Abstract
Copper transport ATPases sustain important roles in homeostasis of heavy metals and delivery of copper to metalloenzymes. The copper transport ATPase from Thermotoga maritima (CopA) provides a useful system for mechanistic studies, due to its heterologous expression and stability. Its sequence comprises 726 amino acids, including the N-terminal metal binding domain (NMBD), three catalytic domains (A, N, and P), and a copper transport domain formed by eight helices, including the transmembrane metal binding site (TMBS). We performed functional characterization and conformational analysis by proteolytic digestion of WT and mutated (NMBD deletion or mutation) T. maritima CopA, comparing it with Archaeoglobus fulgidus CopA and Ca2+ ATPase. A specific feature of T. maritima CopA is ATP utilization in the absence of copper, to form a low-turnover phosphoenzyme intermediate, with a conformation similar to that obtained by phosphorylation with Pi or phosphate analogues. On the other hand, formation of an activated state requires copper binding to both NMBD and TMBS, with consequent conformational changes involving the NMBD and A domain. Proteolytic digestion analysis demonstrates A domain movements similar to those of other P-type ATPases to place the conserved TGES motif in the optimal position for catalytic assistance. We also studied an H479Q mutation (analogous to one of human copper ATPase ATP7B in Wilson disease) that inhibits ATPase activity. We found that, in spite of the H479Q mutation within the nucleotide binding domain, the mutant still binds ATP, yielding a phosphorylation transition state conformation. However, covalent phosphoryl transfer is not completed, and no catalytic turnover is observed.
P-Type ion transport ATPases undergo phosphoryl transfer to a conserved aspartyl residue as an intermediate step in the mechanism of ATP utilization and coupled cation transport (1−3). The enzyme state activated by a specific cation is generally termed E1, yielding E1P following phosphorylation by ATP. Subsequent isomeric transition of E1P to E2P is the energy transduction step that lowers the affinity of the enzyme for the bound cation. Finally, the bound cation undergoes vectorial dissociation, and following hydrolytic cleavage of the phosphoenzyme, the protein regains its ground state (E2).
The P-type ATPase family is divided into five branches termed I−V (4). They include the well-studied PII-type ATPases that are specific for cations such as H+, Na+, K+, and Ca2+ and a PIB subgroup comprising ATPases for transport of heavy metals ions such as Cu+, Cu2+, Zn2+, Pb2+, Cd2+, and Co2+(5). The PIB-type ATPases play important roles in accumulation and tolerance of heavy metals in biological systems (6−8), as well as for their delivery to metalloenzymes (9). Two ATPases of this subgroup serve as copper transporters in humans (10) and are involved in the etiology of Menkes and Wilson diseases (8,11−13). The catalytic mechanism of the PIB-type ATPases is the subject of ongoing studies (14−18).
The overall structure of PII-type ATPases, such as the Ca2+ and Na+/K+ ATPases, has been initially established by sequence analysis (19,20), followed by the determination of their crystal structures (21,22). On the other hand, the atomic models of the PIB-type ATPases have been determined only for the cytoplasmic domains (23−26). Nevertheless, on the basis of the partial sequence homologies (27) and structural analogy (28,29) with PII-type ATPases, three distinct cytoplasmic catalytic domains and eight (rather than ten) transmembrane segments are defined. The cytoplasmic domains include A (“actuator”), P (‘phosphorylation”), and N (“nucleotide binding”) which are shared within the P-type ATPase family, and in fact, their backbone structures are conserved (23−26). A specific feature of the PIB-type ATPases (not present in other P-type ATPases) is the presence of another cytoplasmic domain, an N-terminal metal binding domain (NMBD),1 containing heavy metal binding motifs (CXXC). These motifs may serve as acceptors for chaperone-bound heavy metals (30−32) and may also play a heavy metal-dependent regulatory role in enzyme activity (33,34).
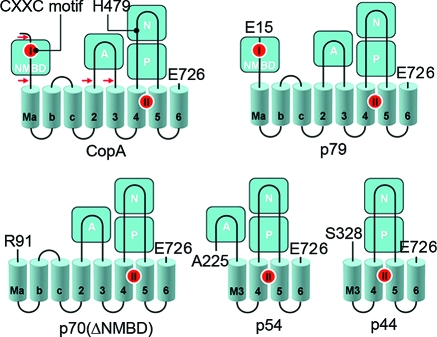
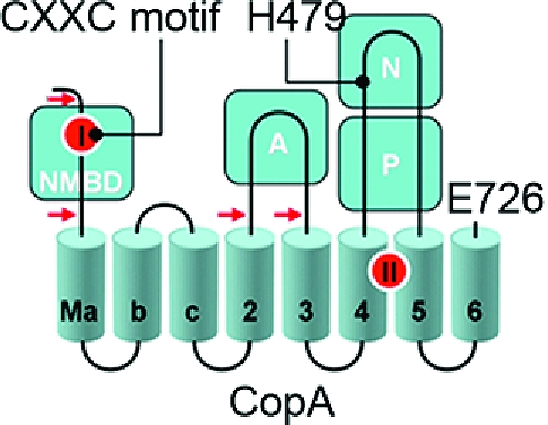
The copper transporting ATPase CopA of Archaeoglobus fulgidus, a thermophilic archaeon, lends itself to heterologous expression, purification, and stability and has provided useful information about the structure and function of PIB-type ATPases (34−36). Expression and purification of a CopA homologue from a thermophilic bacterium, Thermotoga maritima, also provide a useful model for mechanistic studies. As shown in Figure 1, the T. maritima CopA sequence comprises 726 amino acids, beginning with the NMBD that includes one copper binding site (CXXC motif), followed by four closely spaced transmembrane segments (Ma, Mb, Mc, and M2). These are then followed by the cytoplasmic A domain, two transmembrane segments (M3 and M4, numbered to match the nomenclature of PII-type ATPases), a large extramembranous region (first and second segments of the P domain interspaced by the N domain), and the M5 and M6 transmembrane segments. The transmembrane copper transport sites (TMBS) are formed by M4, M5, and M6 (37).
Figure 1.
Folding diagram of the T. maritima CopA sequence. The full-length sequence comprises 726 amino acids, including an N-terminal metal binding domain (NMBD), followed by four closely spaced transmembrane segments (Ma, Mb, Mc, and M2) and an extramembranous A domain. The sequence continues with two (M3 and M4) transmembrane segments, a large extramembranous region, including the N and P domains, and two closely spaced (M5 and M6) transmembrane segments at the carboxyl terminus. Two distinct copper binding sites are colored red on the NMBD and at the transmembrane site (TMBS). On the basis of partial sequence homology with other cation transport ATPases, assignments are made to the actuator domain (A) and the nucleotide binding domain (N) interspaced within the phosphorylation domain (P) sequence where D445 undergoes phosphorylation. The locations of two mutations, in the NMBD (double mutation of C17A and C20A termed the CxxC mutant) and the N domain (H479Q), are indicated in the diagram. The fragments resulting from papain cleavage of the 14 N-terminal residues at the N-terminal end to E15, at the NMBD−Ma link (N-terminal end to R91), at the M2−A domain link (N-terminal end to A225), or at the A domain−M3 link (N-terminal end to S328) are also shown. The arrows point to the cleavage sites. The peptide fragments were identified by amino acid sequencing (38).
We previously reported (38) that limited proteolysis of T. maritima CopA with papain yields four specific fragments (79, 70, 54, and 44 kDa polypeptides, as illustrated in Figure 1). The fragments are produced by truncation of the 14 N-terminal residues (cleavage at the N-terminal side of E15) or cleavage in the NMBD−Ma loop (R91), in the M2−A domain loop (A225), or in the A domain−M3 loop (S328). In this report, we extended this proteolytic study to CopA mutants (mutation in the metal binding CXXC motif or deletion of the NMBD) with the aim of understanding the function of the NMBD in terms of protein conformation. The results define the effects of various ligands such as copper, substrates (i.e., ATP and Pi), and substrate analogues on the digestion patterns of WT, as well as NMBD-deleted or mutated CopA. We then gained an understanding of the roles played by the occupancy of the two alternate binding sites (NMBD and TMBS) by copper and the involvement of the NMBD and A domain in protein conformational transitions within the ATPase cycle. We also characterized the ATP utilization and conformational transitions occurring at low rates in the absence of copper, a specific feature of T. maritima CopA, differing from other P-type ATPases that exhibit an absolute requirement for an activating cation. Finally, we obtained further characterization of the defective ATP utilization produced by the invariant His mutation (H479Q), analogous to those involving the human copper ATPase (ATP7B) in Wilson disease.
Experimental Procedures
Preparations of Proteins
Overexpression of T. maritima CopA in Escherichia coli and purification of the expressed protein were previously described (38). An NMBD-deleted protein and two site-directed mutations were also prepared, as shown in Figure 1. The membrane vesicles obtained from E. coli or the protein purified from them was suspended in a buffer containing 50 mM NaCl, 20% (w/v) glycerol, 0.1% (w/v) n-decyl β-d-maltoside, 0.5 mM DTT, and 50 mM MOPS-NaOH (pH 7.0) and stored at −80 °C. For all experiments, the protein was reduced by adding DTT to a final concentration of 2 mM and incubated for 1 h on ice.
A. fulgidus CopA (33,34) was kindly supplied by J. Argüello. Sarcoplasmic reticulum vesicles containing the SERCA1 isoform of the Ca2+ ATPase were obtained from rabbit skeletal muscle (39).
Preparation of the Lipid Suspension
Twenty microliters of egg yolk phosphatidylcholine (100 mg/mL in chloroform) was dried under vacuum; 200 μL of 10 mM MOPS-triethanolamine (pH 7) was added, and the mixture was sonicated until a nearly clear suspension was obtained. This preparation was used to supplement various reaction mixtures.
ATPase and p-Nitrophenyl Phosphatase Activity Measurements
Steady state ATPase activity was measured in a reaction mixture containing 50 mM MES-triethanolamine (pH 6.0), 30% glycerol, 2.0 mM sodium azide, 0.5 mM DTT, 10 mM cysteine-TRIS (pH 6.0), 5 mM MgCl2, 0.01% (w/v) n-dodecyl β-d-maltoside (DDM), 25 μg/mL phosphatidylcholine, and either 1 mM BCS or added CuCl2 (40 μM in most experiments). Protein was added to yield 25 μg/mL, and the reaction was started by the addition of ATP (2.5 mM), at 40 or 60 °C, depending on the experimental schedule. Colorimetric determination of released inorganic phosphate (Pi) was then conducted in serial samples.
The steady state hydrolytic activity was also measured using 10 mM p-nitrophenyl phosphate (pNPP) as a substrate, in a reaction mixture identical to that used for ATPase, in the absence and presence of 1 mM BCS. The temperature was set at 40 °C, due to the high rate of the reaction with this substrate. Hydrolytic activity was followed by determining the absorption of p-nitrophenol at 410 nm after addition of 4 volumes of 0.5 N NaOH, based on an ε of 18500 M−1 cm−1.
Phosphoenzyme Formation with ATP
The reaction mixture consisted of 50 mM MES-triethanolamine (pH 6.0), 30% glycerol, 0.5 mM DTT, 10 mM cysteine-TRIS (pH 6.0), 5 mM MgCl2, 0.01% DDM, 25 μg/mL phosphatidylcholine, 25−30 μg/mL protein, and either 1 mM BCS or 3−40 μM CuCl2. To 950 μL of reaction mixture (with 1 mM BCS or 40 μM CuCl2) at 60 °C was added 50 μL of 0.5 mM [γ-32P]ATP (25 μM final concentration), and the reaction was quenched at various times with 5 mL of 3% (w/v) trichloroacetic acid (TCA). The quenched samples were passed through 0.45 μm Millipore filters, which were then blotted and processed for radioactivity determination by liquid scintillation counting. When tetrathiomolybdate (copper chelator) was included in the reaction mixture, control samples were obtained in the absence of protein, as the quenching acid produced some precipitation of ATP under these conditions, resulting in collection of radioactivity in the filter which was subtracted from the radioactive signal obtained in the presence of protein. For electrophoretic analysis of the phosphorylated protein, the quenched samples were centrifuged at 5000 rpm for 5 min. The sediments were resuspended in 1 mL of 0.125 N perchloric acid and centrifuged again, and the sediments were dissolved in 2.5% (w/v) lithium dodecyl sulfate, 0.5% (v/v) β-mercaptoethanol, 3% (w/v) sucrose, and 0.1 mg/mL bromophenol blue. An aliquot of the dissolved protein was subjected to gel electrophoresis at pH 6.3, and the gels were finally stained with Coomassie Brilliant Blue R250 or scanned for phosphoimaging.
At the same time, the filtrate was treated with activated charcoal for removal of nucleotide, the charcoal removed by filtration, and the released [32P]Pi in the supernatant isolated by ammonium molybdate isobutanol/benzene extraction as described previously (40). An aliquot was then used for determination of the amount of [32P]Pi by scintillation counting and assessment of ATPase hydrolytic activity in parallel with phosphoenzyme formation.
Phosphoenzyme Assay with Inorganic Phosphate
The reaction mixture consisted of 50 mM MES-triethanolamine (pH 6.0), 20% (w/v) dimethyl sulfoxide, 0.5 mM DTT, 10 mM cysteine-TRIS (pH 6.0), 5 mM MgCl2, 0.01% DDM, 25 μg/mL phosphatidylcholine, and either 1 mM BCS or 40 μM CuCl2. Following incubation with 50 μM [32P]Pi at 60 °C for various periods of time, the samples were quenched and filtered, and the phosphorylated protein on the filters was examined as described above.
Limited Proteolysis with Papain
Proteolysis of CopA by papain was performed as described previously (38) with a minor modification in the composition of the reaction mixture and temperature. Purified CopA (0.5 mg/mL) was digested with 0.5 mg/mL papain (Worthington Biochemical Corp., Lakewood, NJ) at 50 °C in a buffer containing 1 mM MgCl2, 0.1% DDM, 0.5 mg/mL phosphatidylcholine, 0.5 mM DTT, 10 mM cysteine-TRIS (pH 6), 20% glycerol, and 50 mM MES-triethanolamine (pH 6.0) with additives described in distinct figure legends. Phosphate analogues (abbreviated as MgF, AlF, or BeF) were made by adding 2 mM KF to either 0.2 mM MgCl2, 0.2 mM AlCl3, or 0.2 mM Be(NO3)2. When ATP, β,γ-methyleneadenosine 5′-triphosphate (AMPPCP), adenosine 5′-(β,γ-imido)triphosphate (AMPPNP), or Pi was used, MgCl2 was added to reach the corresponding concentrations. Prior to digestion, CopA was reacted with these ligands by incubation at 30 °C for 1 h followed by 50 °C for 5 min. ATP was added just prior to the second incubation at 50 °C to decrease the level of unnecessary hydrolysis. Digestion patterns were assessed by resolving the protein fragments on 12% SDS gels. The gels were stained with Coomassie Brilliant Blue R250.
The activity of papain under the various conditions used in the proteolytic study was measured by using 1 mM N-α-benzoyl-l-Arg-p-nitroanilidine (l-BAPNA) as the substrate. The hydrolysis of l-BAPNA by papain (2−10 μg/mL) was monitored by reading the 405 nm absorption. To avoid interference with the absorption, phosphatidylcholine and BCS were excluded.
Results
Effect of Temperature on Proteolysis
In preliminary experiments, we established that digestion of the synthetic substrate l-BAPNA by papain proceeds linearly for more than 30 min at temperatures varying between 30 and 60 °C, demonstrating that papain is highly stable through this temperature range. In experiments with CopA protein, in which the concentration of papain was adjusted so that the total proteolytic activity was the same at different temperatures, we found that digestion at certain proteolytic sites becomes much faster as the temperature is increased from 30 to 50 °C. In the absence of copper, production of the p79 fragments is very fast and rather insensitive to temperature. On the other hand, the p44 fragment yield, produced by cleavage at the A domain−M3 loop, is dependent on temperature (not shown) and time (Figure 2). This dependence of the cleavage rate at this specific site suggests greater conformational fluctuations around the A domain−M3 loop as the temperature is increased to the optimal level (65 °C) for CopA catalytic activity. It is also likely that a specific conformational state, requiring a high energy of activation, becomes prevalent at higher temperatures.
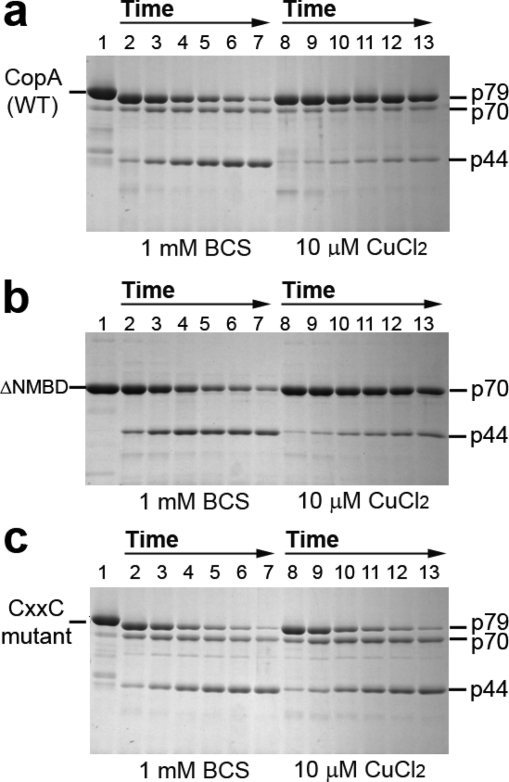
Figure 2.
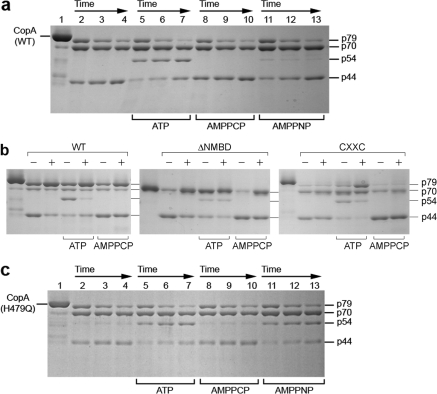
Effect of copper on the digestion pattern of T. maritima CopA WT, ΔNMBD, and CXXC proteins. SDS−PAGE was performed as described in Experimental Procedures. Purified proteins (0.5 mg/mL) of CopA WT (a) and the mutants (b and c) were incubated with 0.5 mg/mL papain at 50 °C in the presence of 1 mM BCS without copper (lanes 2−7) or with 10 μM CuCl2 (lanes 8−13), as indicated at the bottom. The numbers on top of the gels refer to minutes of digestion: lane 1, 0 min; lanes 2 and 8, 2 min; lanes 3 and 9, 5 min; lanes 4 and 10, 10 min; lanes 5 and 11, 15 min; lanes 6 and 12, 20 min; and lanes 7 and 13, 30 min.
Effect of Copper
The ATPase activity of T. maritima CopA is dependent on copper within the micromolar range (40), suggesting that copper binding may produce a conformational change involved in catalytic activation. It is shown in Figure 2a that, in the absence of copper (BCS present), the p79 fragment is first reduced to p70, which appears to remain at a steady state level. Then the p44 is gradually produced. However, the presence of copper at concentrations permitting maximal ATPase activity (i.e., 10 μM) confers significant protection (Figure 2a) of the p79 fragment, whereby only small quantities of p70 and p44 are produced. Thus, copper reduces the susceptibility of both the NMBD−Ma loop (R91) and the A domain−M3 loop (S328) to digestion, suggesting that the NMBD and the A domain fold over and/or change their positions. It should be pointed out that papain activity measurements using l-BAPNA demonstrated that addition of 10 μM CuCl2 produces mild inhibition of papain activity at 50 °C. However, this reduction is much smaller than the observed protection by copper.
If CopA with deletion of the NMBD (ΔNMBD) is used, the intact protein migrates as a p70 band reflecting the NMBD deletion. In the absence of copper, production of the p44 fragment (i.e., cleavage at S328 on the A domain−M3 loop) occurs within a time frame similar to that observed with the WT protein (compare digestion in panels a and b of Figure 2). In the presence of copper, the p70 protein is protected and the yield of p44 fragment reduced, as observed for the WT (Figure 2b). This indicates that the conformational effect is induced by copper binding to the transmembrane copper binding site (TMBS), assuming that the TMBS is the only residual copper binding site in ΔNMBD. In fact, the TMBS was demonstrated to be the only residual (functional) copper binding site following NMBD deletion in A. fulgidus CopA (37).
In the absence of copper, the CXXC mutant, in which the CXXC copper binding motif of the NMBD is disrupted by C17A and C20A mutations, yields the same digestion pattern observed with both WT and ΔNMBD. However, in this case, addition of copper does not produce any protection (Figure 2c), suggesting that the copper-free NMBD (CXXC mutant) inhibits the conformational change induced by the copper binding to the TMBS. It is noteworthy that when the H479Q mutant is used, copper protection occurs (not shown) with the same pattern observed with the WT protein.
Phosphorylation with Pi
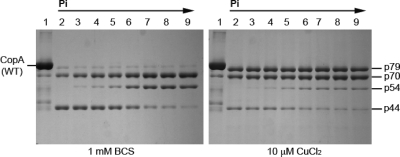
In analogy to the Ca2+ ATPase of the sarcoplasmic reticulum (SERCA1a), T. maritima CopA reacts with inorganic phosphate (Pi) in the absence of copper to yield a phosphoenzyme intermediate in the reverse direction of the catalytic cycle (40). On the basis of the common reaction scheme of P-type ATPases, the resulting phosphointermediate is designated as E2P. We now find that in parallel with the Pi concentration dependence of the phosphorylation reaction with Pi, the digestion pattern by papain changes, indicating specific accumulation of the p70 and p54 fragments, while the p44 band is reduced to a minimal intensity (Figure 3). This Pi effect was less prominent in the presence of copper, indicating that Cu inhibits the Pi-induced conformational change. This is consistent with the previous results of phosphoenzyme measurements following equilibration with saturating [32P]Pi, where the phosphoenzyme level was decreased by copper from 4.0−4.2 nmol/mg (approximately 40% of the enzyme stoichiometry) to 0.30−0.32 nmol/mg. It is noteworthy that the full effect of Pi is not realized in these experiments, since less than half the protein is in the phosphorylated state even in the presence of saturating Pi concentrations while the remaining half is in the covalent E·Pi complex (i.e., Keq ∼ 1).
Figure 3.
Effect of Pi on the digestion pattern of T. maritima CopA with papain. SDS−PAGE was performed as described in Experimental Procedures. Purified CopA (0.5 mg/mL) was incubated with 0.5 mg/mL papain at 50 °C for 30 min in the presence of 1 mM BCS (no copper) or 10 μM CuCl2, in the presence of various concentrations of Pi as indicated by the numbers above the gels: lane 2, 0 mM; lane 3, 0.5 mM; lane 4, 1 mM; lane 5, 2 mM; lane 6, 5 mM; lane 7, 10 mM; and lane 8, 20 mM. Lane 1 contained a nondigested control.
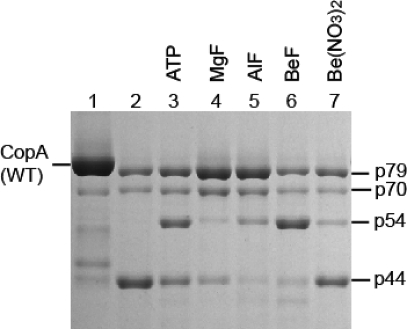
AlF binds to CopA yielding a stable complex which is protected from papain digestion (38). We now find that, in the absence of copper, not only AlF but also MgF and, much more prominently, BeF produce conformational effects which are featured by the accumulation of p79 and/or p54 fragments (Figure 4). BeF has a striking effect as it enhances the decrease in the level of p79 into p54 instead of p44 (compare lanes 4 and 5 to lane 6 in Figure 4). Beryllium nitrate alone is much less effective (Figure 4, lane 7), demonstrating the fluoride requirement for realizing this conformational effect. It is apparent that the digestion pattern observed with BeF presents analogies to that produced by Pi. Therefore, the BeF·CopA complex formed in the absence of copper may be considered an analogue of the phosphoenzyme obtained with Pi. In fact, it is known that BeF binds to SERCA1, producing a stable analogue of the phosphoenzyme (E2P) ground state (41,42).
Figure 4.
Effect of phosphate analogues on the digestion pattern of T. maritima CopA. SDS−PAGE was performed as described in Experimental Procedures. Purified CopA (0.5 mg/mL) was incubated with 0.5 mg/mL papain at 50 °C for 30 min in the presence of 1 mM BCS (no copper added), in the absence or presence of 2 mM ATP, or following preincubation with 0.2 mM MgF, 0.2 mM AlF, 0.2 mM BeF, or 0.2 mM Be(NO3)2.
Phosphorylation with ATP
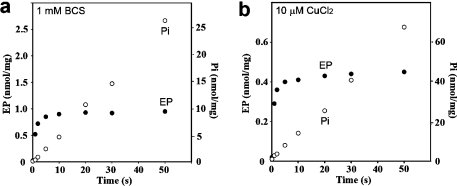
ATP, in the absence of copper, produces a digestion pattern nearly identical to that observed following treatment with Pi or BeF, enhancing the level of the p54 fragment (Figure 4, lane 3). This suggests that, in the absence of copper, CopA utilizes ATP to assume a state similar to E2P, even though most P-type ATPases utilize ATP only in the presence of transported ions. Such an unexpected feature of T. maritima CopA has also been suggested in our previous study using phosphoenzyme measurements (40). Since those experiments were performed with purified CopA protein, we have now obtained new measurements using the membrane-bound protein (instead of purified protein) obtained directly from expression host cells, to rule out possible effects of exposure to detergent and purification procedures. It is shown in Figure 5a that this protein forms a phosphoenzyme by utilizing ATP in the absence of copper, reaching a steady state level of nearly 1 nmol/mg of protein, while hydrolytic cleavage of Pi occurs at a rate of 31 nmol mg−1 min−1. In the presence of copper (Figure 5b), the phosphoenzyme level reaches only 0.45 nmol/mg, while the rate of Pi production is higher (80 nmol mg−1 min−1). It is clear that the effect of copper is to stimulate hydrolytic cleavage, whereby the steady state level of the phosphoenzyme intermediate is reduced. These experiments confirm that phosphoenzyme formation by utilization of ATP can indeed be realized even in the absence of copper, although the conformation of the phosphoenzyme may be different in the absence versus the presence of copper. It should be pointed out that we previously demonstrated by ICP-mass spectrometry that copper is in fact removed from CopA by 1 mM BCS (40). In the experiments presented here, to possibly ensure a more effective copper removal, we used 10 mM BCS or 1 mM tetrathiomolybdate. In either case, we observed no reduction in the level of phosphoenzyme formation by utilization of ATP.
Figure 5.
Effect of copper on formation and subsequent processing of the phosphoenzyme upon addition of ATP to CopA in membrane vesicles obtained directly from expression host cells. The reaction was started by the addition of 25 μM [γ32P]ATP to membrane vesicles including T. maritima CopA (25 μg/mL) in the presence of 1 mM BCS (a) or 25 μM CuCl2 (b) at 60 °C (see Experimental Procedures for details).
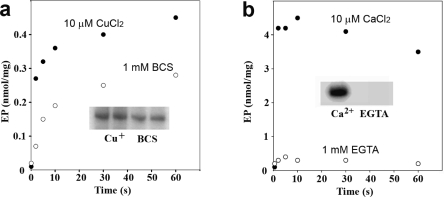
To further evaluate the validity of these findings, we extended our measurements to other comparable cation transport ATPases. It is shown in Figure 6a that the copper ATPase from the thermophilic A. fulgidus(34,35) displays a different behavior under the same assay conditions, whereas the phosphoenzyme level obtained in the presence of copper is higher than that obtained in the absence of copper (BCS present). We also performed the measurments using the assay conditions identical to those reported previously (pH 7.5 and 40 °C used in refs (34) and (35)), confirming that the phosphoenzyme levels in the presence of copper are significantly higher (0.96−1.0 nmol/mg), due to a lower degree of hydrolytic cleavage at this pH and temperature. On the other hand, the low phosphoenzyme levels obtained in the absence of copper remain exactly the same (0.3 nmol/mg). Finally, comparative experiments with Ca2+ ATPase (SERCA1) revealed an even more different behavior, yielding high phosphoenzyme levels soon after addition of ATP in the presence of Ca2+ (Figure 6b), but no phosphoenzyme at all in the absence of Ca2+(43). Therefore, we conclude that the ability to form phosphoenzyme by utilization of ATP in the absence of activating cation (i.e., copper) is a specific, or at least more pronounced, feature of T. maritima CopA.
Figure 6.
Effect of copper or calcium on formation of the phosphoenzyme upon addition of ATP to A. fulgidus CopA or Ca2+ ATPase (SERCA1a). Specific cation dependence of phosphoenzyme formation by A. fulgidus CopA (a) and SERCA1a Ca2+ ATPase (b), to be compared with that of T. maritima CopA (shown in Figure 5). (a) The reaction was started by the addition of 25 μM [γ-32P]ATP to 25 μg/mL CopA in the presence of 3 μM CuCl2 (●) or in the absence (○) of copper (1 mM BCS present) at 60 °C. (b) The reaction was started by the addition of 25 μM [γ-32P]ATP to 25 μg/mL SERCA1a in the presence of 50 μM CaCl2 (●) or in the absence (○) of calcium (1 mM EGTA present) at 25 °C. Autoradiograms showing steady state levels of the radioactive phosphoenzyme are shown as insets.
We further explored the effects of ATP and its nonhydrolyzable analogues (AMPPCP and AMPPNP) on the enzyme conformations of WT, ΔNMBD, and CXXC CopA. It should be noted that purified CopA protein utilizes ATP in the absence of copper to yield nearly stoichiometric levels (8−10 nmol/mg) of phosphoenzyne (40). AMPPCP and, to a large extent, AMPPNP have no effect on the digestion patterns (Figure 7a), indicating that the confomational effect requires γ-phosphate transfer rather than nucleotide binding. This again underscores the formation of E2P by utilization of ATP (Figure 4).
Figure 7.
Effect of ATP, AMPPCP, and AMPPNP on the digestion pattern of T. maritima CopA WT, ΔNMBD, and CXXC and H479Q mutant proteins. SDS−PAGE was performed as described in Experimental Procedures. Purified CopA protein (0.5 mg/mL) was incubated with 0.5 mg/mL papain at 50 °C in the absence and presence of nucleotides. (a and c) Time course digestion experiments with CopA WT (a) and H479Q (c) were performed in the absence of copper (1 mM BCS present). Nucleotides were added as indicated in the figure. The time of digestion was 10 min for lanes 2, 5, 8, and 11; 20 min for lanes 3, 6, 9, and 12; and 30 min for lanes 4, 7, 10, and 13. (b) Digestion of CopA WT, ΔNMBD, and CXXC was performed for 30 min in the presence of either 10 μM CuCl2 (+) or 1 mM BCS (−), with or without 2 mM ATP or 2 mM AMPPCP as indicated.
If CopA is phosphorylated with ATP in the presence of copper, the p54 band is not observed and the p79 band is prominent (Figure 7b), with a pattern similar to that produced by copper in the absence of ATP. This may be attributed to a E1-P conformation, although, due to hydrolytic cleavage of the phospoenzyme, a significant level of CopA may be in the E1 state under steady state conditions.
When we extended our studies with ATP to the mutants, we found that the NMBD deletion renders the ATP effect less prominent and insensitive to copper (Figure 7b). This demostrates the interdependence of the ATP-induced conformational change and the presence of the NMBD. On the other hand, a significant effect of ATP (i.e., formation of p54) is obtained with the CXXC mutatant (Figure 7b). In this case, copper shows an effect, indicating that the residual copper binding site (TMBS) is occupied by bound copper and is involved in the observed effect. Considering that the previous finding that the CXXC mutant is fully phosphorylated in the presence of copper and ATP (40), we conclude that the protein assumes a copper-bound phosphoenzyme conformation, commonly termed E1P in the field of P-type ATPases. This suggests that the lack of catalytic turnover observed with this mutant (ref (40); also shown in Figure 8a) is due to a deficiency in processing of E1P to subsequent conformational states.
Figure 8.
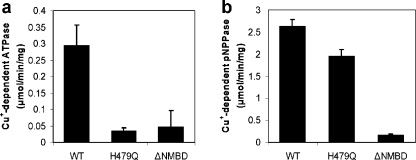
Copper-dependent ATPase and p-nitrophenyl phosphatase activities. ATPase activity (a) and p-nitrophenyl phosphatase (pNPPase) activity (b) of CopA WT, H479Q, and ΔNMBD proteins were measured in the presence or absence of copper. The Cu+-dependent activities were obtained by subtracting the activity in the absence of copper from that in its presence. These measurements were obtained at 40 °C, due to the high pNPPase activity. The average and standard deviation (n = 3) are shown.
Finally, we performed additional studies on the H479Q mutant, mimicking one of the most common mutations (H1044Q) of human copper ATPase (ATP7B) in Wilson disease (44). It is noteworthy that the H479Q mutant is not phosphorylated by ATP either in the absence or in the presence of copper (40). We were therefore surprised to find that ATP produces an effect on the digestion pattern of the H479Q mutant (Figure 7c) similar to that produced in the WT protein (Figure 7a). We checked again whether the H479Q protein is phosphorylated using high ATP concentrations (1 mM) as used for the proteolytic study and found no significant level of acid stable phosphoenzyme. On the basis of these findings, we concluded that, at least in the absence of copper, ATP interacts with the H479Q mutant and produces a conformational effect similar to that observed for the WT even though the phosphoryl transfer reaction does not proceed to completion (see Discussion). An additional observation of interest is that the conformational effects of AMPPCP and AMPPNP are more prominent in the H479Q mutant than in WT CopA (compare panels a and c of Figure 7).
These findings indicate that the H479Q mutant catalytic defect is caused by unfavorable positioning of ATP and may be compensated by use of other phosphate donor substrates commonly used for P-type ATPases. Therefore, we examined the ability of CopA WT and the mutants to hydrolyze p-nitrophenyl phosphate (pNPP), which was reported to be utilized by A. fulgidus CopA (45). Under the same conditions that were used for the ATPase reaction, T. maritima CopA showed copper-dependent pNPPase activity, inasmuch as it undergoes approximately 75% inhibition in the presence of 1 mM BCS. Interestingly, the H479Q mutation affects only slightly the copper-dependent pNPPase activity, while it inhibits strongly the ATPase activity. In contrast, the NMBD deletion affects both copper-dependent pNPPase and ATPase activity.
Discussion
Digestion of CopA with papain occurs with specific patterns that provide information about protein conformational states in solution, without constraints by crystallization requirements. In fact, proteolytic digestion patterns with papain have provided useful information even on conformational states of human copper ATPase ATP7B (46), as well as bacterial CopA (38). Using the method of limited proteolysis established in our previous study, we demonstrate here how the specific mutations in the NMBD and the invariant His (H479) cause a catalytic defect by comparing ligand effects on protein conformation. The effects detected by proteolytic analysis are prevalent on sites near the region between the cytoplasmic domains and transmembrane segments. In fact, the cleavage sites yielding p70, p54, and p44 are located at the NMBD−Ma loop (R91), the M2−A domain loop (A225), and the A domain−M3 loop [S328 (Figure 1)]. Therefore, the ligand-induced digestion patterns observed in our experiments are related to protein conformation changes around the A domain and the NMBD. The importance of conformational changes in this area, and related A domain movements, was demonstrated in the Ca2+ ATPase (SERCA) reaction cycle, as they influence P and N domain interactions, promote long-range effects on cation binding sites, and place the TGES motif in an ideal position for catalytic assistance in the hydrolytic cleavage of the aspartyl phosphate (47). Relevant to our observations, structural evidence for A domain homologies in CopA and Ca2+ ATPase (25) and for the interaction of the NMBD with the catalytic headpiece of CopA (28) has been reported. We find that the effects of ligand on conformational states are much more prominent at high temperatures, near the catalytic optimum of T. maritima CopA (65 °C), likely due to temperature-dependent domain fluctuation and organization.
One of the important changes in the digestion pattern is due to the effect of copper, which is very clear at 60 °C but hardly noticeable at 30 °C (38). When recombinant CopA (83 kDa) is digested in the absence of copper (BCS present), p70 and p44 fragments are formed through cleavage at the NMBD−Ma (R91) and A domain−M3 (S328) loops (Figure 1). On the other hand, copper binding to WT CopA (when binding to both the NMBD and TMBS is available) causes significant protection of the p79 fragment, i.e., R91 site (Figure 2a). This enzyme state is designated as Cu·Ea1 in Scheme 1, where the conformations with the Cu-bound NMBD and Cu-free NMBD are represented as Ea and Eb, respectively, to indicate a high-turnover “activated” state, as opposed to a low-turnover “basal” state (40). It is of interest that the ΔNMBD mutant, p70, is protected by copper (Figure 2b), indicating that protection of A225 can be produced by copper occupancy at TMBS, even in the absence of the NMBD. In contrast, the copper-induced protection is not observed for the CXXC mutant (Figure 2c), suggesting that the presence of the Cu-free NMBD interferes with functionally relevant transitions. Such a conformational role of the NMBD is consistent with a recent study by cryoelectron microscopy (28), providing structural evidence of the proximity of the NMBD and the catalytic headpiece in A. fulgidus CopA.
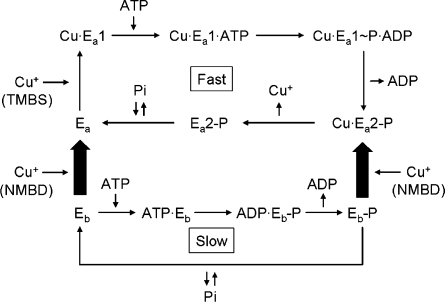
Scheme 1. Reaction Scheme Based on the Functional (40) and Conformational (this paper) Characterization of T. maritima CopA.
The enzyme can be phosphorylated by either ATP or Pi in the absence of copper, yielding Eb-P undergoing slow turnover. However, the active (Ea) conformation undergoing fast turnover is only obtained following binding of Cu+ to both NMBD and TMBS. Interaction of NMBD with the catalytic headpiece and A domain movements play important roles in the sequential reactions of the fast cycle. It is possible that the copper occupancy of the NMBD remains throughout the cycle, while copper bound to the TMBS is released within each cycle. See Discussion.
As previously reported, the CXXC mutant strongly inhibits ATPase activity (ref (40); also shown in Figure 8). The catalytic deficiency in this mutant is then related to its inability to assume the digestion-resistant state upon binding of Cu+ to TMBS. It is of interest that while activation of SERCA (Ca·E1 formation) is due to the cooperative calcium binding to two neighboring transmembrane sites (21,48), activation of CopA occurs through a more complex mechanism triggered by copper binding at two distant sites (NMBD and TMBS).
Another important aspect of our observations relates to the conformational states of the phosphoenzyme intermediate. From the experimental point of view, a most convenient method for obtaining enzyme phosphorylation is equilibration of CopA with inorganic phosphate (Pi) in the absence of copper (40). Under these conditions, the phosphoenzyme intermediate is obtained in the reverse direction of the hydrolytic reaction at the end of the catalytic cycle. We now find that in parallel with the Pi concentration dependence of the phosphorylation reaction, the digestion pattern by papain changes to yield prominent p70 and p54 bands, while the p44 band is reduced to a minimal intensity (Figure 3). As these changes in digestion pattern are related to the susceptibility of the M2−A domain loop (A225) and the A domain−M3 loop (S328), it is apparent that CopA phosphorylation with Pi induces movement of the A domain. The same effect, but more pronounced, is obtained with BeF (Figure 4), suggesting that the BeF·CopA complex assumes a state analogous to that obtained with Pi. It should be noted that the BCS used for these experiments removes bound copper ions from both the NMBD and TMBS (40), suggesting that the phosphoenzyme obtained with Pi, and the BeF analogue thereof, corresponds to Eb-P in Scheme 1.
An unexpected feature of T. maritima CopA is its ability to form the phosphoenzyme intermediate by ATP utilization even in the absence of copper; direct evidence of removal of copper by BCS was obtained using ICP-MS (40). In fact, addition of copper reduces rather than increases the phosphoenzyme level, due to catalytic activation of its hydrolytic cleavage. The resulting phosphoenzyme demonstrates a digestion pattern analogous to those obtained by Pi and BeF (Figures 4 and 7a). This catalytic property is in clear contrast with those of other P-type ATPases that are generally phosphorylated only in the presence of specific cations undergoing transport. We have now performed comparative experiments on ATP utilization in the absence or presence of transported cations using P-type ATPases from different biological sources (SERCA1, A. fulgidus CopA, and T. maritima CopA), demonstrating that the ATP utilization in the absence of transported cations is unique, or at least greater, for T. maritima CopA. We may consider, in this regard, the broad family of haloacid dehalogenases (49), including the P-type ATPases. Members of this family that utilize phosphosubstrates, such as phosphoserine phosphatase (41), share the catalytic mechanism of the phosphoryl transfer reaction which is triggered by proper positioning of the bound substrate and coordinating Mg2+, with no need for long-range activation by specific cations. In the case of SERCA, and possibly most members of the PII-type ATPase family, the proper positioning of bound ATP is allowed only in Ca·E1, obtained through long-range effects of bound calcium (43). This requirement is underpinned by the interdomain cross-linking nature of the bound ATP and mechanical cooperation between cytoplasmic ATPase domains (A, N, and P) and the transmembrane Ca2+ transport domain (50). The requirement appears to be less stringent for T. maritima CopA, at least for formation of a low-turnover phosphoenzyme intermediate.
The involvement of copper in the sequential conformational transitions required for fast turnover in T. maritima CopA is revealed by comparing the behavior of WT, ΔNMBD, and CXXC proteins. The ΔNMBD protein is protected just by the copper occupancy of the TMBS (Figure 2). On the other hand, in the CXXC mutant, copper binding to the TMBS cannot yield the Cu·Ea1 conformation, due to interference by the copper-free NMBD (Figure 2). Therefore, in the WT protein, the active conformation (Cu·Ea1 in Scheme 1) can be obtained only following occupancy of both NMBD and TMBS copper sites. It is most interesting that the CXXC mutant can be phosphorylated with ATP, and in this case, protection of copper from proteolytic digestion is observed (Figure 7b). However, the phosphoenzyme does not undergo turnover, and no significant ATPase activity is observed (40). Therefore, the copper-bound NMBD is required for the transition to subsequent states in the reaction cycle. It is evident that proper interaction of the NMBD with the catalytic headpiece (28) is very important in this regard.
A final point of interest is related to the functional consequence of the H479Q mutation mimicking one of the most common mutations (H1044Q) of the human copper ATPase (ATP7B) in Wilson disease (44). It is noteworthy that a Phe and an Arg involved in nucleotide stabilization in PII-type ATPases are missing in the CopA sequence. Therefore, an invariant His within the nucleotide binding site of the PIB-ATPases, including CopA, is likely to sustain an important role in stabilization of the adenosine moiety and possibly α- and β-phosphates (23,24,26). In fact, the H479Q mutation produces a drastic inhibition of ATPase activity through specific interference with phosphoenzyme formation by ATP utilization, without affecting phosphorylation with Pi(40) or pNPPase activity (Figure 8). The interesting finding is that ATP can interact with the H479Q mutant in the absence of copper, producing a conformational effect quite similar to that obtained with BeF on the WT CopA. Therefore, following the H479Q substitution, ATP binds yielding the conformation of the phosphoenzyme analogue, but covalent transfer of γ-phosphate does not occur.
In fact, the nonhydrolyzable ATP analogue AMPPNP produces an effect similar to that of ATP, while AMPPCP does not (Figure 7c). This may be related to the AMPPCP PXP bond angle (117°), which is significantly different from that in ATP (128.7°) or AMPPNP [127.2° (51)]. This underlines the importance of the γ-phosphate positioning for the conformational effect. It is possible that limited restraint at the nucleotide site and significant motional freedom due to the H479 mutation allow favorable positioning of the γ-phosphate at the phosphorylation site, yielding a conformational analogue of the phosphoenzyme. However, inadequate stabilization of bound ATP interferes with covalent phosphoryl transfer, possibly by distortion of the environment around γ-phosphate and the lack of ADP stabilization as a leaving group. These observations on the comparative behavior of five different substrates (ATP, AMPPCP, AMPPNP, Pi, and pNPP) define the requirements for conformational change and phosphoryl transfer in CopA.
Acknowledgments
We are thankful to Dr. Jose Argüello for the kind gift of A. fulgidus CopA.
This work was supported by NHLBI Grant RO1-69830 from the National Institutes of Health. Y.H. was supported in part by a fellowship from Astellas Foundation for Research on Metabolic Disorders.
Funding Statement
National Institutes of Health, United States
Footnotes
Abbreviations: NMBD, N-terminal metal binding domain; TMBS, transmembrane binding site; DDM, n-dodecyl β-d-maltoside; BCS, bathocuproine disulfonate; pNPP, p-nitrophenyl phosphate; AMPPCP, β,γ-methyleneadenosine 5′-triphosphate; AMPPNP, adenosine 5′-(β,γ-imido)triphosphate; l-BAPNA, N-α-benzoyl-l-Arg-p-nitroanilidine; SERCA, sarcoendoplasmic reticulum Ca2+ ATPase.
References
- Pedersen P. L.; Carafoli E. (1987) Ion motive ATPases. I. Ubiquity, properties, and significance to cell function. Trends Biochem. Sci. 12, 146–150. [Google Scholar]
- Lutsenko S.; Kaplan J. H. (1995) Organization of P-type ATPases: Significance of structural diversity. Biochemistry 34, 15607–15613. [DOI] [PubMed] [Google Scholar]
- Mo̷ller J. V.; Juul B.; le Maire M. (1996) Structural organization, ion transport, and energy transduction of P-type ATPases. Biochim. Biophys. Acta 1286, 1–51. [DOI] [PubMed] [Google Scholar]
- Axelsen K. B.; Palmgern M. G. (1998) Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 46, 84–101. [DOI] [PubMed] [Google Scholar]
- Argüello J. M. (2003) Identification of ion-selectivity determinants in heavy-metal transport P1B-type ATPases. J. Membr. Biol. 195, 93–108. [DOI] [PubMed] [Google Scholar]
- Williams L. E.; Pittman J. K.; Hall J. L. (2000) Emerging mechanisms for heavy metal transport in plants. Biochim. Biophys. Acta 1465, 104–126. [DOI] [PubMed] [Google Scholar]
- Silver S. (1996) Bacterial resistances to toxic metal ions. Gene 179, 9–19. [DOI] [PubMed] [Google Scholar]
- Bull P. C.; Cox D. W. (1994) Wilson disease and Menkes disease: New handles on heavy-metal transport. Trends Genet. 10, 246–252. [DOI] [PubMed] [Google Scholar]
- Petris M. J.; Strausak D.; Mercer J. F. (2000) The Menkes copper transporter is required for the activation of tyrosinase. Hum. Mol. Genet. 9, 2845–2851. [DOI] [PubMed] [Google Scholar]
- Lutsenko S.; Barnes N. L.; Bartee M. Y.; Dmitriev O. Y. (2007) Function and regulation of human copper-transporting ATPases. Physiol. Rev. 87, 1011–1046. [DOI] [PubMed] [Google Scholar]
- Petrukhin K; et al. (1994) Characterization of the Wilson disease gene encoding a P-type copper transporting ATPase: Genomic organization, alternative splicing, and structure/function predictions. Hum. Mol. Genet. 3, 1647–1656. [DOI] [PubMed] [Google Scholar]
- Bull P. C.; Thomas G. R.; Rommens J. M.; Forbes J. R.; Cox D. W. (1993) The Wilson disease gene is a putative copper transporting P-type ATPase similar to the Menkes gene. Nat. Genet. 5, 327–337. [DOI] [PubMed] [Google Scholar]
- Vulpe C.; Levinson B.; Whitney S.; Packman S.; Gitschier J. (1993) Isolation of a candidate gene for Menkes disease and evidence that it encodes a copper-transporting ATPase. Nat. Genet. 3, 7–13. [DOI] [PubMed] [Google Scholar]
- Solioz M.; Odermatt A. (1995) Copper and Silver Transport by CopB-ATPase in Membrane Vesicles of Enterococcus hirae. J. Biol. Chem. 270, 9217–9221. [DOI] [PubMed] [Google Scholar]
- Okkeri J.; Haltia T. (1999) Expression and mutagenesis of ZntA, a zinc-transporting P-type ATPase from Escherichia coli. Biochemistry 38, 14109–14116. [DOI] [PubMed] [Google Scholar]
- Sharma R.; Rensing C.; Rosen B. P.; Mitra B. (2000) The ATP hydrolytic activity of purified ZntA, a Pb(II)/Cd(II)/Zn(II)-translocating ATPase from Escherichia coli. J. Biol. Chem. 275, 3873–3878. [DOI] [PubMed] [Google Scholar]
- Rensing C.; Mitra B.; Rosen B. P. (1997) The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc. Natl. Acad. Sci. U.S.A. 94, 14326–14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K. J.; Yoon K. P.; Lynn A. R. (1992) ATP-dependent cadmium transport by the cadA cadmium resistance determinant in everted membrane vesicles of Bacillus subtilis. J. Bacteriol. 174, 116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLennan D. H.; Brandl C. J.; Korczak B.; Green N. M. (1985) Amino-acid sequence of a Ca2+ + Mg2+-dependent ATPase from rabbit muscle sarcoplasmic reticulum, deduced from its complementary DNA sequence. Nature 316, 696–700. [DOI] [PubMed] [Google Scholar]
- Shull G. E.; Schwartz A.; Lingrel J. B. (1985) Amino-acid sequence of the catalytic subunit of the (Na+ + K+)ATPase deduced from a complementary DNA. Nature 316, 691–695. [DOI] [PubMed] [Google Scholar]
- Toyoshima C.; Nakasako M.; Nomura H.; Ogawa H. (2000) Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature 405, 6787647–655. [DOI] [PubMed] [Google Scholar]
- Morth J. P; et al. (2007) Crystal structure of the sodium-potassium pump. Nature 450, 1043–1049. [DOI] [PubMed] [Google Scholar]
- Dmitriev O; et al. (2006) Solution structure of the N-domain of Wilson disease protein: Distinct nucleotide-binding environment and effects of disease mutations. Proc. Natl. Acad. Sci. U.S.A. 103, 5302–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazinsky M. H.; Mandal A. K.; Argüello J. M.; Rosenzweig A. C. (2006) Structure of the ATP binding domain from the Archaeoglobus fulgidus Cu+-ATPase. J. Biol. Chem. 281, 11161–11166. [DOI] [PubMed] [Google Scholar]
- Sazinsky M. H.; Agarwal S.; Argüello J. M.; Rosenzweig A. C. (2006) Structure of the actuator domain from the Archaeoglobus fulgidus Cu+-ATPase. Biochemistry 45, 9949–9955. [DOI] [PubMed] [Google Scholar]
- Lübben M; et al. (2007) Sulfate acts as phosphate analog on the monomeric catalytic fragment of the CPx-ATPase CopB from Sulfolobus solfataricus. J. Mol. Biol. 369, 368–385. [DOI] [PubMed] [Google Scholar]
- Solioz M.; Vulpe C. (1996) CPx-type ATPases: A class of P-type ATPases that pump heavy metals. Trends Biochem. Sci. 21, 237–241. [PubMed] [Google Scholar]
- Wu C. C.; Rice W. J.; Stokes D. L. (2008) Structure of a copper pump suggests a regulatory role for its metal-binding domain. Structure 16, 976–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintalapati S.; Al Kurdi R.; Terwisscha van Scheltinga A. C.; Kühlbrandt W. (2008) Membrane Structure of CtrA3, a Copper-transporting P-type-ATPase from Aquifex aeolicus. J. Mol. Biol. 378, 581–595. [DOI] [PubMed] [Google Scholar]
- Walker J. M; et al. (2004) The N-terminal metal-binding site 2 of the Wilson’s Disease Protein plays a key role in the transfer of copper from Atox1. J. Biol. Chem. 279, 15376–15384. [DOI] [PubMed] [Google Scholar]
- Cobine P. A; et al. (2002) Copper transfer from the Cu(I) chaperone, CopZ, to the repressor, Zn(II)CopY: Metal coordination environments and protein interactions. Biochemistry 41, 5822–5829. [DOI] [PubMed] [Google Scholar]
- Multhaup G.; Strausak D.; Bissig K. D.; Solioz M. (2001) Interaction of the CopZ copper chaperone with the CopA copper ATPase of Enterococcus hirae assessed by surface plasmon resonance. Biochem. Biophys. Res. Commun. 288, 172–177. [DOI] [PubMed] [Google Scholar]
- Tsivkovskii R.; MacArthur B. C.; Lutsenko S. (2001) The Lys1010-Lys1325 fragment of the Wilson’s disease protein binds nucleotides and interacts with the N-terminal domain of this protein in a copper-dependent manner. J. Biol. Chem. 276, 2234–2242. [DOI] [PubMed] [Google Scholar]
- Mandal A. K.; Cheung W. D.; Argüello J. M. (2002) Characterization of a thermophilic P-type Ag+/Cu+-ATPase from the extremophile Archaeoglobus fulgidus. J. Biol. Chem. 277, 7201–7208. [DOI] [PubMed] [Google Scholar]
- Mandal A. K.; Argüello J. M. (2003) Functional roles of metal binding domains of the Archaeoglobus fulgidus Cu+-ATPase CopA. Biochemistry 42, 11040–11047. [DOI] [PubMed] [Google Scholar]
- Rice W. J.; Kovalishin A.; Stokes D. L. (2006) Role of metal-binding domains of the copper pump from Archaeoglobus fulgidus. Biochem. Biophys. Res. Commun. 348, 124–131. [DOI] [PubMed] [Google Scholar]
- González-Guerrero M.; Eren E.; Rawat S.; Stemmler T. L.; Argüello J. M. (2008) Structure of the two transmembrane Cu+ transport sites of the Cu+-ATPases. J. Biol. Chem. 283, 29753–29759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori Y.; Majima E.; Tsuda T.; Toyoshima C. (2007) Domain organization and movements in heavy metal ion pumps: Papain digestion of CopA, a Cu+-transporting ATPase. J. Biol. Chem. 282, 25213–25221. [DOI] [PubMed] [Google Scholar]
- Eletr S.; Inesi G. (1972) Phospholipid orientation in sarcoplasmic membranes: Spin-label ESR and proton MNR studies. Biochim. Biophys. Acta 282, 174–179. [DOI] [PubMed] [Google Scholar]
- Hatori Y; et al. (2008) Intermediate phosphorylation reactions in the mechanism of ATP utilization by the copper ATPase (CopA) of Thermotoga maritima. J. Biol. Chem. 283, 22541–22549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H; et al. (2001) BeF3− acts as a phosphate analog in proteins phosphorylated on aspartate: Structure of a BeF3− complex with phosphoserine phosphatase. Proc. Natl. Acad. Sci. U.S.A. 98, 8525–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima C.; Norimatsu Y.; Iwasawa S.; Tsuda T.; Ogawa H. (2007) How processing of aspartylphosphate is coupled to lumenal gating of the ion pathway in the calcium pump. Proc. Natl. Acad. Sci. U.S.A. 104, 19831–19836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inesi G.; Almendares J. (1968) Interaction of fragmented sarcoplasmic reticulum with 14C-ADP, 14C-ATP, and 32P-ATP. Effect of Ca2+ and Mg2+. Arch. Biochem. Biophys. 126, 733–735. [DOI] [PubMed] [Google Scholar]
- Tsivkovskii R.; Efremov R. G.; Lutsenko S. (2003) The role of the invariant His-1069 in folding and function of the Wilson’s disease protein, the human copper-transporting ATPase ATP7B. J. Biol. Chem. 278, 13302–13308. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Mandal A. K.; Bredeston L. M.; Gonzalez-Flecha F. L.; Arguello J. M. (2007) Activation of Archaeoglobus fulgidus Cu+-ATPase CopA by cysteine. Biochim. Biophys. Acta 1768, 495–501. [DOI] [PubMed] [Google Scholar]
- Leonhardt K.; Gebbardt R.; Mossner J.; Lutsenko S.; Huster D. (2009) Functional interactions of CU-ATPase ATP7B with cisplatin and the role of ATP7B in cells resistance to the drug. J. Biol. Chem. 284, 7793–7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima C.; Inesi G. (2004) Structural basis of ion pumping by Ca2+-ATPase of the sarcoplasmic reticulum. Annu. Rev. Biochem. 73, 269–292. [DOI] [PubMed] [Google Scholar]
- Clarke D. M.; Loo T. W.; Inesi G.; MacLennan D. H. (1989) Location of high affinity Ca2+-binding sites within the predicted transmembrane domain of the sarcoplasmic reticulum Ca2+-ATPase. Nature 339, 476–478. [DOI] [PubMed] [Google Scholar]
- Burroughs A. M.; Allen K. N.; Dunaway-Mariano D.; Aravind L. (2006) Evolutionary genomics of the HAD superfamily: Understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J. Mol. Biol. 361, 1003–1034. [DOI] [PubMed] [Google Scholar]
- Toyoshima C.; Mizutani T. (2004) Crystal structure of the calcium pump with a bound ATP analogue. Nature 430, 529–535. [DOI] [PubMed] [Google Scholar]
- Yount R. G.; Babock D.; Ballantyne W.; Ojala D. (1971) Imidodiphosphate, and Adenosine Triphosphate Analog containing a P-N-P linkage. Biochemistry 10, 2484–2489. [DOI] [PubMed] [Google Scholar]