Abstract
Because the deletion of self-reactive T cells is incomplete, thymic development of natural Foxp3+CD4+ regulatory T (Treg) cells is required for preventing autoimmunity. However, the role of T cell receptor (TCR) specificity in thymic Treg cell development remains controversial. To address this issue, we generated a transgenic line expressing a naturally occurring Treg cell-derived TCR. Surprisingly, efficient thymic Treg cell development occurred only when the antigen-specific Treg cell precursors were present at low clonal frequency (<1%) within a normal thymus. Using retroviral vectors and bone marrow chimeras, we observed similar behavior with two other Treg cell-derived TCRs. These data demonstrate that thymic Treg cell development is a TCR-instructive process involving a niche which can be saturable at much lower clonal frequencies than the niche for positive selection.
Introduction
The adaptive immune system generates an enormous diversity of antigen receptors for the recognition of a wide variety of pathogens. However, the cost of this diversity is that some receptors can recognize self-antigens and cause autoimmunity. For T cells, one mechanism that reduces this problem is the deletion (negative selection) of self-reactive developing thymocytes. It is now clear that the deletion of self-reactive cells is incomplete, since T cells with autoimmune potential are readily observed in a number of experimental models.
Self-reactive T cells that evade negative selection are now known to be controlled by naturally arising Foxp3+ regulatory T (Treg) cells, which comprise ∼10% of peripheral CD4+ cells1. Importantly, Treg cells are required throughout life to preserve immune tolerance and prevent fatal autoimmunity2,3. While it is likely that thymic and extra-thymic Treg cell development are both important4–6, it appears that peripheral Treg cell development alone is not sufficient to establish self-tolerance. For example, autoimmunity was observed after the interruption of thymic Treg cell development in classic experiments using neonatal thymectomy7, or the adoptive transfer of peripheral cells depleted of Treg cells into lymphopenic hosts8. Thus, the generation of Treg cells in the thymus is crucial for maintaining immune homeostasis.
Since Treg cells represent a minor subset of CD4+ T cells, a central question has been how Treg cells are selected from the developing thymocyte population. One hypothesis is that Treg cells may be selected based on their recognition of self-antigens; this process would presumably promote the suppression of responses to self, rather than foreign, antigens. This hypothesis is consistent with the observation that Treg cells were not found in T cell receptor (TCR) transgenic mice expressing receptors specific for foreign antigens9, and that the Treg and non-Treg TCR repertoires are largely different in murine models10–13. A role for self-antigen recognition by Treg cells is also supported by the observation that TCR transgenic cells developed into Treg cells only after encounter with their cognate antigen—which is expressed by a second transgene—in the thymus6,14,15.
An alternative hypothesis is that Treg cell selection occurs largely via a TCR-independent process that produces an initial Treg TCR repertoire that is similar to the conventional T cell repertoire. Although this initial repertoire may subsequently undergo other selective modifications, this model predicts that many conventional and regulatory T cells will share identical TCR specificities. The overlap in antigen specificity could provide regulation of both self-reactive and pathogen-reactive cells. This hypothesis was supported by the study of a TCR transgenic system16 different from those described above6,14,15. It was observed that the increased percentage of Treg cells associated with higher amounts of cognate antigen in vivo did not correlate with an increased overall number of antigen-specific Treg cells, but rather a reduction in the size of the transgenic non-Treg cell population16. It was therefore suggested that TCR-independent expression of Foxp3 rendered those Treg cells more resistant to TCR-dependent negative selection, a conclusion not reached by the aforementioned studies6,14,15. Additionally, studies of CD4−CD8− double negative (DN) thymocyte populations have also revealed that large DN2 cells preferentially generates a higher frequency of CD4+CD8− single positive (CD4SP) Treg cells. This finding supports a TCR-independent model of Foxp3 induction, since any selection of Treg precursors into the DN2 subset would occur prior to the somatic rearrangements that generate TCR molecules in developing T cells17. Finally, another study reported that the TCR repertoires of Treg and non-Treg cells are mostly overlapping, arguing for a TCR-independent model of Treg cell selection18.
TCR transgenic mice have been invaluable in dissecting the CD4 versus CD8 fate decision during thymic development. However, it is unclear if TCR transgenic models that enforce TCR engagement with cognate antigen are representative of Treg cell development driven by naturally-arising Treg cell-derived TCRs, as natural Treg cell selecting ligands have not been identified. Even though thymic Treg cell development has been described for well over a decade, Treg cell development in a TCR transgenic line using a naturally arising Treg TCR has not yet been reported. We therefore generated a TCR transgenic line expressing a natural Treg cell-derived TCR identified from our studies of the Treg cell TCR repertoire in polyclonal TCRβ chain transgenic mice. Remarkably, we found that thymic Treg cell development was virtually undetectable when thymocytes expressing this Treg cell-derived TCR were present at high clonal frequency, but that Treg cell development markedly improved at clonal frequencies below 1%. However, a non-Treg cell-derived TCR and the OTII TCR were unable to induce Treg cell development even at low clonal frequencies. These results therefore suggest that thymic Treg cell development is a TCR-instructive process dependent on a small selecting niche.
Results
Few thymic Treg cells in Treg TCR transgenic mice
To study the role of TCR specificity in thymic Treg cell development, we decided to utilize the classic approach of TCR transgenesis. However, it is difficult to select a natural Treg cell TCR from the normal Treg cell population, as one must first determine the distribution of this TCR within the Treg and non-Treg cell subsets. Thus, rather than randomly selecting a natural Treg cell TCR from a fully polyclonal population, we chose TCRs identified in our ongoing studies of the Treg cell TCR repertoire19,20. Because the fully polyclonal TCR repertoire, with an estimated 1014 diverse TCR sequences, cannot be studied experimentally at the individual TCR level, we utilized TCRβ chain transgenic mice to restrict the variability of the TCR repertoire to only the TCRα chain11. These mice are also heterozygous at the Tcra locus such that only one TCR can be expressed per cell to affect TCR-dependent cell-fate decisions. From our database of TCRs, we selected the TCR clones G113 and B8, which are strongly associated with the Treg cell and naïve TCR data sets, respectively (Table 1). G113 appears to be self-reactive based on its ability to induce proliferation of peripheral T cells after adoptive transfer into lymphopenic and non-lymphopenic hosts. In contrast, B8 does not exhibit self-reactivity in these assays11,12.
Table 1.
In vivo context of the TCRs used in this study.
| % in periphery | % in thymus | ||||||
|---|---|---|---|---|---|---|---|
| Clone | CDR3 Sequence | Foxp3− CD44hi | Foxp3− CD44lo | Foxp3+ | Foxp3− | Foxp3+ | |
| “Treg” | G25 | AASADYSNNRLT | 0.07 | 0.00 | 1.05 | 0.00 | 2.93 |
| R19 | AASSGTYQR | 0.02 | 0.03 | 0.64 | 0.00 | 0.31 | |
| G113 | AARLNNNNAPR | 0.00 | 0.00 | 0.59 | 0.00 | 0.31 | |
| “Naïve” | B8 | AASEDNNNAPR | 0.12 | 3.27 | 0.12 | 1.13 | 0.00 |
| Number of TCRs in data set | 5947 | 6003 | 5973 | 1149 | 955 | ||
The frequency of individual TCRs, as identified by their CDR3 amino acid sequence, was obtained from our database of TCR sequences isolated from thymic and peripheral T cell subsets in TCli TCRβ transgenic Foxp3gfp Tcra+/− mice19,20. TCR sequences were pooled from 3–4 experiments, each comprised of several mice. Of note, the presence of B8 TCR sequences in the peripheral Foxp3+ and CD44hi data sets may arise from contamination of those subsets with CD44lo cells. Although our FACS purity is over 95% for those subsets, the major contaminants are CD44lo cells, which constitute the largest peripheral CD4+ T cell subset. In contrast, the presence of contaminants in the CD44lo subset is much less likely, as we typically achieve 99% sort purity in this population. TCR transgenic lines generated for this study are indicated in italics. For reference, the highest frequency non-Treg cell TCR in the thymus is found at a frequency in the aggregate data set of 2.0%, representing an estimated clone size of 0.2% in the CD4SP subset (10% Vα2+ × 2% frequency of TCR in that subset).
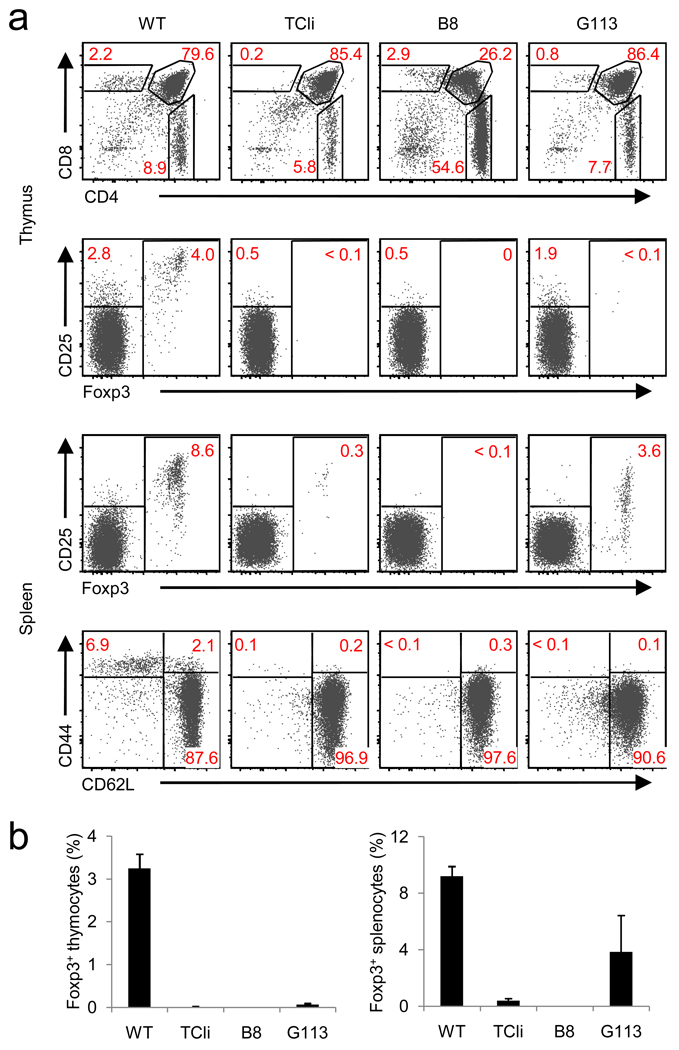
As expected, we did not observe thymic or peripheral Treg cell development in the B8 TCR transgenic line when crossed onto the Rag1 −/− background (Fig. 1). Surprisingly, virtually no thymic Treg cells were found in G113 TCR transgenic Rag1−/− mice, although a small percentage of peripheral Foxp3+ cells was seen (Fig. 1). In addition, we tested 5 other Treg cell-derived TCRs present in our thymic and peripheral Treg cell TCR database using retroviral vectors and bone marrow (BM) chimeras. Similar to our results with G113 TCR transgenic mice, we found peripheral, but essentially no thymic, Treg cells (data not shown). Thus, we were unable to show that thymic Treg cell development could be directed by TCRs derived from naturally arising Treg cells in a TCR transgenic setting.
Figure 1.
Flow cytometric characterization of TCR transgenic lines. (a) FACS plots of thymic and splenic T cells from 6-week old TCR transgenic Foxp3 gfp Rag1 −/− mice expressing TCRs described in Table 1 (B8 and G113). Wild-type Foxp3 gfp (WT), and TCli αβ–TCR transgenic -Foxp3 gfp Rag1 −/− mice (origin of the TCRβ chain for B8 and G113) are shown as controls. (b) Summary of flow cytometric data. The frequency of Foxp3+ T cells in the CD4SP thymic (left) or CD4+ splenic (right) subset are shown (mean ± s.d., n=3). Data were obtained from 3 independent experiments.
These data appeared contradictory, as the G113 TCR was associated with the Treg cell subset within the normal polyclonal repertoire, but when expressed as a transgene G113 did not appear to drive Treg cell development. This paradox led us to ask whether Treg cell development was itself influenced by the polyclonality of the thymus. Our studies of thymic Treg cell development suggested that cytokines present in the thymic environment may play an important role19,21,22. We therefore asked if the presence of a polyclonal population could generate an environment permissive for thymic Treg cell development of G113 T cells. To test this hypothesis, we injected G113 thymocytes into wild-type (WT) congenic thymuses and followed their development. In this setting, approximately 10% of CD4+CD8− (CD4SP) G113 cells became Foxp3+ within three days after transfer (Fig. 2a). This percentage represents a 100-fold increase in the frequency of Foxp3-expressing cells compared with the monoclonal G113 TCR transgenic setting. Thus, this finding suggested that the environment of a normal polyclonal thymus is essential for G113 thymocytes to efficiently develop into Treg cells.
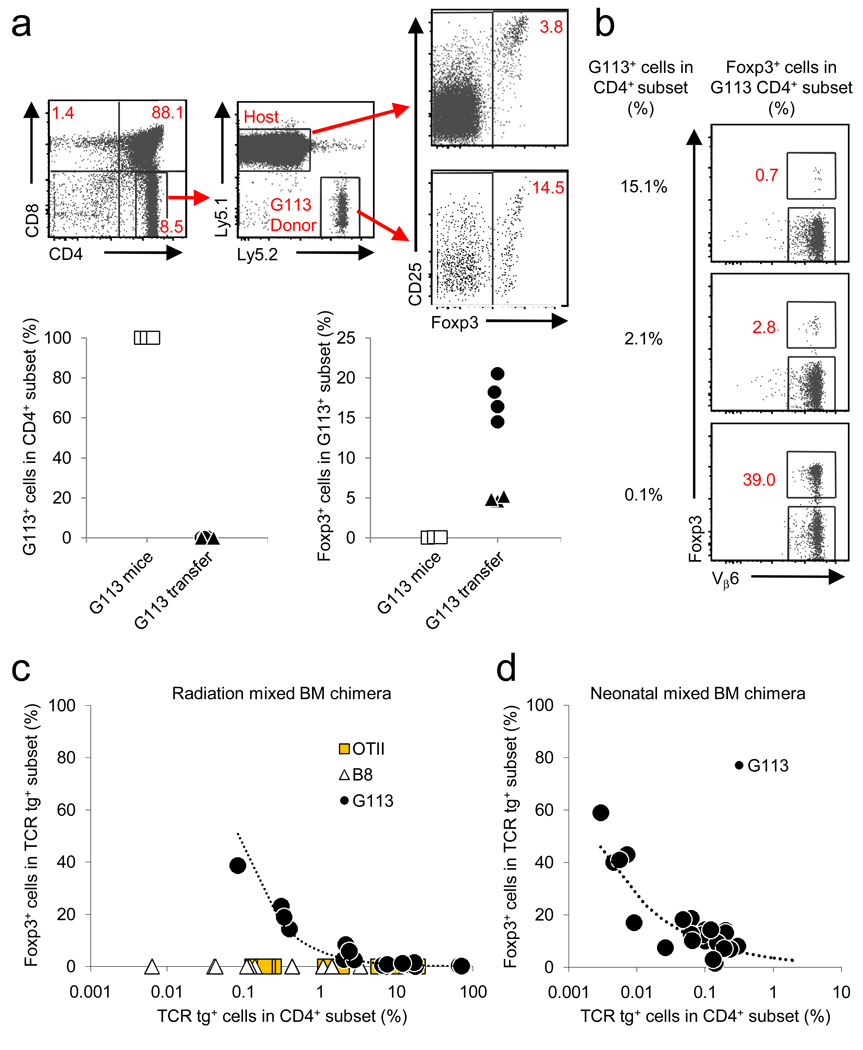
Figure 2.
An inverse relationship between TCR transgenic cell frequency and thymic Treg cell development. (a) G113 thymocytes (107) were intrathymically injected into congenic CD45.1 hosts, and analyzed by flow cytometry on day 3. The analysis scheme is illustrated in the FACS plots. The charts below show the percentage of CD45.2+ G113 cells in the CD4SP subset (left), and the percentage of Foxp3+ cells within the G113 population (right). Data from G113 transgenic Rag1 −/− mice (G113 mice) are shown for reference. The exact P-value from the Wilcoxon rank sum test between the percentage of Foxp3+ cells in the two settings is 0.017. Each symbol represents data from individual recipients from 2 independent experiments. (b–d) G113 BM was mixed with congenically marked wild-type BM at various ratios and injected into irradiated adult wild-type recipients or non-irradiated neonatal CD45.1 Foxp3 gfp recipients as described in the Methods. (b) Representative dot plots are gated on G113 CD45.2+CD45.1−CD4SP thymocytes from the radiation BM chimeras. The G113 TCR incorporates a Vβ6 chain. (c,d) Graphs show the frequency of G113 cells in the CD4SP subset versus the frequency of Foxp3+ cells. Each symbol represents an individual recipient. For radiation BM chimeras, 4 independent experiments were performed. For neonatal BM chimeras, 3 independent experiments were performed. Using a linear mixed model, we detected a significant difference between OTII or B8 and G113 (P < 0.01), but not between B8 and OTII (P = 0.41).
Foxp3+ cell frequency inversely correlates with clonal frequency
We next generated radiation BM chimeras in which G113 transgenic and congenically marked wild-type BM were mixed in different ratios in order to vary the frequency of transgenic cells within the polyclonal thymocyte population. A ratio of 1:1 or 1:5 (G113:WT) BM generated only a minor increase in thymic G113 Treg cell development (Fig. 2b). However, when the ratio of G113:WT BM was decreased to 1:50, we observed a marked increase in the efficiency of G113 Treg cell development. In this setting, up to 40% of CD4SP G113 thymocytes expressed Foxp3 (Fig. 2b–c). Coincident with the increase in Foxp3+ cells, the frequency of CD25hiFoxp3− thymocytes also increased (data not shown), indicating an increase in Treg cell precursors as well19. We observed no Foxp3+ CD4+CD8+ double positive (DP) thymocytes (data not shown), which have been observed in some TCR and cognate antigen double transgenic models23. Although Treg cell development from wild-type BM occurred normally in these mixed chimeras (Supplementary Fig. 1 online), we wanted to exclude the possibility that total body irradiation was selectively augmenting G113 Treg cell development via thymic lymphopenia or other mechanisms. Injection of G113 BM into non-irradiated congenically-marked neonatal hosts also resulted in efficient G113 Treg cell development (Fig. 2d). Thus, these data demonstrate that the efficiency of Foxp3+ Treg cell development is inversely related to the frequency of G113 thymocytes.
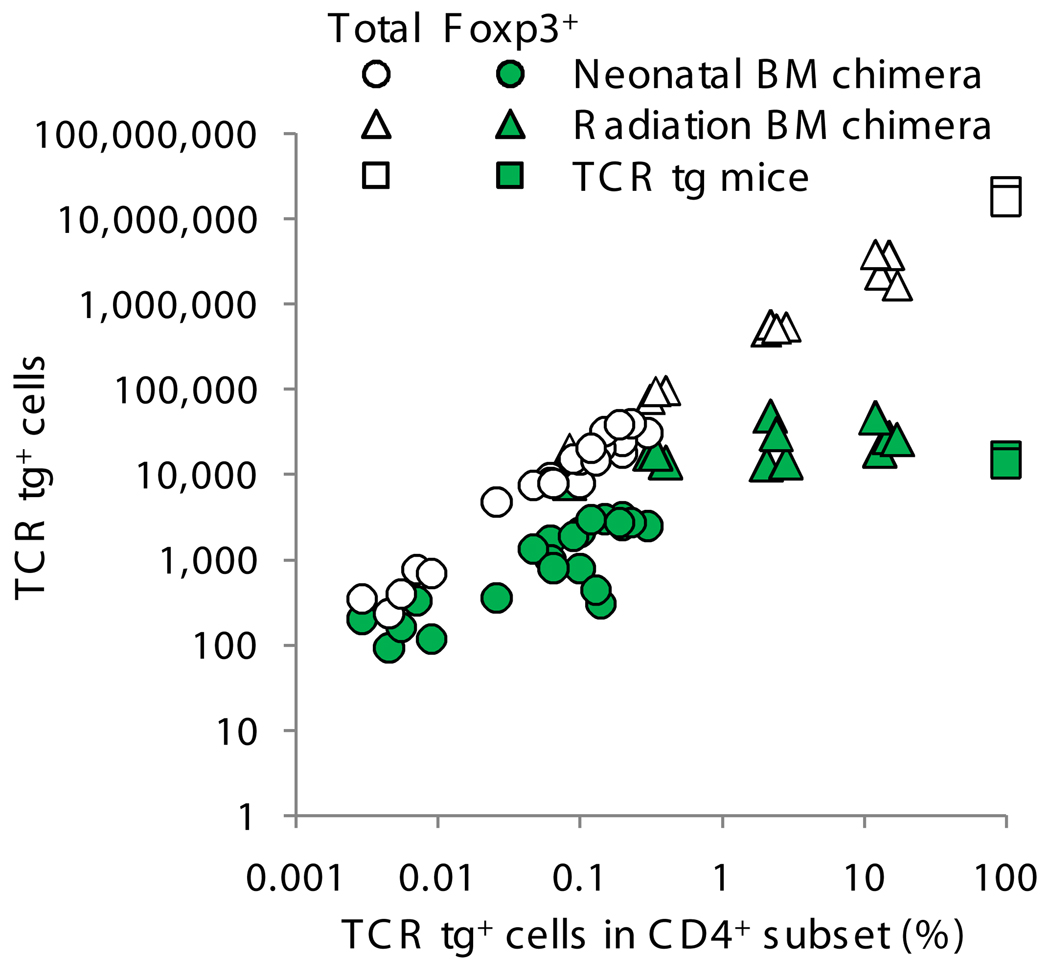
We also plotted the absolute number of G113 Foxp3+CD4SP and G113 total CD4SP cells (Fig. 3). Because it is very difficult to achieve low levels of chimerism using radiation BM chimeras, as it is to obtain really high levels of chimerism with neonatal BM chimeras, we combined the data in one figure. However, we segregated the data to clearly show the technique used to obtain each data point, because we cannot exclude the possibility that small differences might be related to the age of the recipients or the use of radiation. Curiously, this analysis revealed that the number of thymic Foxp3+ cells reaches a plateau with increasing clonal frequency, suggesting that the development of G113 Treg cells is saturable. In contrast, the total number of G113 cells in the CD4SP stage does not plateau, suggesting that the positively selecting niche for G113 is substantially larger than that for Treg cell development.
Figure 3.
Foxp3+ G113 Treg cell generation is saturable. Graph shows the absolute number of Foxp3+CD4SP cells from TCR transgenic mice and from irradiation and neonatal BM chimeras (Fig. 2c–d). Numbers were derived from the total number of thymocytes multiplied by the frequency of Foxp3+ G113 CD4SP T cells. Also shown is the total number of TCR transgenic CD4SP thymocytes in each mouse. Each symbol represents data from an individual mouse from 7 independent experiments.
In contrast to the behavior of G113 cells, development of Foxp3+ B8 cells was not observed even when the frequency of B8 cells was diminished in mixed BM chimeras to frequencies comparable to those with which we observed G113 Treg cell development (Fig. 2c). Lack of Treg cell development was also observed in OTII TCR transgenic cells (Fig. 2c). Because of the distinct behavior of G113 compared with B8 and OTII cells in these mixed BM chimeras, these data argue against the hypothesis that stable expression of Foxp3 can occur via a TCR-independent mechanism (Supplementary Fig. 2 online).
Minor role for peripheral Treg cell conversion
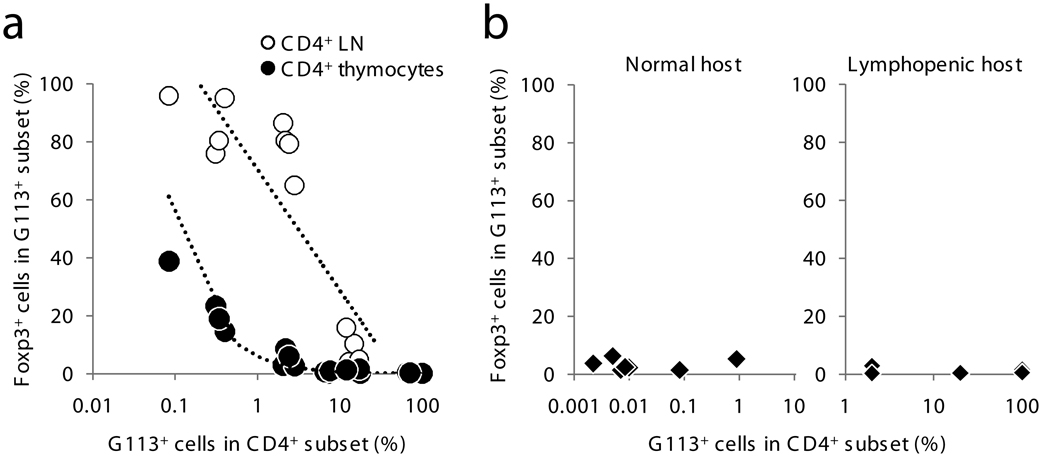
Although the frequency of G113 Foxp3+ cells in these mixed chimeras was typically less than 50% in the CD4SP subset, the frequency of G113 Treg cells was higher in the periphery than in the thymus (Fig. 4a). G113 T cells were preferentially found in the axillary-inguinal lymph nodes (Supplementary Fig. 3 online), consistent with our previous findings12. The difference between the thymic and peripheral frequency of Foxp3+ cells may reflect either the presence of Foxp3− Treg cell precursors within the thymic CD4SP subset, or could imply an important role for peripheral development of G113 Treg cells. To test whether Foxp3− G113 T cells undergo peripheral conversion, we adoptively transferred varying numbers of naïve CD44loFoxp3− peripheral G113 T cells into normal or lymphopenic hosts. Less than 5% of G113 T cells underwent peripheral conversion into Foxp3+ cells after 3 weeks (Fig. 4b). In addition, unlike thymic Treg cell development, the clonal frequency of G113 T cells did not markedly affect the frequency of cells which became Foxp3+ in the periphery. In contrast, intrathymic injection of G113 thymocytes resulted in ∼10% of G113 CD4SP cells expressing Foxp3 in just three days (Fig. 2a), suggesting that Foxp3− G113 cells in the CD4SP subset likely includes Treg cell precursors. Consistent with this hypothesis, Foxp3− G113 CD4SP thymocytes predominantly displayed a less mature HSAhi CD62Llo surface phenotype, whereas Foxp3+ G113 CD4SP thymocytes showed a more mature HSAlo CD62Lhi phenotype (Supplementary Fig. 4 online). Peripheral conversion may therefore explain the small frequency of Treg cells observed in G113 TCR transgenic mice (Fig. 1), but cannot account for the high frequency of peripheral G113 Treg cells arising within a polyclonal population (Fig. 4a). Thus, these data suggest that peripheral G113 Treg cells arise almost exclusively due to thymic Treg cell differentiation in the normal polyclonal TCR setting.
Figure 4.
Peripheral versus thymic G113 Treg cell development. (a) Graph shows the frequency of G113 TCR transgenic T cells which are Foxp3+ in the thymus and in the pooled axillary and inguinal lymph nodes (LN). The data are indexed to the percent of TCR transgenic cells in the CD4SP subset in radiation mixed BM chimeras (Fig. 2b–c). The Wilcoxon signed rank test for paired samples gives a P-value of < 0.01 between the thymus and LN. Data were obtained from 2 independent experiments. (b) FACS-purified peripheral Foxp3− G113 T cells were intravenously transferred into congenic wild-type or lymphopenic Tcrb −/− hosts as described in the Methods. The frequency of Foxp3+ cells was analyzed after 3 weeks by flow cytometry. Of note, T cells using the naÏve TCR B8 did not undergo conversion when transferred into lymphopenic hosts (data not shown). Each symbol represents data from an individual mouse from 3 independent experiments.
Clonal frequency impacts other aspects of thymic development
Although we focused on Treg cell development during the course of these studies, we found that the frequency of TCR transgenic cells can affect other parameters of thymic development. The TCR transgenic lines described here show a strong preference to generate CD4SP cells (Fig. 1). When embedded as a small fraction of developing thymocytes, the G113, TCli, and OTII TCRs retained their preference for CD4+ T cell development. However, in contrast, the B8 TCR lost its preference for CD4 when present at low frequencies, concomitant with the development of a sizable number of CD4−CD8+ single positive (CD8SP) T cells (Supplementary Fig. 5 online). This phenomenon has also been reported for AND, another well-known CD4 TCR transgenic line24. Also consistent with previous observations, the efficiency of positive selection of TCR transgenic thymocytes also appeared to improve with reduced clonal frequency, based on the decreased frequency of cells at the DP stage (Supplementary Fig. 5,6 online)25,26.
No coincident negative selection or proliferation
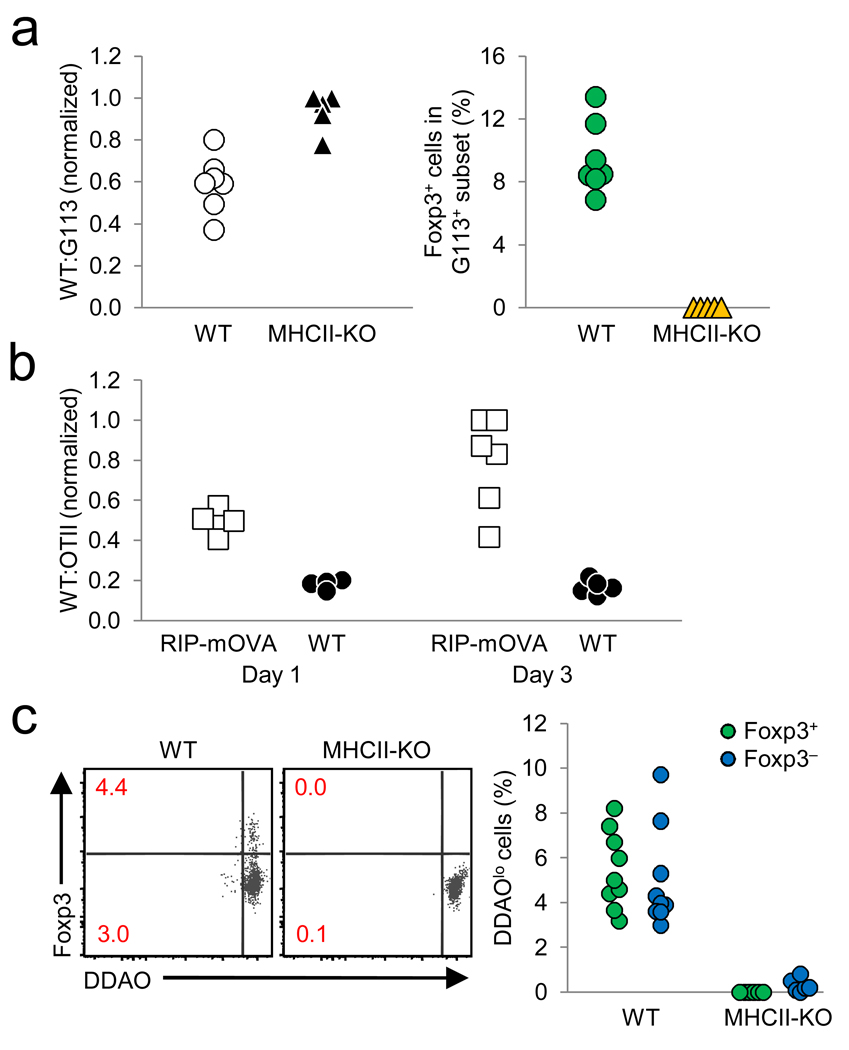
Previous studies of thymic Treg cell development in transgenic models observed negative selection coincident with Treg cell development14. Analysis of CD4 and CD8 expression by flow cytometry did not clearly reveal a correlation between negative selection and Treg cell development, as the relative frequency of G113 CD4SP cells increased with decreasing clonal frequency (Supplementary Fig. 5,6 online). However, this increased frequency of CD4SP cells may be due to increased efficiency of positive selection as mentioned above. We therefore adoptively transferred G113 transgenic Rag1 −/− and Thy1.1-marked wild-type thymocytes into wild-type versus MHC class II-deficient thymuses. G113-induced Treg cell development required the expression of MHC class II on host cells (Fig. 5a). However, the ratio between the injected wild-type and G113 transgenic cells remained comparable regardless of the presence or absence of host MHC class II molecules. In contrast, the ratio of co-injected WT:OTII TCR transgenic Rag1 −/− cells increased markedly after transfer into RIP-mOVA-expressing hosts, demonstrating that negative selection can be observed as early as one day after intrathymic transfer (Fig. 5b). Although we cannot exclude the possibility that negatively selecting signals can be received by G113 TCR transgenic cells prior to intrathymic transfer, evidence for extensive negative selection coincident with G113 TCR-mediated Treg cell selection was not observed within this experimental setting.
Figure 5.
No evidence for proliferation or negative selection during TCR-dependent G113 Treg cell development. (a) Wild-type and G113 Rag1 −/− thymocytes (1:1 ratio) were intrathymically injected into wild-type (WT) or MHC class II-deficient hosts. After three days, the ratio of remaining WT:G113 CD4SP cells (left), and the frequency of Foxp3+ G113 CD4SP cells (right) were determined by flow cytometry. The donor G113 cells were discriminated by DDAO labeling, CD45.2, and Thy1.2 expression; the co-injected wild-type cells by DDAO labeling, CD45.2, and Thy1.1 expression; and the host cells by CD45.1 expression. Each symbol represents an individual recipient. Data from 2 independent experiments are shown (3–4 WT and 2–3 MHC class II-deficient hosts per experiment), and were normalized by dividing the WT:TCR tg ratio with the highest value for a given experiment. (b) Whole thymocytes from OTII Rag1 −/− mice and Thy1.1 wild-type mice were mixed at a 1:1 ratio and intrathymically injected into congenically marked wild-type or RIP-mOVA mice and analyzed 1 and 3 days post-injection by flow cytometry. Data are from 2 independent experiments (5 WT and 5–6 RIP-mOVA recipients per experiment). (c) Sorted Thy1.1 wild-type and G113 Rag1 −/− HSAhi CD25lo Foxp3− CD4SP cells (5×105 total at 1:1 ratio) were DDAO labeled and intrathymically injected into wild-type or MHC class II-deficient recipients, and analyzed by flow cytometry on day 3. Numbers in dot plots indicate the frequency of DDAOlo cells for the Foxp3+ and Foxp3− CD4SP cells; these frequencies are summarized in the graph on the right. Data are from 2 independent experiments using 9 WT and 6 MHC class II-deficient hosts.
Increased self-reactivity of the G113 TCR to peripheral antigens was previously inferred based on the observation of proliferation of G113 cells after adoptive transfer into non-lymphopenic hosts12. It was therefore possible that the G113 TCR merely facilitated the expansion of cell populations which had upregulated Foxp3 expression in a TCR-independent manner. To increase the number of observable Foxp3+ events after intrathymic transfer, we used sorted HSAhiFoxp3− CD4SP G113 thymocytes. To visualize cell division, the cells were labeled with DDAO, a CFSE analogue compatible with the Foxp3gfp reporter. Virtually no DDAO dilution of thymocytes was observed in MHC class II-deficient hosts, although a small subset did appear to proliferate in wild-type hosts (Fig. 5c). Importantly, we observed that the majority of Foxp3+ cells did not dilute DDAO, suggesting that efficient thymic Treg cell development can occur in the absence of marked thymocyte population expansion.
Other TCRs facilitate niche-dependent Treg cell development
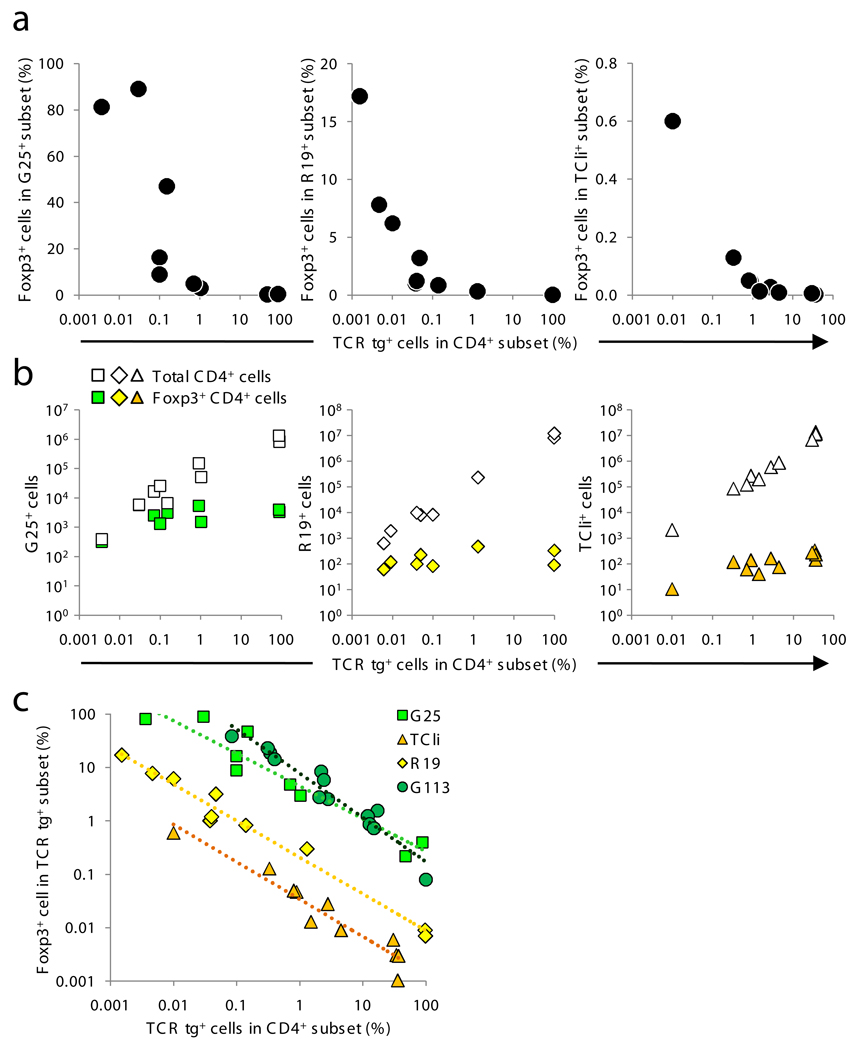
To generalize the observations made with the G113 Treg TCR, we asked whether other TCRs showed a similar relationship between clonal frequency and Treg cell development. We examined the TCli TCR26, which was isolated from a fully polyclonal repertoire based on its reactivity to the foreign antigen human CLIP (class II-associated invariant chain peptide). Although it is unknown whether this TCR is preferentially associated with the Treg or non-Treg cell subsets in a normal polyclonal setting, the TCli TCR transgenic line showed a very small frequency of peripheral Treg cells, but essentially no thymic [JB11]Treg cells, on a Rag1 −/− background (Fig. 1). Interestingly, thymic Treg cell generation was also enhanced by decreasing the frequency of TCli TCR cells in a polyclonal setting, although Treg cell development was far less efficient in comparison with G113 T cells (Fig. 6a). This suggests that reactivity to foreign antigen does not preclude TCR-dependent Treg cell selection, and that TCRs can vary widely in the efficiency with which they induce Treg cell differentiation.
Figure 6.
Varying efficiencies of TCR-dependent Treg cell development. (a–b) G25 and R19 (Table 1) were retrovirally expressed in Foxp3 gfp Rag1 −/− BM and mixed with CD45.1 Foxp3 gfp BM at varying ratios. In the case of TCli, TCli-αβ TCR transgenic Foxp3 gfp Rag1 −/− BM was used. (a) Thymocytes were analyzed by flow cytometry as in Fig. 2b–c. (b) The absolute numbers of Treg and CD4SP thymocytes were plotted as in Fig. 3. Each symbol represents data from an individual mouse from 2 independent experiments (n=10–12). (c) Data are plotted as in Fig. 2c, except the frequency of Foxp3+ cells is on a log scale. Using a linear mixed model taking the percentage of CD4SP cells into consideration, we detected significant differences in pair wise comparisons between G113, R19, and TCli (P < 0.01), but not G113 and G25 (P = 0.08). The geometric means of percent Foxp3+ cells (±s.e.m.) for each TCR are: G113 (6.5 ± 1.0), G25 (4.3 ± 0.8), R19 (0.2 ± 0.04), and TCli (0.03 ± 0.006).
Furthermore, we produced retroviral BM chimeras using two additional Treg TCRs, G25 and R19 (Table 1). We transduced either the G25 or R19 TCRα chains along with the fixed TCli TCRβ chain into Rag1 −/− Foxp3 gfp BM cells. These cells were adoptively transferred along with congenically-marked wild-type BM into lethally irradiated hosts, again at various ratios. Like G113, G25 and R19 Treg cell-derived TCRs more efficiently induced Treg cell development when present at lower frequencies within the polyclonal population. The degree of Treg cell development ranged from virtually undetectable when the frequency of Treg cell TCR-expressing cells was high, to 20–80% Foxp3+ when the frequency was below 1% (Fig. 6a). Like G113 (Fig. 3), the absolute number of Treg cells generated by G25 and R19 TCRs also reached a plateau (Fig. 6b). In contrast to G25 and R19, the naïve T cell-derived TCR B8 was unable to induce thymic Treg cell development in BM chimeras (data not shown), consistent with its behavior in TCR transgenic mice (Fig. 2c). Thus, these data with two additional Treg TCRs support the notion that the Treg selecting niche is saturable and much smaller than the niche for positive selection.
The Treg cell-derived TCRs examined here exhibited widely differing (100-fold or more) relative efficiencies for Treg cell selection (Fig. 6c). These data suggest that the notion of Treg versus non-Treg cell-derived TCRs is overly simplistic, and that the observed overlap between the Treg and non-Treg cell-derived TCR repertoires11–13,27 may be comprised of TCRs that are inefficient at inducing Treg cell development. Importantly, all Treg-derived TCRs (G113, G25, and R19) showed a similar relationship between their efficiency of inducing Treg cell development and their fractional composition within the polyclonal T cell population. Thus, a reciprocal relationship between the frequency of CD4SP thymocytes expressing an individual TCR and the frequency of those cells expressing Foxp3 may be a general feature of Treg cell development.
Discussion
Our in vivo analysis of the development of T cells expressing Treg cell-derived TCRs within the context of a polyclonal thymic environment generated several important conclusions. First, thymic Treg cell development involves a TCR-instructive step. Second, not all TCR transgenic models will recapitulate TCR-driven thymic Treg cell development. Third, thymic selection of Treg cells often involves a saturable TCR-dependent niche. Finally, the capacity of the Treg cell selection niche may be much smaller than that of positive selection.
These data support a model for thymic Treg cell development in which TCR-derived signals induce or maintain Foxp3 expression. Although the phrase “stochastic-selective” has previously been used in thymic Treg cell development16, we prefer to use the term “TCR-independent” instead of “stochastic” as certain aspects of TCR-driven Treg cell development may have probabilistic elements (e.g. the likelihood that a given thymocyte will interact with a selecting antigen-presenting cell). We also use Foxp3 as a readout representing the completion of Treg cell development as it appears late in the developmental process and is required for Treg cell phenotypic stability28–30. We found no evidence to support the previously proposed model in which TCR-independent generation of Foxp3+ cells renders those cells resistant to TCR-dependent negative selection, as no Foxp3+ OTII or B8 cells were found. We also found no evidence for TCR-driven expansion of Foxp3+ cells generated via TCR-independent processes, as Treg cell development was observed in the absence of cell division. Since it appeared from the above analyses that TCR specificity does not modify events post-Foxp3 induction, we reasoned that TCR-dependent signals may be important prior to or coincident with induction of stable Foxp3 expression. At this time, we cannot exclude the possibility that TCR-independent mechanisms induce Foxp3 in a rare subset of cells below the limit of our detection, with subsequent TCR-dependent signals required to maintain Foxp3 expression. A simpler and more direct model is that TCR-dependent signals trigger a developmental pathway that results in Foxp3 expression. Since the TCR-dependent step in both of these models either directly or indirectly results in the induction or stabilization of Foxp3, these data support a TCR-instructive model of Treg cell development14,31.
In addition, these findings suggest that TCR-driven thymic Treg cell development, unlike positive selection or CD4 versus CD8 lineage commitment, may not be easily visualized using TCR transgenic models. While we cannot exclude potential bias from the TCRβ chain used, we have not encountered a naturally-arising Treg cell TCR capable of efficiently inducing thymic Treg cell development in a monoclonal setting. Although these data do not exclude the possibility that some natural Treg cell-derived TCRs can facilitate thymic Treg cell development in TCR transgenic mice, it is clear that the lack of thymic Foxp3+ cells in TCR transgenic mice is not necessarily indicative of the TCR’s inability to induce Treg cell development under conditions of low clonal frequency, such as those found in the normal polyclonal setting.
We propose that thymic Treg cell development is governed by a saturable TCR-specific mechanism. Our initial hypothesis for the inability of G113 thymocytes to undergo Treg cell development in the TCR transgenic setting was that polyclonal thymocytes were required to provide an appropriate cytokine environment. However, we found that decreasing G113, G25, and R19 T cells to 0.1% of the CD4SP population led to markedly more efficient Treg cell development than when G113, G25 and R19 cells were present at 1% of the CD4SP population. The corresponding increase of wild-type thymocytes from 99% to 99.9% would not likely alter the overall thymic environment for Treg cell development. Indeed, no marked differences in the frequency of Treg cells emerging from wild-type BM were observed over a broad range of G113:WT BM ratios. Furthermore, a previous study found that a 1:1 mixture of interleukin-2 (IL-2)-deficient:WT BM was sufficient to restore normal Treg cell development in IL-2-deficient thymocytes32. Rather, these data support the alternative possibility that decreasing the frequency of thymocytes with identical antigen specificity is crucial for thymic Treg cell development. In fact, even though the percentage of G113 Treg cells decreased markedly with increasing clonal frequency, the absolute number of Treg cells reached a plateau. One possible explanation for the inverse relationship between clonal frequency and the efficiency of Treg cell development is intraclonal competition for peptide-MHC complexes. T cells of the same specificity are known to compete for ligands during peripheral homeostasis and normal immune responses33–35. A TCR-specific niche for positive selection of thymocytes has also been described25,26. Ligand competition could occur via antigen presenting cell blockade36,37, or via the removal of peptide-MHC complexes from antigen presenting cells by antigen-specific T cells38. However, preliminary two-photon imaging studies of mixed bone-marrow chimeras did not show clustering of CFP-labeled G113 thymocytes, suggesting that antigen presenting cell blockade is not mediating intraclonal competition (J.L.B., C.S.H., and M. Miller (Washington University, St. Louis); data not shown). Although future experiments will be required to determine the exact mechanism through which clonal frequency influences thymic Treg cell differentiation, the current studies suggest that intraclonal competition for ligand may be an important step in Treg cell development.
Finally, the capacity of the positively selecting niche for thymocytes with a given TCR specificity appears to be much greater than that for Treg cell development. The clonal frequencies of transgenic cells in the thymus that allow for efficient Treg cell development are an order of magnitude lower than those permissive for positive selection, as measured in mixed BM chimeras24–26. These findings are consistent with the fact that positive selection but not Treg cell development can be consistently and easily visualized in the monoclonal setting. This differential niche size is also illustrated by the observation that the number of Treg cells plateaus around 10,000 for G113 T cells, whereas the number of positively selected CD4SP G113 T cells can be 1,000 fold greater.
Our data demonstrate that a low frequency of Foxp3+ cells in a TCR transgenic setting may not be indicative of the TCR’s potential to direct Treg cell development. This finding suggests that previously generated TCR transgenic strains may need to be re-evaluated for potential mismatch between the Treg cell selecting niche size and the high clonal frequencies in TCR transgenic mice. We hypothesize that TCRs isolated based on pathogenic self-reactivity may be efficient at facilitating thymic Treg cell development at low clonal frequencies, even if Foxp3+ cells are not present in the monoclonal setting39,40. It is also possible that these TCRs do not efficiently induce Treg cell development, and may be found on both Treg and non-Treg cells in the polyclonal setting. However, preferential expansion of the non-Treg cell effector population may occur under pro-inflammatory immunization conditions. We also speculate that negative selection will be enhanced with decreasing clonal frequency, given the observed changes in the efficiency of Treg cell development and positive selection. Thus, the mechanisms by which the immune system controls the cell fate of developing T cells expressing self-reactive TCRs may need re-examination within the context of their natural clonal frequency.
Methods
Mice and reagents
Foxp3 gfp 41, OTII42, RIP-mOVA43, and TCli TCRβ and TCR-αβ transgenic mice have been described26. B6.SJL (CD45.1), Tcrb −/−, and Tcra −/− mice were obtained from the Jackson Laboratory. TCR transgenic lines were generated by cloning the cDNA for B8 and G113 TCRα chains into the VA-hCD2 expression vector44, which was then co-injected with the TCli TCRβ chain in the pTβ vector26. TCR transgenic mice did not show any overt signs of autoimmunity, even on a Rag1 −/− background. Amounts of TCRβ expressed in mature CD4SP and peripheral CD4+ T cells was comparable to amounts of TCRβ proteins expressed on wild-type thymocytes, as determined by flow cytometry (data not shown). Animals were housed in a specific-pathogen-free animal facility and were used according to protocols approved by the Institutional Animal Care and Use Committee (IACUC) at Washington University in Saint Louis School of Medicine. Monoclonal antibodies were purchased from eBioscience, Biolegend, and Becton Dickenson: CD4 (RM4-5), CD8 (53–6.7), CD45.1 (A20), CD45.2 (104), Thy1.1 (HIS51), Thy1.2 (53–2.1), CD25 (PC61), HSA (M1/69), CD62L (MEL-14), TCR Vβ6 (RR4-7). The Cell-Trace Far-Red DDAO dye was purchased from Invitrogen. The BM culture cytokines recombinant mouse interleukin-3 (rmIL-3), recombinant human interleukin-6 (rhIL-6), and recombinant mouse stem cell factor (rmSCF) were purchased from Peprotech.
Bone marrow chimeras
All BM cells were T cell-depleted by labeling cells with biotinylated anti-CD4 and anti-CD8 and anti-biotin microbeads, followed by magnetic cell separation using an AutoMACS (Miltenyi Biotech). TCR transgenic Foxp3 gfp Rag1 −/− BM was mixed with CD45.1 Foxp3 gfp BM at 1:1, 1:10, and 1:50 ratios. Ten million cells were injected into lethally irradiated CD45.1 hosts (1050 rads). After 6 weeks, the thymus, peripheral lymph nodes, and spleen were analyzed by flow cytometry (FACSAria, Becton Dickenson). Neonatal chimeras were generated via the intraperitoneal injection of 3×106 TCR transgenic BM cells into 2-day old mice, and were analyzed 6 weeks after injection by flow cytometry. Some mice received a second injection 3 days later. Because of the experimentally induced low frequency of chimerism in some samples, the entire thymus was routinely analyzed. First, ∼2×106 events were recorded to obtain the frequency of CD45.2+ TCR transgenic and CD45.1+ wild-type cells. Next, the remaining thymocytes were analyzed at high speed (20–30,000 events/second), recording only the CD45.2+ CD45.1− events. With an electronic abort rate of around 3%, we estimate that over 90% of observable thymic events originating from the TCR transgenic donor were collected (excluding doublets by FSC-H vs FSC-W and SSC-H vs SSC-W comparisons).
Retroviral bone marrow chimeras
As previously described45, donor Foxp3 gfp Rag1 −/− mice were conditioned with 5-FU prior to BM harvest. After 2 days of culture in rmIL-3 (10 ng/mL), rhIL-6 (20 ng/mL), and rmSCF (100 ng/mL), BM cells underwent two spin-fections on consecutive days with MigR1-derived retrovirus expressing the G25 or R19 TCRα chain, as well as the fixed TCli TCRβ chain downstream of an internal ribosomal entry site. On the fourth day of culture, the BM cells were mixed with CD45.1 Foxp3 gfp BM from 5-FU treated mice at ratios of 1:0, 1:1, 1:10 and injected into lethally irradiated CD45.1 hosts (1050 rads). Mice were analyzed by flow cytometry 6 weeks later.
Assessment of peripheral conversion in vivo
For non-lymphopenic hosts, 1.5 × 106 or 1.5 × 105 FACS purified Foxp3− CD4+ CD45.2+ CD44lo T cells from pooled spleen and lymph nodes of TCR transgenic Foxp3 gfp Rag1 −/− mice were intravenously injected into congenic CD45.1 hosts. Acquisition of Foxp3 and other markers by flow cytometry was assessed after 3–4 weeks. For lymphopenic hosts, 3.5×104 or 3.5×105 TCR tg cells were mixed with 1.8×106 wild-type CD4+ filler cells to achieve varying levels of chimerism, and intravenously injected into Tcrb −/− hosts. As a baseline at 100%, 3.5 × 104 cells were injected without wild-type filler cells. Pooled cells from several secondary lymphoid organs were analyzed for acquisition of Foxp3 by flow cytometry after 19–21 days.
Assessment of negative selection in vivo
Whole wild-type Thy1.1 thymocytes were mixed at a 1:1 ratio with Thy1.2 Rag1 −/− TCR transgenic thymocytes (G113 or OTII) and stained with DDAO prior to intrathymic injection of 2×107 cells into congenically marked WT, RIP-mOVA, or MHCII-KO hosts. TCR transgenic thymocytes were analyzed by flow cytometry on day 3 and as indicated.
Supplementary Material
Acknowledgements
We would like to thank N. Santacruz and J. Hunn for expert technical assistance; K. Murphy, W. Yokoyama, P. Allen, and W. Swat (Washington University); J. Scott-Brown (U. of Colorado); and J.Fontenot (Biogen Idec); for helpful discussions and critical review of the manuscript. Supported by the Arthritis Foundation, Burroughs Wellcome Fund, and National Institutes of Health (C.S.H.).
Footnotes
The authors have no conflicts of interest to report.
References
- 1.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Lahl K, et al. Selective depletion of Foxp3+ regulatory T cells induces a scurfy-like disease. J. Exp. Med. 2007;204:57–63. doi: 10.1084/jem.20061852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 2007;8:191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- 4.Apostolou I, von Boehmer H. In vivo instruction of suppressor commitment in naive T cells. J. Exp. Med. 2004;199:1401–1408. doi: 10.1084/jem.20040249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curotto de Lafaille MA, et al. Adaptive Foxp3+ regulatory T cell-dependent and -independent control of allergic inflammation. Immunity. 2008;29:114–126. doi: 10.1016/j.immuni.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 6.Knoechel B, Lohr J, Kahn E, Bluestone JA, Abbas AK. Sequential development of interleukin 2-dependent effector and regulatory T cells in response to endogenous systemic antigen. J. Exp. Med. 2005;202:1375–1386. doi: 10.1084/jem.20050855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakaguchi S, Sakaguchi N. Thymus and Autoimmunity: Capacity of the Normal Thymus to Produce Pathogenic Self-Reactive T Cells and Conditions Required for their Induction of Autoimmune Disease. J. Exp. Med. 1990;172:537–545. doi: 10.1084/jem.172.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Powrie F, Leach MW, Mauze S, Caddle LB, Coffman RL. Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic instestinal inflammation. Int. Immunol. 1993;5:1461–1471. doi: 10.1093/intimm/5.11.1461. [DOI] [PubMed] [Google Scholar]
- 9.Itoh M, et al. Thymus and autoimmunity: Production of CD25+ CD4+ naturally anergic and suppressive T cells as a key function of the thymus in maintaining immunologic self-tolerance. J. Immunol. 1999;162:5317–5326. [PubMed] [Google Scholar]
- 10.Wong J, et al. Adaptation of TCR repertoires to self-peptides in regulatory and nonregulatory CD4+ T cells. J. Immunol. 2007;178:7032–7041. doi: 10.4049/jimmunol.178.11.7032. [DOI] [PubMed] [Google Scholar]
- 11.Hsieh C-S, et al. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity. 2004;21:267–277. doi: 10.1016/j.immuni.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Hsieh CS, Zheng Y, Liang Y, Fontenot JD, Rudensky AY. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat. Immunol. 2006;7:401–410. doi: 10.1038/ni1318. [DOI] [PubMed] [Google Scholar]
- 13.Pacholczyk R, Ignatowicz H, Kraj P, Ignatowicz L. Origin and T cell receptor diversity of Foxp3+CD4+CD25+ T cells. Immunity. 2006;25:249–259. doi: 10.1016/j.immuni.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 14.Jordan MS, et al. Thymic selection of CD4+ CD25+ regulatory T cells induced by an agonist self-peptide. Nat. Immunol. 2001;2:301–306. doi: 10.1038/86302. [DOI] [PubMed] [Google Scholar]
- 15.Apostolou I, Sarukhan A, Klein L, von Boehmer H. Origin of regulatory T cells with known specificity for antigen. Nat. Immunol. 2002;3:756–763. doi: 10.1038/ni816. [DOI] [PubMed] [Google Scholar]
- 16.Van Santen H-M, Benoist C, Mathis D. Number of T reg cells that differentiate does not increase upon encounter of agonist ligand on thymic epithelial cells. J. Exp. Med. 2004;200:1221–1230. doi: 10.1084/jem.20041022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pennington DJ, et al. Early events in the thymus affect the balance of effector and regulatory T cells. Nature. 2006;444:1073–1077. doi: 10.1038/nature06051. [DOI] [PubMed] [Google Scholar]
- 18.Pacholczyk R, et al. Nonself-antigens are the cognate specificities of foxp3(+) regulatory T cells. Immunity. 2007;27:493–504. doi: 10.1016/j.immuni.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lio CW, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity. 2008;28:100–111. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lathrop SK, Santacruz NA, Pham D, Luo J, Hsieh CS. Antigen-specific peripheral shaping of the natural regulatory T cell population. J. Exp. Med. 2008 doi: 10.1084/jem.20081359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malek TR, Yu A, Vincek V, Scibelli P, Kong L. CD4 regulatory T cells prevent lethal autoimmunity in IL-2Rbeta-deficient mice. Implications for the nonredundant function of IL-2. Immunity. 2002;17:167–178. doi: 10.1016/s1074-7613(02)00367-9. [DOI] [PubMed] [Google Scholar]
- 22.Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J. Immunol. 2007;178:280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- 23.Cabarrocas J, et al. Foxp3+ CD25+ regulatory T cells specific for a neo-self-antigen develop at the double-positive thymic stage. Proc. Natl. Acad. Sci. USA. 2006;103:8453–8458. doi: 10.1073/pnas.0603086103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Canelles M, Park ML, Schwartz OM, Fowlkes BJ. The influence of the thymic environment on the CD4-versus-CD8 T lineage decision. Nat. Immunol. 2003;4:756–764. doi: 10.1038/ni953. [DOI] [PubMed] [Google Scholar]
- 25.Huesmann M, Scott B, Kisielow P, von Boehmer H. Kinetics and efficacy of positive selection in the thymus of normal and T cell receptor transgenic mice. Cell. 1991;66:533–540. doi: 10.1016/0092-8674(81)90016-7. [DOI] [PubMed] [Google Scholar]
- 26.Wong P, Goldrath AW, Rudensky AY. Competition for specific intrathymic ligands limits positive selection in a TCR transgenic model of CD4+ T cell development. J. Immunol. 2000;164:6252–6259. doi: 10.4049/jimmunol.164.12.6252. [DOI] [PubMed] [Google Scholar]
- 27.Liu VC, et al. Tumor evasion of the immune system by converting CD4+CD25-T cells into CD4+CD25+ T regulatory cells: role of tumor-derived TGF-beta. J. Immunol. 2007;178:2883–2892. doi: 10.4049/jimmunol.178.5.2883. [DOI] [PubMed] [Google Scholar]
- 28.Hill JA, et al. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27:786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 29.Gavin MA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–775. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 30.Lin W, et al. Regulatory T cell development in the absence of functional Foxp3. Nat. Immunol. 2007;8:359–368. doi: 10.1038/ni1445. [DOI] [PubMed] [Google Scholar]
- 31.Feuerer M, et al. Enhanced thymic selection of FoxP3+ regulatory T cells in the NOD mouse model of autoimmune diabetes. Proc. Natl. Acad. Sci. USA. 2007;104:18181–18186. doi: 10.1073/pnas.0708899104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tai X, Cowan M, Feigenbaum L, Singer A. CD28 costimulation of developing thymocytes induces Foxp3 expression and regulatory T cell differentiation independently of interleukin 2. Nat. Immunol. 2005;6:152–162. doi: 10.1038/ni1160. [DOI] [PubMed] [Google Scholar]
- 33.Troy AE, Shen H. Cutting edge: homeostatic proliferation of peripheral T lymphocytes is regulated by clonal competition. J. Immunol. 2003;170:672–676. doi: 10.4049/jimmunol.170.2.672. [DOI] [PubMed] [Google Scholar]
- 34.Moses CT, Thorstenson KM, Jameson SC, Khoruts A. Competition for self ligands restrains homeostatic proliferation of naive CD4 T cells. Proc. Natl. Acad. Sci. USA. 2003;100:1185–1190. doi: 10.1073/pnas.0334572100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hataye J, Moon JJ, Khoruts A, Reilly C, Jenkins MK. Naive and memory CD4+ T cell survival controlled by clonal abundance. Science. 2006;312:114–116. doi: 10.1126/science.1124228. [DOI] [PubMed] [Google Scholar]
- 36.Tang Q, et al. Visualizing regulatory T cell control of autoimmune responses in nonobese diabetic mice. Nat. Immunol. 2006;7:83–92. doi: 10.1038/ni1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia Z, et al. Competition for antigen determines the stability of T cell-dendritic cell interactions during clonal expansion. Proc. Natl. Acad. Sci. USA. 2007;104:4553–4558. doi: 10.1073/pnas.0610019104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romagnoli P, Hudrisier D, van Meerwijk JP. Molecular signature of recent thymic selection events on effector and regulatory CD4+ T lymphocytes. J. Immunol. 2005;175:5751–5758. doi: 10.4049/jimmunol.175.9.5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.You S, Slehoffer G, Barriot S, Bach JF, Chatenoud L. Unique role of CD4+CD62L+ regulatory T cells in the control of autoimmune diabetes in T cell receptor transgenic mice. Proc. Natl. Acad. Sci. USA. 2004;101(Suppl 3):14580–14585. doi: 10.1073/pnas.0404870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olivares-Villagomez D, Wang Y, Lafaille JJ. Regulatory CD4+ T Cells Expressing Endogenous T Cell Receptor Chains Protect Myelin Basic Protein-specific Transgenic Mice from Spontaneous Autoimmune Encephalomyelitis. J. Exp. Med. 1998;188:1883–1894. doi: 10.1084/jem.188.10.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fontenot JD, et al. Regulatory T cell lineage specification by the forkhead transcription factor Foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 42.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha-and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell. Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 43.Kurts C, et al. Constitutive class I-restricted exogenous presentation of self antigens in vivo. J. Exp. Med. 1996;184:923–930. doi: 10.1084/jem.184.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhumabekov T, Corbella P, Tolaini M, Kioussis D. Improved version of a human CD2 minigene based vector for T cell-specific expression in transgenic mice. J. Immunol. Methods. 1995;185:133–140. doi: 10.1016/0022-1759(95)00124-s. [DOI] [PubMed] [Google Scholar]
- 45.Gough PJ, Raines EW. Gene therapy of apolipoprotein E-deficient mice using a novel macrophage-specific retroviral vector. Blood. 2003;101:485–491. doi: 10.1182/blood-2002-07-2131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.