3-lux expression.
To confirm the structure of E. coli generated AIP, an agr activation assay was performed. For this test, we utilized a reporter strain called ROJ143 that is unable to produce AIP but can respond to exogenously provided AIP (24). In the presence of AIP, bioluminescence is generated through induction of the lux operon that is under the control of the agr P3 promoter. The same samples were prepared as described above and this time added to the ROJ143 reporter. Gratifyingly, when supernatants were tested from E. coli grown with pAIP1, bioluminescence was induced (Fig. 2B). Similar to the inhibition bioassay, the highest levels of AIP were detected in samples induced with 0.01% arabinose, and no AIP was detectable in the absence of arabinose inducer or in the pBAD18 control samples. As anticipated, the control AIP sample from SH1001 with pAgrD1 resulted in strong bioluminescence, and the pAgrD7 negative control did not result in lux expression (Fig. 2B). To further test the E. coli samples for activation of the agr system, we performed a different agr activation bioassay that monitors the induction of the cascade at an early time point in the S. aureus growth phase (23). For this test, E. coli was grown with plasmids pAIP1 or pBAD18 control with and without arabinose inducer. The activation assay was performed as reported previously (23), and similar to the lux results, the supernatant prepared from E. coli carrying pAIP1 and grown with arabinose successfully induced the agr system (data not shown). Importantly, agr activation assays are more stringent than the inhibition assay (23), suggesting that E. coli heterologous production is resulting in the correct AIP structure. Altogether, these findings allow for the examination of the AIP biosynthetic processing steps using E. coli as an expression host.
AgrB Cleavage Assay
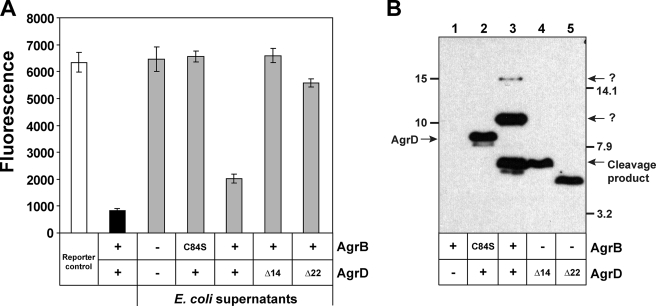
To begin investigating AIP biosynthesis, we adapted and modified the AgrB cleavage assay developed by Zhang et al. (13). Initially, the agrB and agrD genes were separated and cloned onto the pCOLADuet (Novagen) vector for tandem expression in E. coli. This plasmid contains two separated cloning regions, each driven by identical T7 promoters, allowing for similar levels of each protein to be produced while also facilitating downstream genetic manipulations. The agrD gene was cloned in the 5′ expression region with an N-terminal His6 tag, and the agrB gene with a fused N-terminal T7 tag was cloned into the 3′ expression region. As a control, we also built additional constructs without the agrD gene and a construct with an AgrB C84S catalytic mutation, which is reported to lack cleavage activity (11).
E. coli ER2566 containing each of these plasmids was induced with isopropyl β-d-thiogalactopyranoside, and both cells and supernatant were saved for further analysis. Supporting our results in Fig. 2, functional AIP was detected by inhibition bioassay only when wild type agrB and agrD were co-expressed (Fig. 3A). To monitor AgrB endopeptidase activity, cell lysates were prepared and separated using SDS-PAGE, and then gels were immunoblotted using anti-His monoclonal antibody. In a control lysate expressing only AgrB, no bands were observed in the immunoblot (Fig. 3B, lane 1). When His6-AgrD was co-expressed with the AgrB C84S mutant, a single band corresponding to full-length His6-AgrD (8 kDa) was observed (Fig. 3B, lane 2). If the agrB gene was removed from the plasmid, the same result was obtained (data not shown). When functional AgrB was provided, the His6-AgrD propeptide was processed to a smaller product that matched the exact size of a size standard composed of His6-AgrD with the C-terminal 14 residues removed (Δ14). An additional His6-AgrD size standard without the 8-residue AIP sequence or the 14-residue C-terminal tail (Δ22) migrates as a smaller band on the immunoblot. In the AgrB cleavage reaction, some additional bands were also apparent on the immunoblot (Fig. 3B, lane 3). A prominent band at ∼10 kDa is always observed. The identity of this band is unknown, but it is not present in reactions with the AgrB C84S mutant, suggesting its formation is dependent on functional AgrB. A couple of other faint background bands are also apparent, including one at ∼15 kDa. The identities of these bands are unknown, but their intensity is significantly reduced compared with the other detectable bands. Overall, the results confirm that AgrB has cysteine-dependent endopeptidase activity that removes the AgrD 14-residue C-terminal tail. Through numerous repeated tests, we have found that the cleavage assay is a robust, reproducible indicator of AgrB catalytic function that will be used throughout this study.
FIGURE 3.
Development of an AgrB cleavage assay. A, agr inhibition bioassay. White bar indicates reporter strain grown with supplements. Black bar is SH1001 with plasmid pAgrD1 as a positive control. All gray-shaded bars are E. coli cell-free supernatants with empty vector or different agrB and agrD constructs as indicated. Constructs Δ14 and Δ22 are AgrD truncations from the C terminus. B, immunoblot of E. coli cell lysates with the same agrB and agrD constructs. Blots were probed with antibody for the AgrD N-terminal His6 tag. Bands corresponding to full-length AgrD and cleavage product are shown, and the AgrD Δ14 truncation serves as a size standard for the cleavage product. The bands present at 10 and 15 kDa are unknown and labeled with question marks.
AgrD C-terminal Region Is Required for AIP Biosynthesis
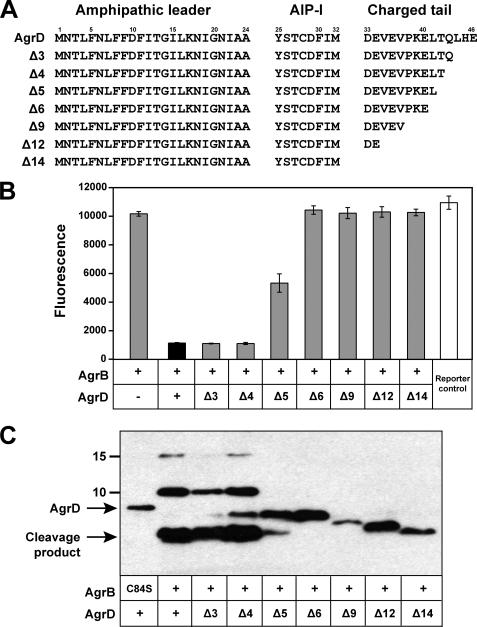
We focused our initial investigation into AIP biosynthesis on molecular characterization of AgrD. Previous studies demonstrated that the N-terminal amphipathic helix is essential (12), but the role of the C-terminal region is unclear. Interestingly, the C-terminal region is negatively charged (5 of 14 residues are acidic) and is the most conserved portion of AgrD among Staphylococcus species (25), suggesting an important role in AIP production. To investigate the requirement for this region, truncations of the AgrD C-terminal tail were constructed by sequentially deleting terminal residues (Fig. 4A). Each agrD deletion mutant was expressed on a plasmid in tandem with wild type agrB in an S. aureus Δagr deletion mutant (SH1001). Cell-free supernatants were prepared and tested for AIP levels using the agr inhibition bioassay (Fig. 4B). Deletion of three or four C-terminal residues did not have a measurable effect on AIP production. When five residues were deleted, a significant decrease in AIP production was evident. Deletion of six or more residues resulted in no detectable AIP being produced. When the agrD truncations were expressed in E. coli, similar results were obtained in the inhibition bioassay (data not shown). These results indicate that the first 9 residues of the AgrD C-terminal tail are essential for production of AIP, and for optimal production at least 10 residues are required.
FIGURE 4.
Effect of AgrD C-terminal truncations on AIP production and AgrB endopeptidase activity. A, graphic of AgrD C-terminal truncations. B, AIP production for each truncation (gray bars) expressed in SH1001 (Δagr::Tet). Cell-free supernatants were prepared and tested in the agr inhibition bioassay. For controls, full-length AgrD is shown as a black bar, and the reporter control is shown as a white bar. As an additional control, a construct without AgrD was also included. C, AgrB cleavage of His6-AgrD truncations. Each AgrD construct was co-expressed in E. coli with AgrB and immunoblotted with anti-His monoclonal antibody. For comparative purposes, the immunoblot lanes were aligned with AIP bioassay results in B.
Although we have demonstrated that the AgrD C-terminal region is essential, the requirement for this region for cleavage by AgrB is unknown. Each AgrD truncation was cloned into the pCOLADuet vector to generate a plasmid that dually expressed AgrB and each His6-AgrD truncation construct. The constructed plasmids were transformed into E. coli ER2566 along with controls, and the AgrB cleavage assay was performed. As shown in Fig. 4C, when three or four residues were deleted from the AgrD C terminus, the peptide was still a substrate for cleavage by AgrB. When five residues were deleted, only partial cleavage was apparent, and when six or more residues were deleted, no cleavage by AgrB was observed. Altogether, these results parallel the AIP bioassay results (Fig. 4B), suggesting the failure to secrete AIP is because of a defect in AgrB endopeptidase activity.
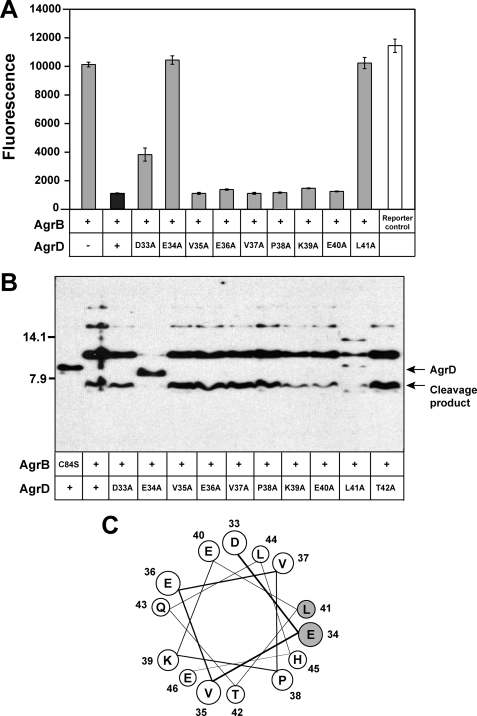
AgrD Residues Glu-34 and Leu-41 Are Essential
Knowing the first nine AgrD C-terminal residues are critical for AIP biosynthesis, alanine substitutions were generated across this region to determine which of the residues were required for proper function. Each construct was tested in the same manner as the truncations above, and we observed that many of the alanine substitutions did not result in a measurable defect in AIP production (Fig. 5A). Mutations in two of the residues, AgrD glutamate 34 or leucine 41, resulted in a substantial decrease in AIP production. Mutation of aspartate 33 also reduced AIP levels, although not to the extent of the Glu-34 and Leu-41 mutations. When comparing with other staphylococcal AgrD sequences, Asp-33, Glu-34, and Leu-41 are three of the most highly conserved residues (25).
FIGURE 5.
Effect of AgrD point mutations on AIP production and AgrB endopeptidase activity. A, AIP production for each mutant (gray bars) expressed in SH1001 (Δagr::Tet). Cell-free supernatants were prepared and tested in the agr inhibition bioassay. For controls, full-length AgrD is shown as a black bar, and the reporter control is shown as a white bar. As an additional control, a construct without AgrD was also included. B, AgrB cleavage of each AgrD C-terminal point mutant. Each AgrD construct was co-expressed in E. coli with AgrB and immunoblotted with anti-His monoclonal antibody. C, helical wheel of AgrD C-terminal 14 residues. Residues are labeled with numbers that coordinate with the position in full-length AgrD. The Glu-34 and Leu-41 residues are shaded gray.
The AgrD point mutations were also expressed in E. coli to test for AgrB endopeptidase activity. A mutation in threonine 42 was included in the analysis. In the immunoblot assay, only AgrD mutants E34A and L41A were defective in cleavage by AgrB (Fig. 5B). These same two mutants also failed to generate AIP (Fig. 5A), demonstrating parallels between AgrB cleavage and AIP production. The identities of the additional background bands in the L41A mutant lane are unknown. The D33A mutant did not display a phenotype in the cleavage assay, although this mutant produced less AIP than wild type. These results demonstrate that AgrD residues Glu-34 and Leu-41 are essential for both AgrB endopeptidase activity and AIP production.
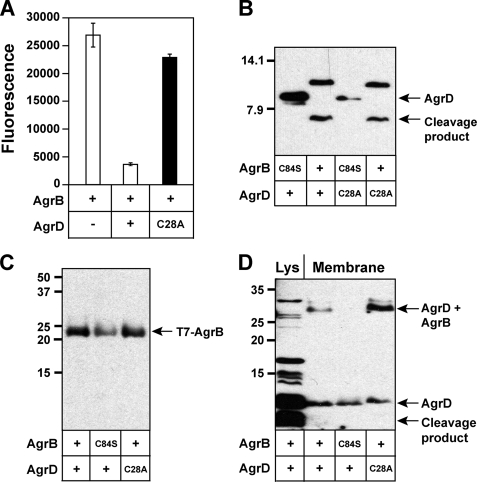
Characterization of the AgrD Cysteine Residue
The sequence of AgrD contains only one cysteine residue at position 28. In the final AIP structure, the cysteine residue is part of the cyclic thiolactone ring (Fig. 1). Considering the importance of the residue, we anticipated it would be essential for AIP biosynthesis, but it is not clear at what stage of the pathway the cysteine residue is required. To investigate this question, a cysteine-to-alanine (C28A) mutation was constructed and tested for AIP production. As anticipated, the expression of an AgrD C28A mutation did not result in detectable AIP levels (Fig. 6A). To see if the C28A mutation affected AgrB endopeptidase activity, the mutation was tested in the cleavage reaction. Interestingly, no phenotype was detectable in the immunoblot (Fig. 6B), indicating the AgrD C28A peptide was a normal substrate for AgrB. These findings were important because they demonstrated the mutant was defective at another step in AIP biosynthesis, presumably downstream of the AgrB cleavage reaction.
FIGURE 6.
Characterization of AgrD cysteine mutant and detection of AgrD bound to AgrB. A, AgrD C28A mutation (black bar) was constructed and expressed in SH1001 (Δagr::Tet) along with controls (white bars). Cell-free supernatants were prepared and tested in the agr inhibition bioassay. B, AgrB cleavage of AgrD C28A mutant along with wild type control. C, E. coli membrane samples expressing different agrB and agrD constructs were prepared and immunoblotted for T7 tag on the N terminus of AgrB. D, using the same membrane preparations as in C, the samples were immunoblotted for the His6 tag on the N terminus of AgrD. As a control, a cell lysate of E. coli co-expressing His6-AgrD and AgrB was included in the 1st lane and is labeled Lys.
Identification of a Structure with AgrD Bound to AgrB
We speculated the C28A mutation may allow an investigation into formation of the AIP cyclic thiolactone ring, which is step 3 in our proposed model of AIP biosynthesis (Fig. 1B). If the model is correct, AgrD amino acid residues Met-1 to Met-33 would be linked to the AgrB Cys-84 residue as an enzyme-bound thioester. We speculate that following formation of this structure, the thioester bond would be susceptible to attack by the AgrD Cys-28 nucleophile. The resulting thioester exchange would release the cyclic AIP moiety with an attached N-terminal amphipathic helix. By using the AgrD C28A mutant, the proposed AgrD-AgrB intermediate might be stabilized, making the structure detectable in immunoblots.
To investigate the formation of the intermediate, His6-AgrD was co-expressed in the presence of T7-tagged AgrB in E. coli. Membrane fractions of the cells were purified and initially tested for the presence of T7-AgrB. As shown in Fig. 6C, T7-AgrB is readily detectable in immunoblots of membrane preparations with anti-T7 tag antibody. We found the enrichment of the membranes to be essential for detection, as no T7-AgrB was detectable in cell lysates (data not shown).
To monitor the fate of AgrD, the same membrane samples were immunoblotted with anti-His antibody (Fig. 6D). We observed the appearance of a new band running ∼30 kDa in the lane with His6-AgrD C28A and T7-AgrB. This size matches the approximate combined molecular mass of T7-AgrB (23.2 kDa) and His6-AgrD-Δ14 (5.5 kDa), which was predicted to be 28.7 kDa. Interestingly, the new band was also observed in samples with wild type His6-AgrD, although at a lower intensity. When the AgrB catalytic cysteine was mutated (C84S), the band was not observed, suggesting this cysteine residue is essential for formation of the AgrD-AgrB structure. A similar result was obtained when His6-AgrD C28A was co-expressed with T7-AgrB C84S (data not shown). In each case, unbound His6-AgrD was also present in the membrane samples. As a control, E. coli lysate co-expressing His6-AgrD and T7-AgrB was included to show the location of full-length His6-AgrD and the cleavage product. The immunoblot bands of this cleavage reaction were more compressed and intense compared with Fig. 3B due to the different SDS-PAGE running conditions and longer film exposure (see “Experimental Procedures”). Also, the AgrD-AgrB band is not visualized in the sample showing the control cell lysate. When cell lysates are prepared through sample boiling, we do not observe T7-AgrB in immunoblots (data not shown), and for this reason, the AgrD-AgrB band was not expected. Of additional note, the membrane samples shown in Fig. 6D were prepared at pH 6 using MES buffer. When membrane preparations were repeated at pH 7.5 using HEPES, we failed to consistently detect the structure (data not shown). Overall, the use of the His6-AgrD C28A mutation resulted in higher levels of AgrD-AgrB structure and more reproducible detection than when the experiments were performed with wild type His6-AgrD. We believe these results provide experimental evidence of an AgrD-AgrB structure that might be an important intermediate in the AgrB catalytic mechanism.
DISCUSSION
The studies presented herein provide new insights into the S. aureus AIP biosynthetic mechanism. We demonstrate that AIP can be produced using E. coli as a heterologous host, opening doors to a more detailed investigation into essential AgrB and AgrD regions. Molecular characterization demonstrated that residues Glu-34 and Leu-41 of the AgrD C-terminal tail are essential for AgrB endopeptidase activity and AIP biosynthesis. Importantly, evidence of a structure with AgrD bound to AgrB was presented, and the formation of the structure was dependent on the catalytic cysteine of AgrB.
We overcame a significant technical hurdle through the heterologous expression of the agrBD genes in E. coli and successful production of AIP type I. This important finding demonstrated that only AgrD and AgrB are required for AIP production in other hosts and simultaneously allowed the use of E. coli as an expression host. Other conserved enzymes, such as type I signal peptidase, are present in all bacteria and do not have to be provided to complete the pathway (14). In support of this argument, the expression of agrBD in Bacillus subtilis also allowed detectable AIP production (data not shown). To the best of our knowledge, this study is the first report of functional AIP being produced in a heterologous host, which makes available more powerful genetic tools for future studies on related peptide signals. Because of the lack of information, it is not clear why Zhang et al. (13) were unable to use E. coli as a host.
The truncation and point mutation studies confirm that the C-terminal tail of AgrD is playing an important role in AIP production. When more than five residues are removed from the C terminus, cleavage of the AgrD tail is completely prevented (Fig. 4C), and no AIP production was detectable (Fig. 4B). We postulate the C-terminal tail may play a role in mediating a peptide-protein interaction with AgrB, and such an interaction would be advantageous as a precursor to the AgrB cleavage step. The topology map of AgrB predicts numerous positively charged residues located in the cytoplasmic loops of AgrB that could be sites of interaction with AgrD (13). In support of this proposal, both the Asp-33 and Glu-34 residues are required for AIP production, although the Asp-33 residue was not essential for AgrB endopeptidase activity. The importance of Leu-41 suggests other hydrophobic interactions are also playing a critical role.
Although the N terminus of AgrD is known to be an amphipathic α-helix (12), it is possible that the entire AgrD peptide adopts an α-helical conformation. Secondary structure analysis using the PredictProtein server suggests AgrD is over 60% α-helical (26), with the N-terminal and C-terminal regions displaying the strongest α-helical signature. Interestingly, analysis of the central AIP region suggested these residues possess the least amount of secondary structure. When the C-terminal 14 residues are oriented on a helical wheel, residues Glu-34 and Leu-41 both reside on the same α-helical face in close proximity (Fig. 5C). Thus, it is possible these two essential residues make key contacts with AgrB that are necessary to initiate the sequence of events to AIP production. Further studies will be necessary to evaluate the AgrD and AgrB interaction mechanism that serves as a precursor to cleavage.
A key finding in this report was the identification of a structure with AgrD bound to AgrB. In a previous study, Qiu et al. (11) noted a faint band in an AgrD processing immunoblot at ∼25 kDa. The authors speculated it could be AgrD bound to AgrB, but no further analysis was performed. By using the AgrD C28A mutation in this study, the formation of the structure was stabilized, allowing its consistent detection through immunoblots. The dependence of the structure formation on the AgrB Cys-84 catalytic cysteine is important, as it supports the proposal that AgrB is a cysteine endopeptidase. The appeal of this reaction mechanism is that the C-terminal carboxylate group on the methionine 33 residue of AgrD is activated, setting the stage for nucleophilic attack by AgrD cysteine 28. Presumably, once Cys-28 attacks the activated carbon, thioester exchange proceeds, allowing formation of the cyclic thiolactone ring on AIP (Fig. 1B). Without the cysteine nucleophile on AgrD, no AIP was produced (Fig. 6A), supporting the critical nature of this residue. Also, AgrD Cys-28 was not required for the AgrB cleavage reaction (Fig. 6B), indicating Cys-28 has a role in either thiolactone ring formation or AIP secretion. Whether the thioester exchange is spontaneous or facilitated through peptide-protein interactions is a question that will require further analysis.
Our attempts to characterize the AgrD-AgrB structure were met with limited success. Detection of the structure is pH-dependent, which supports our proposal that the AgrD peptide is held as a thioester on AgrB Cys-84. Thioester bonds are base labile, and the preparation of membrane samples at lower pH appeared to stabilize the structure (Fig. 6D). However, our attempts to drive the AgrD peptide from AgrB using thioester sensitive reagents were not successful. For instance, the addition of excess hydroxylamine and dithiothreitol did not significantly affect the level of detectable AgrD-AgrB structure (data not shown), perhaps suggesting the bound peptide is protected in the AgrB active site. Alternatively, this result might suggest the reaction mechanism is different or perhaps more complex than proposed (Fig. 1B). Thus, a demonstration that the structure is a catalytic intermediate in the AgrB enzyme mechanism will require more in-depth analysis.
Although we have taken steps forward in our understanding of AIP production, aspects of the biosynthetic pathway still remain unknown. Once the AIP cyclic thiolactone ring is generated, the predicted intermediate structure needs to be transported to the outer face of the membrane. Considering no other proteins besides AgrB and AgrD are required for heterologous generation of AIP, it seems likely that AgrB can perform the transport step. In support of this idea, there are regions removed from the AgrB active site that are divergent among AgrB types but important for group-specific AIP production, suggesting these distal regions could have a role in transport (27). Once the AIP intermediate structure is moved to the outside face, type I signal peptidase SpsB is available for removal of the N-terminal leader and completes the pathway (14).
Cyclic peptide signals are emerging as a common structure utilized by diverse Gram-positive bacteria to regulate cellular events. Enterococcus faecalis and Lactobacillus plantarum produce cyclic lactone and thiolactone peptide signals, respectively, and possess chromosomal loci with parallels to the S. aureus agr system (28, 29). Listeria monocytogenes secretes a functional peptide signal (30), although the structure is unknown, and through bioinformatics mining, other Gram positives, such as members of the genera Clostridia and Bacillus, also appear to contain agr-like regions. Alignments of AgrB homologues have been performed, and they are all integral membrane proteins with a cysteine residue in a similar position as the Cys-84 in S. aureus AgrB (31). The AgrD sequences also display similarities with an N-terminal amphipathic region and C-terminal charged region. When the signal structure is known, it is always encoded in the middle of the AgrD-like sequence (31). Based on our observations with S. aureus AgrD, it seems probable that the charged C-terminal regions on the diverse AgrD homologues could be critical for signal biosynthesis. Similarly, the internal cysteine (or serine for E. faecalis) present on the AgrD homologues, and the conserved cysteine in AgrBs, should be essential for the signal biosynthetic pathway.
The results reported herein provide further insight on molecular and biochemical details of S. aureus AIP biosynthesis. Integral to the advancement of the work was the development of heterologous hosts for AIP production, allowing the use of improved molecular tools. More specifically, our results demonstrate that the AgrD C-terminal region is critical for AgrB activity and AIP production. We also provide evidence of an AgrD-AgrB bound structure that may be an important intermediate in the AIP biosynthetic mechanism. The results of this study and the developed approaches could be employed to investigate the diverse cyclic peptide signals produced by other Gram-positive bacteria.
Supplementary Material
Acknowledgments
We thank Paul Williams for the ROJ143 reporter strain. We thank Jeffrey Kavanaugh for technical assistance and comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant AI078921 from the NIAID. This work was also supported by an American Heart Association beginning grant-in-aid.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- AIP
- autoinducing peptide
- Cam
- chloramphenicol
- Tet
- tetracycline
- TSB
- tryptic soy broth
- MES
- 4-morpholineethanesulfonic acid
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1.Lowy F. D. (1998) N. Engl. J. Med. 339, 520–532 [DOI] [PubMed] [Google Scholar]
- 2.Novick R. P., Geisinger E. (2008) Annu. Rev. Genet. 42, 541–564 [DOI] [PubMed] [Google Scholar]
- 3.Dunman P. M., Murphy E., Haney S., Palacios D., Tucker-Kellogg G., Wu S., Brown E. L., Zagursky R. J., Shlaes D., Projan S. J. (2001) J. Bacteriol. 183, 7341–7353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Booth M. C., Cheung A. L., Hatter K. L., Jett B. D., Callegan M. C., Gilmore M. S. (1997) Infect. Immun. 65, 1550–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdelnour A., Arvidson S., Bremell T., Rydén C., Tarkowski A. (1993) Infect. Immun. 61, 3879–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park J., Jagasia R., Kaufmann G. F., Mathison J. C., Ruiz D. I., Moss J. A., Meijler M. M., Ulevitch R. J., Janda K. D. (2007) Chem. Biol. 14, 1119–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bubeck Wardenburg J., Patel R. J., Schneewind O. (2007) Infect. Immun. 75, 1040–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boles B. R., Horswill A. R. (2008) PLoS Pathog. 4, e1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koenig R. L., Ray J. L., Maleki S. J., Smeltzer M. S., Hurlburt B. K. (2004) J. Bacteriol. 186, 7549–7555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novick R. P., Ross H. F., Projan S. J., Kornblum J., Kreiswirth B., Moghazeh S. (1993) EMBO J. 12, 3967–3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu R., Pei W., Zhang L., Lin J., Ji G. (2005) J. Biol. Chem. 280, 16695–16704 [DOI] [PubMed] [Google Scholar]
- 12.Zhang L., Lin J., Ji G. (2004) J. Biol. Chem. 279, 19448–19456 [DOI] [PubMed] [Google Scholar]
- 13.Zhang L., Gray L., Novick R. P., Ji G. (2002) J. Biol. Chem. 277, 34736–34742 [DOI] [PubMed] [Google Scholar]
- 14.Kavanaugh J. S., Thoendel M., Horswill A. R. (2007) Mol. Microbiol. 65, 780–798 [DOI] [PubMed] [Google Scholar]
- 15.Datsenko K. A., Wanner B. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schenk S., Laddaga R. A. (1992) FEMS Microbiol. Lett. 73, 133–138 [DOI] [PubMed] [Google Scholar]
- 17.Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995) J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forsyth R. A., Haselbeck R. J., Ohlsen K. L., Yamamoto R. T., Xu H., Trawick J. D., Wall D., Wang L., Brown-Driver V., Froelich J. M., C. K. G., King P., McCarthy M., Malone C., Misiner B., Robbins D., Tan Z., Zhu Zy Z. Y., Carr G., Mosca D. A., Zamudio C., Foulkes J. G., Zyskind J. W. (2002) Mol. Microbiol. 43, 1387–1400 [DOI] [PubMed] [Google Scholar]
- 19.Urban A., Neukirchen S., Jaeger K. E. (1997) Nucleic Acids Res. 25, 2227–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schägger H. (2006) Nat. Protoc. 1, 16–22 [DOI] [PubMed] [Google Scholar]
- 21.Douville K., Price A., Eichler J., Economou A., Wickner W. (1995) J. Biol. Chem. 270, 20106–20111 [DOI] [PubMed] [Google Scholar]
- 22.Cregg K. M., Wilding I., Black M. T. (1996) J. Bacteriol. 178, 5712–5718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malone C. L., Boles B. R., Horswill A. R. (2007) Appl. Environ. Microbiol. 73, 6036–6044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen R. O., Winzer K., Clarke S. R., Chan W. C., Williams P. (2008) J. Mol. Biol. 381, 300–309 [DOI] [PubMed] [Google Scholar]
- 25.Novick R. P. (2003) Mol. Microbiol. 48, 1429–1449 [DOI] [PubMed] [Google Scholar]
- 26.Rost B., Yachdav G., Liu J. (2004) Nucleic Acids Res. 32, W321–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L., Ji G. (2004) J. Bacteriol. 186, 6706–6713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama J., Cao Y., Horii T., Sakuda S., Akkermans A. D., de Vos W. M., Nagasawa H. (2001) Mol. Microbiol. 41, 145–154 [DOI] [PubMed] [Google Scholar]
- 29.Sturme M. H., Nakayama J., Molenaar D., Murakami Y., Kunugi R., Fujii T., Vaughan E. E., Kleerebezem M., de Vos W. M. (2005) J. Bacteriol. 187, 5224–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riedel C. U., Monk I. R., Casey P. G., Waidmann M. S., Gahan C. G., Hill C. (2009) Mol. Microbiol. 71, 1177–1189 [DOI] [PubMed] [Google Scholar]
- 31.Nakayama J., Chen S., Oyama N., Nishiguchi K., Azab E. A., Tanaka E., Kariyama R., Sonomoto K. (2006) J. Bacteriol. 188, 8321–8326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kreiswirth B. N., Löfdahl S., Betley M. J., O'Reilly M., Schlievert P. M., Bergdoll M. S., Novick R. P. (1983) Nature 305, 709–712 [DOI] [PubMed] [Google Scholar]
- 33.Horsburgh M. J., Aish J. L., White I. J., Shaw L., Lithgow J. K., Foster S. J. (2002) J. Bacteriol. 184, 5457–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.