Abstract
KISS1 was first discovered as a metastasis suppressor, but also plays crucial roles in the onset of puberty. The KISS1 gene encodes a secreted protein of 145 amino acids that exhibits no sequence similarity with any known proteins. KISS1 protein is proteolytically processed to generate a number of so-called kisspeptins (KP), the most well characterized is known as KP-54 or metastin. KP-54 is carboxy-terminally amidated and binds to and activates the KISS1 receptor (KISS1R). The current studies were undertaken in order to determine structure of KP-54 using nuclear magnetic resonance and circular dichroism. KP-54 is mostly disordered both in water and in trifluoroethanol/water mixed solvent, with no structural motifs. In sodium dodecyl sulfate micelles, KP-54 remains mostly disordered except for a small increase in helical propensity (from 3.7% in water to 9.9% in micelles). Despite this apparent lack of structure, KP-54 is biologically active. The intrinsic disorder of KP-54 may confer advantages in its ability to recognize and bind a wide range of target proteins.
Keywords: Metastasis suppressor, KISS1, GPR54, Kisspeptin
Introduction
Metastasis suppressors are a growing class of molecules that, when re-expressed in metastatic tumor cells, block the formation of metastases without blocking orthotopic tumor growth (reviewed in [1, 2]). The KISS1 metastasis suppressor gene was originally discovered in 1996 by comparing mRNA expression in metastasis-suppressed chromosome 6-melanoma cell hybrids to their metastatic counterparts [3]. Subsequently, KISS1 was mapped to chromosome 1q32 [4], but its expression was found to be regulated by CRSP3 which maps to 6q32 via a TXNIP intermediate [5]. KISS1 re-expression in melanoma [3, 6], breast [7] or ovarian [8] cancer cell lines significantly blocks metastasis while not significantly affecting growth of tumor cells implanted at orthotopic sites.
Minimal structural or functional insights regarding KISS1 related to its mechanism of action were apparent by simply examining the primary amino acid sequence [3]. A putative signal peptide (Fig. 1) suggested that KISS1 was secreted, but efforts to identify nascent KISS1 in cell culture conditioned medium were unsuccessful. Since KISS1 contains a PEST (proline, glutamate, serine, threonine and aspartate-rich sequence), which often targets proteins for ubiquitination and proteosomal degradation, the short half-life of nascent KISS1 was partly explained. Additional presumptive protein kinase C, cAMP and tyrosine phosphorylation sites and an N-myristoylation site have yet to be validated in multiple studies.
Fig. 1.

Amino acid Sequence of KISS1 and KP-54 (bold) reveal a signal peptide (underlined) and mostly hydrophobic composition. Relatively high density of proline residues in KISS1 and KP-54 are also noted
A mechanism of action for KISS1 was enigmatic until, in 2001, three laboratories independently identified a polypeptide derived from KISS1 as the ligand for an orphan G-protein coupled receptor {GPR54, AXOR12, hOT7T175 [9–11], which hereafter will be called the KISS1 receptor (KISS1R)}. Ohtaki et al. [11] first reported that an internal 54 amino acid polypeptide, which they originally termed metastin, was amidated and bound KISS1R. Kotani et al. [9] discovered that additional KISS1-derived peptides could be identified in serum and cell culture medium and termed them kisspeptins (KP). KP-54 is identical to metastin, but two smaller peptides, KP13 and KP10, representing the C-terminal 13 and 10 amino acids of KP-54, respectively, can also bind KISS1R and initiate signaling. Nash et al. [12] recently showed that KISS1 processing into KP occurs outside the cell and that autocrine signaling may not be required for metastasis suppression.
Goldberg et al. [13] as well as Nash et al. [12] showed that the metastasis suppressed melanoma cells expressing KISS1 were capable of completing antecedent steps of the metastatic cascade, but failed to colonize ectopic sites. Their findings imply that KISS1/KP therapy could be used to maintain disseminated cells in a dormant state [12, 14]. While administration of native proteins/peptides is feasible, stability is an issue [15]. Likewise, more potent agonists or pharmacologically more desirable variants could potentially be developed if structural features of KPs were well defined. To realize this potential, additional structural information regarding KP-54, the natural ligand for KISS1R would be helpful. Therefore, the purpose of this study was to determine the structure of KP-54.
Materials and methods
Attempts at producing significant amounts of KISS1 and KP-54 using bacterial expression were unsuccessful; so, all of the studies reported here were performed on synthetic KP-54. The amidated KP-54 peptide was synthesized at the UAB Center for Aging Peptide Synthesis Core Facility using an automated peptide synthesizer (Model PS3; Protein Technologies, Inc., Tucson, AZ) using standard F-moc procedures for solid phase peptide synthesis. KP-54 was purified by preparative high pressure liquid chromatography (HPLC, Beckman, Fullerton, CA) using a Varian preparative column (250 cm length, 2.5 cm inner diameter) with a water/acetonitrile/0.1% trifluoroacetic acid (TFA) solvent system. The synthetic KP-54 was analyzed for purity using SDS–PAGE [16] and confirmed by analytical HPLC to be >95% pure. The peptide was then characterized by Ion-Spray mass spectral analysis (PE SCIEX, API III Plus LC/MS/MS System, Concord, ON, Canada).
Separate quantities of the peptide were dissolved in 50 mM sodium phosphate (pH 5.0) buffer, containing either a mixture of 90% H2O/10% D2O or 100% D2O along with 0.02% sodium azide, 0.1 mM deuterated EDTA (Cambridge Isotope Laboratories; Andover, MA) and a trace amount of a protease inhibitor cocktail (Sigma–Aldrich, St. Louis, MO).
Nuclear magnetic resonance spectroscopy was performed using 500 MHz or 600 MHz Bruker Avance systems (DRX; Billerica, MA), equipped with TXI probes. Silicon Graphics SGI O2 workstations, running XWIN-NMR v3.5 (Bruker) were used to collect and preliminarily analyze the data. The experiments conducted included: 1D proton Nuclear magnetic resonance spectrometry (NMR), 2D TOCSY (70 ms mixing time), 2D NOESY (100, 200, and 250 ms mixing times), ROESY, and 2D COSY (phase sensitive, and double quantum filtered). Pre-saturation and WATERGATE pulses were used for water suppression.
For the 2D experiments, 32 scans, 4,096 points, and 1,024 increments were used. The experiments were conducted at room temperature (298°K) to 313°K, using VT control, and nitrogen gas to maintain a temperature. The sample containing the protonated solvent was used in the 2D NOESY, ROESY, TOCSY, and PH-COSY experiments, while the sample using the deuterated solvent was used in separate 2D NOESY, TOCSY, and DQF-COSY experiments. Pre-saturation with minimal power was used for the sample dissolved in the deuterated solvent to remove residual solvent signals.
Nuclear magnetic resonance spectrometry data sets were processed using NMRPIPE [17]. All NMR data were processed using a sine bell window function with a 30° shift in both dimensions. The data sets were also zero filled as needed, and baseline correction enabled. Linear prediction was not used. The data were converted to XEASY format for assignment [18], using SPSCAN [18], and resonance assignments were made using XEASY.
Circular dichroism spectra were recorded using an AVIV Model 62 DS(Lakewood, NJ), with the sample chamber being saturated with nitrogen gas. Adjustments to the sample's pH were made with 1.0 microliter aliquots of 0.1 M HCl or 0.1 M NaOH as needed. Wavelengths ranging from 190 to 250 nm were scanned. The blank buffer was used to standardize the baseline, and subtracted from the actual measured spectrum. Secondary structure calculations were conducted using DICHROPROT and SOMCD [19].
To probe the effect of membrane association on the structure of KP-54, solutions of KP-54 in both DPC (dodecylphosphocholine) and SDS (sodium dodecyl sulfate) micelles were prepared. Both DPC and SDS were used above their critical micellar concentrations and at 100-fold molar excess over the peptide to ensure a single peptide per micelle. The CD spectra were recorded, and analyzed as described above.
Results and discussion
The therapeutic potential of KISS1 and kisspeptin(s) for controlling metastasis is great. In addition to the anti-metastatic activities, KP-54 binding and activation of KISS1R is the molecular trigger to initiate puberty in a variety of species [20–23]. As a result, kisspeptin(s) could also be useful for correcting some cases of idiopathic hypogonadotropic hypogonadotropism. However, achieving these potentials in either pathology will be aided by knowing the structure of the active kisspeptin(s) so that agonists can be more systematically developed.
Based upon primary amino acid sequence information, unprocessed KISS1 and KP-54 are both predominantly hydrophilic (Fig. 1), which is consistent with being found in physiologic solutions. The putative signal sequence (amino acids 1–19) has been confirmed both functionally-by detection of KP in human serum [9–11, 20] - and in experimental model systems [12]. Therefore, circular dichroism and NMR analyses were initiated to assess structure of the presumptive mature KISS1R ligand.
Secondary structure prediction algorithms such as SCRATCH [24] had predicted that the first half of the sequence in KP-54 would be disordered with small stretches of “strand” and “turn” regions in the second half of the sequence especially between residues 30–40, but with no well defined helical regions. The circular dichroism spectroscopy data in water (Fig. 2) is consistent with this structural prediction and confirmed that the protein, under solubilized conditions, exhibits greater than 90% random coil content, with no significant β sheet or α-helical content. Varying pH from 5.0 to 9.0 did not affect the CD spectrum. Even when KP-54 was solubilized in a 60% trifluoroethanol solution, there was no change in secondary structure content, confirming that the protein did, indeed, favor a near-complete random coil content under solution conditions. To probe if KP-54 would adopt a stable structure when associated with a membrane, the CD data were recorded on KP-54 in separate solutions containing zwitterionic DPC micelles and negatively charged SDS micelles. The analysis of the CD data (Fig. 3) shows that there is a very slight increase in the total helix content of the peptide in the DPC and in SDS micelles (11.07% in DPC, 12.7% in SDS and 3.7% in buffer), but nevertheless the peptide remains largely disordered even in the presence of the micelles.
Fig. 2.
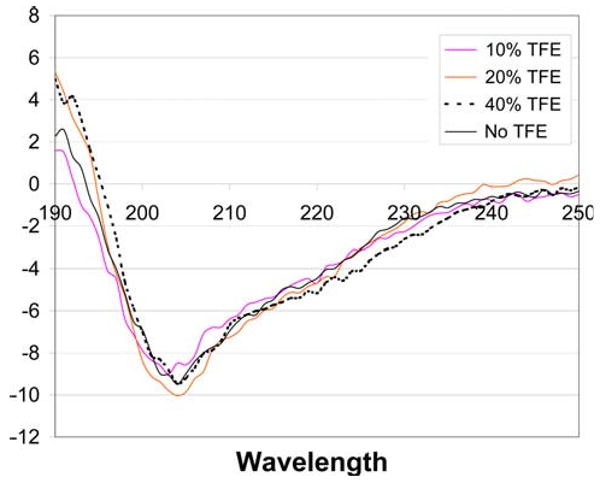
Circular dichroism spectra of KP-54 in water and water/trifluoroethanol solvent mixtures
Fig. 3.
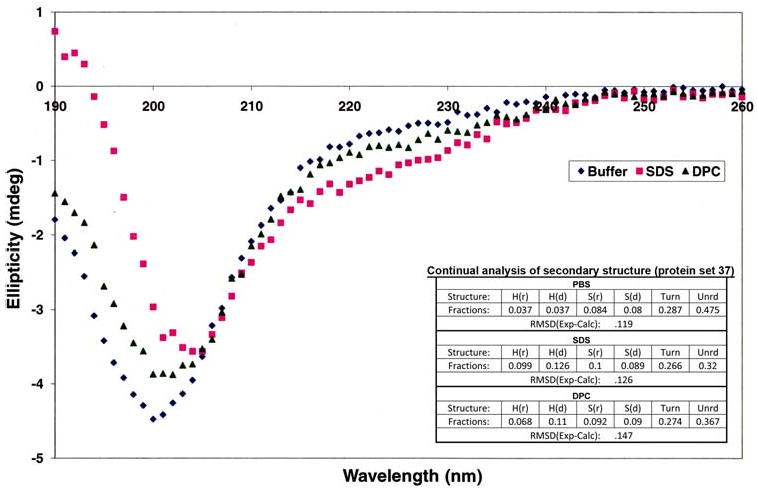
Circular dichrosim spectra and analysis of KP-54 in SDS and DPC micelles. r Regular, d distorted, Unrd unordered
Nuclear magnetic resonance spectroscopy further confirmed the lack of any significant secondary structural elements in water. The NMR spectra were unremarkable; some peaks in the TOCSY and NOESY spectra were either missing or were significantly overlapping, thus making it difficult to unambiguously assign the peaks using the standard sequential assignment methodology [25]. Figure 4 shows a partial assignment of peaks in the structured region in the TOCSY spectrum that were based on sequential NOESY connectivities that could be unambiguously identified. The use of a non-polar solvent mixture that was supposed to mimic transmembrane environment [26], consisting of a 2:2:1 ratio of chloroform/MeOH/H2O, did not affect NMR data. Increasing the non-polar solvent content to a ratio of 9:9:2 chloroform/MeOH/H2O also did not result in any differences in NMR data, confirming that the protein does not undergo folding in more non-polar solvents.
Fig. 4.
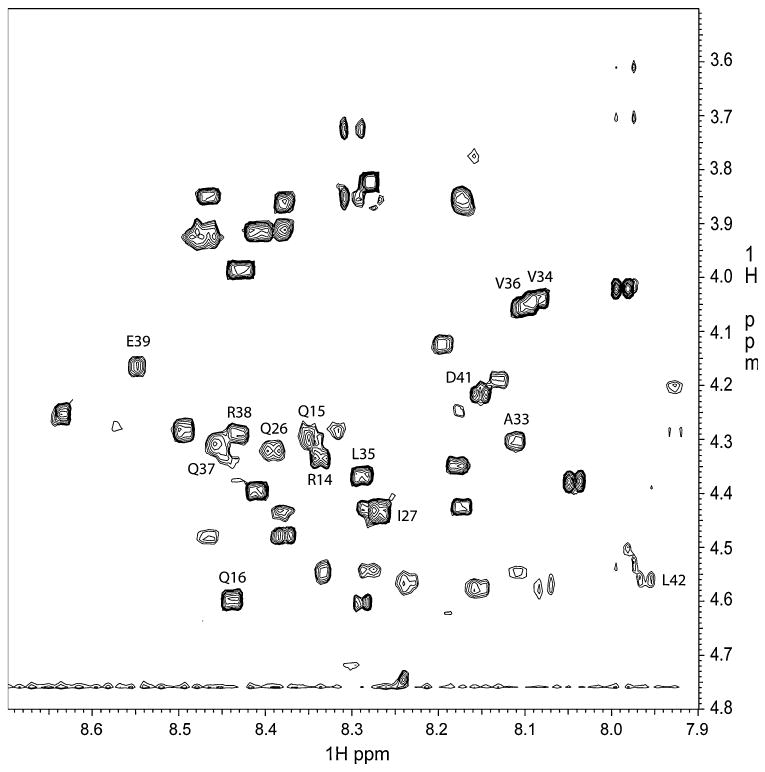
Finger print region in the TOCSY spectrum of KP-54/water showing some assignments
In the NOESY data on the peptide dissolved in water, there were only four weak NH(i) → NH(i + 1) contacts that were observed. These are identified in Fig. 5. NMR experiments (TOCSY and NOESY) were also performed on KP-54/SDS-micelle sample at a slightly elevated temperature (313 K) where the peaks were slightly sharper. The NOESY experiments with 200 and 400 ms mixing times only revealed a few more NH(i)-NH(i + 1) NOE contacts compared with the peptide in water sample, and consistent with the CD data on the peptide dissolved in the micelles.
Fig. 5.
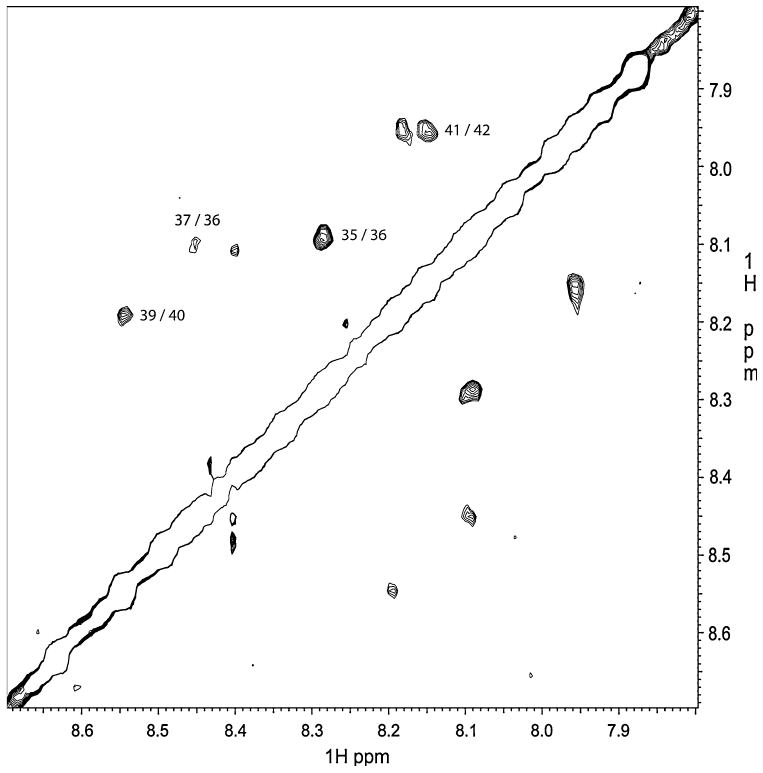
NOESY spectrum showing the three weak NH(i) → NH(i + 1) connectivities observed for KP-54/water sample
The lack of distinguishable structural elements raises several important issues regarding KP-54 function. First, the lack of defined structure would imply that there may be some promiscuity for KP-54 with regard to ligand receptor interactions. Although we cannot rule out the existence of another KISS1 receptor, we and others have utilized multiple methods (e.g., failure of iodinated ligand to bind KISS1R non-expressing cells) and failed to identify one. Second, an as yet undefined, post-cleavage modification could alter KP-54 so that the polypeptide has one or more regions with structure, which would lend specificity to ligand-receptor interactions. Third, structure could be conferred by KP-54 interacting with another protein, which alters structure so that specificity of binding to KISS1R is possible. To date, amidation of the C-terminal portion of KP-54 has been observed and increases KISS1R binding [11]; however, the amidation is not required. We know of no systematic studies that evaluate interactions of KP-54 with other proteins. Preliminary yeast two-hybrid genetic screens undertaken in our laboratory have identified putative non-KISS1R binding partners; however, they have not yet been validated in vivo.
The lack of a well-defined protein structure is not unprecedented in physiological signaling and cancer-related processes [27]. For example, the central 75% of BRCA1 is largely disordered, but functions as a scaffold for various intermolecular interactions crucial for DNA repair [28]. TC-1, a protein implicated in the development of thyroid cancer, has a completely disordered state under physiological conditions [29]. The disordered state of KP-54 could explain the diversity of functions of KPs.
In a recent paper from Orsini et al. [30], KP-13 in detergent (sodium dodecyl sulfate) micelles at 298°K revealed that the last seven residues adopted a stable helical structure. Our NMR measurements on the KP-54/SDS-micelle sample also revealed some sequential NH(i)-NH(i + 1) NOE contacts consistent with the findings of Orsini et al. except that because of the elevated temperature (313°K) to obtain sharper spectra in the micelles and the increased peptide length (54 amino acids vs. 13 amino acids) employed in our NMR studies these NOESY contacts were weak due to increased segmental flexibility.
The data presented here represent the first reported attempt to determine the structure of the predominant ligand for the KISS1 receptor. Although the lack of structural information is disappointing, the findings provide important insights that may help refine future studies into the anti-metastatic and neuro-endocrine functions of KISS1.
Acknowledgments
The authors gratefully acknowledge support from the U.S. Public Health Service grants 1P30-CA 13148, CA87728, and 1S10RR021064-01A1, as well as the National Foundation for Cancer Research-Center for Metastasis Research. NMR measurements were performed at the NMR Core Facility of the UAB Comprehensive Cancer Center. The authors thank Dr. Palgunachari Mayakonda for peptide synthesis, and Dr. Mike Jablonsky for access to the CD spectrometer in the UAB Chemistry Department. The authors also thank Dr. Craig Smith for valuable discussions about this work.
Abbreviations
- KISS1R
KISS1 receptor, also known as GPR54, AXOR12 or hOT7T175
- KP
Kisspeptin, peptide derived from KISS1
- HPLC
High pressure liquid chromatography
- TFA
Trifluoroacetic acid
- EDTA
Ethylenediaminetetraacetic acid
- D2O
Deuterated water
- CD
Circular dichroism
- NMR
Nuclear magnetic resonance spectrometry
- MeOH
Methanol
Contributor Information
Ronald Shin, Comprehensive Cancer Center, The University of Alabama at Birmingham, Birmingham, AL 35294, USA.
Danny R. Welch, Comprehensive Cancer Center, The University of Alabama at Birmingham, Birmingham, AL 35294, USA, Departments of Pathology, The University of Alabama at Birmingham, 1670 University Blvd. Room VH-G019, Birmingham, AL 35294-00194, USA DanWelch@uab.edu, Department of Cell Biology, The University of Alabama at Birmingham, Birmingham, AL 35294, USA, Department of Pharmacology & Toxicology, The University of Alabama at Birmingham, Birmingham, AL 35294, USA, National Foundation for Cancer Research – Center for Metastasis Research, The University of Alabama at Birmingham, Birmingham, AL 35294, USA
Vinod K. Mishra, Department of Medicine, The University of Alabama at Birmingham, Birmingham, AL 35294, USA
Kevin T. Nash, Departments of Pathology, The University of Alabama at Birmingham, 1670 University Blvd. Room VH-G019, Birmingham, AL 35294-00194, USA
Douglas R. Hurst, Departments of Pathology, The University of Alabama at Birmingham, 1670 University Blvd. Room VH-G019, Birmingham, AL 35294-00194, USA, National Foundation for Cancer Research – Center for Metastasis Research, The University of Alabama at Birmingham, Birmingham, AL 35294, USA
N. Rama Krishna, Comprehensive Cancer Center, The University of Alabama at Birmingham, Birmingham, AL 35294, USA, Department of Biochemistry & Molecular Genetics, The University of Alabama at Birmingham, Community Health Services Building B31, Birmingham, AL 35294-2041, USA nrk@uab.edu.
References
- 1.Bodenstine TM, Welch DR. Metastasis suppressors and the tumor microenvironment. Cancer Microenviron. 2008;1:1–11. doi: 10.1007/s12307-008-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stafford LJ, Vaidya KS, Welch DR. Metastasis suppressors genes in cancer. Int J Biochem Cell Biol. 2008;40:874–891. doi: 10.1016/j.biocel.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 3.Lee JH, Miele ME, Hicks DJ, et al. KiSS-1, a novel human malignant melanoma metastasis-suppressor gene. J Natl Cancer Inst. 1996;88:1731–1737. doi: 10.1093/jnci/88.23.1731. [DOI] [PubMed] [Google Scholar]
- 4.West A, Vojta PJ, Welch DR, Weissman BE. Chromosome localization and genomic structure of the KiSS-1 metastasis suppressor gene (KISS1) Genomics. 1998;54:145–148. doi: 10.1006/geno.1998.5566. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg SF, Miele ME, Hatta N, et al. Melanoma metastasis suppression by chromosome 6: evidence for a pathway regulated by CRSP3 and TXNIP. Cancer Res. 2003;63:432–440. [PubMed] [Google Scholar]
- 6.Lee JH, Welch DR. Identification of highly expressed genes in metastasis-suppressed chromosome 6/human malignant melanoma hybrid cells using subtractive hybridization and differential display. Int J Cancer. 1997;71:1035–1044. doi: 10.1002/(SICI)1097-0215(19970611)71:6<1035∷AID-IJC20>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 7.Lee JH, Welch DR. Suppression of metastasis in human breast carcinoma MDA-MB-435 cells after transfection with the metastasis suppressor gene, KiSS-1. Cancer Res. 1997;57:2384–2387. [PubMed] [Google Scholar]
- 8.Jiang Y, Berk M, Singh LS, et al. KiSS1 suppresses metastasis in human ovarian cancer via inhibition of protein kinase C alpha. Clin Exp Metastasis. 2005;22:369–376. doi: 10.1007/s10585-005-8186-4. [DOI] [PubMed] [Google Scholar]
- 9.Kotani M, Detheux M, Vandenbogaerde A, et al. The metastasis suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J Biol Chem. 2001;276:34631–34636. doi: 10.1074/jbc.M104847200. [DOI] [PubMed] [Google Scholar]
- 10.Muir AI, Chamberlain L, Elshourbagy NA, et al. AXOR12: a novel human G protein-coupled receptor, activated by the peptide KiSS-1. J Biol Chem. 2001;276:28969–28975. doi: 10.1074/jbc. M102743200. [DOI] [PubMed] [Google Scholar]
- 11.Ohtaki T, Shintani Y, Honda S, et al. Metastasis suppressor gene KiSS1 encodes peptide ligand of a G-protein-coupled receptor. Nature. 2001;411:613–617. doi: 10.1038/35079135. [DOI] [PubMed] [Google Scholar]
- 12.Nash KT, Phadke PA, Navenot JM, et al. KISS1 metastasis suppressor secretion, multiple organ metastasis suppression, and maintenance of tumor dormancy. J Natl Cancer Inst. 2007;99:309–321. doi: 10.1093/jnci/djk053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldberg SF, Harms JF, Quon K, Welch DR. Metastasis-suppressed C8161 melanoma cells arrest in lung but fail to proliferate. Clin Exp Metastasis. 1999;17:601–607. doi: 10.1023/A:10067 18800891. [DOI] [PubMed] [Google Scholar]
- 14.Eccles SA, Welch DR. Metastasis: recent discoveries and novel treatment strategies. Lancet. 2007;369:1742–1757. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell AW, McCluskey J, Rossjohn J. More than one reason to rethink the use of peptides in vaccine design. Nat Rev Drug Discov. 2007;6:404–414. doi: 10.1038/nrd2224. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 18.Bartels C, Xia TH, Billeter M, Guntert P, Wuthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 19.Unneberg P, Merelo JJ, Chacon P, Moran F. SOMCD: method for evaluating protein secondary structure from UV circular dichroism spectra. Proteins. 2001;42:460–470. doi: 10.1002/1097-0134(20010301)42:4<460∷AID-PROT50>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 20.Gianetti E, Seminara S. Kisspeptin and KISS1R: a critical pathway in the reproductive system. Reproduction. 2008;136:295–301. doi: 10.1530/REP-08-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohkura S, Uenoyama Y, Yamada S, et al. Physiological role of metastin/kisspeptin in regulating gonadotropin-releasing hormone (GnRH) secretion in female rats. Peptides. 2009;30:49–56. doi: 10.1016/j.peptides.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 22.Arai AC. The role of kisspeptin and GPR54 in the hippocampus. Peptides. 2009;30:16–25. doi: 10.1016/j.peptides.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 23.Tena-Sempere M. Timeline: the role of kisspeptins in reproductive biology. Nat Med. 2008;14:1196. doi: 10.1038/nm1108-1196. [DOI] [PubMed] [Google Scholar]
- 24.Cheng J, Randall AZ, Sweredoski MJ, Baldi P. SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res. 2005;33:W72–76. doi: 10.1093/nar/gki396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wuthrich K. NMR of proteins and nucleic acids. Wiley; New York: 1986. [Google Scholar]
- 26.Rastogi VK, Girvin ME. 1H, 13C, and 15 N assignments and secondary structure of the high pH form of subunit c of the F1F0 ATP synthase. J Biomol NMR. 1999;13:91–92. doi: 10.1023/A:100837 9624478. [DOI] [PubMed] [Google Scholar]
- 27.Sickmeier M, Hamilton JA, LeGall T, et al. DisProt: the database of disordered proteins. NAR. 2007;35:D786–793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaiser OJ, Ball LJ, Schmieder P, et al. Solution structure, backbone dynamics, and association behavior of the C-terminal BRCT domain from the breast cancer-associated protein BRCA1. Biochemistry. 2004;43:15983–15995. doi: 10.1021/bi049550q. [DOI] [PubMed] [Google Scholar]
- 29.Sunde M, McGrath KCY, Young L, et al. TC-1 is a novel tumorigenic and natively disordered protein associated with thyroid cancer. Cancer Res. 2004;64:2766–2773. doi: 10.1158/0008-5472.CAN-03-2093. [DOI] [PubMed] [Google Scholar]
- 30.Orsini MJ, Klein MA, Beavers MP, Connolly PJ, Middleton SA, Mayo KH. Metastin (KiSS-1) mimetics identified from peptide structure-activity relationship-derived pharmacophores and directed small molecule database screening. J Med Chem. 2007;50:462–471. doi: 10.1021/jm0609824. [DOI] [PubMed] [Google Scholar]


