Abstract
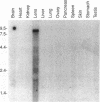
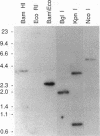
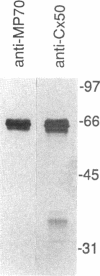
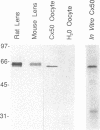
The crystalline lens is an attractive system to study the biology of intercellular communication; however, the identity of the structural components of gap junctions in the lens has been controversial. We have cloned a novel member of the connexin family of gap junction proteins, Cx50, and have shown that it is likely to correspond to the previously described lens fiber protein MP70. The N-terminal amino acid sequence of MP70 closely matches the sequence predicted by the clone. Cx50 mRNA is detected only in the lens, among the 12 organs tested, and this distribution is indistinguishable from that of MP70 protein. A monoclonal antibody directed against MP70 and an anti-Cx50 antibody produced against a synthetic peptide identify the same proteins on western blots and produce identical patterns of immunofluorescence on frozen sections of rodent lens. We also show that expression of Cx50 in paired Xenopus oocytes induces high levels of voltage-dependent conductance. This indicates that Cx50 is a functional member of the connexin family with unique physiological properties. With the cloning of Cx50, all known participants in gap junction formation between various cell types in the lens are available for study and reconstitution in experimental systems.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrio L. C., Suchyna T., Bargiello T., Xu L. X., Roginski R. S., Bennett M. V., Nicholson B. J. Gap junctions formed by connexins 26 and 32 alone and in combination are differently affected by applied voltage. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8410–8414. doi: 10.1073/pnas.88.19.8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassnett S., Duncan G. The influence of pH on membrane conductance and intercellular resistance in the rat lens. J Physiol. 1988 Apr;398:507–521. doi: 10.1113/jphysiol.1988.sp017054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti E. L., Dunia I., Bentzel C. J., Vermorken A. J., Kibbelaar M., Bloemendal H. A portrait of plasma membrane specializations in eye lens epithelium and fibers. Biochim Biophys Acta. 1976 Dec 14;457(3-4):353–384. doi: 10.1016/0304-4157(76)90004-6. [DOI] [PubMed] [Google Scholar]
- Benedetti E. L., Dunia I., Bloemendal H. Development of junctions during differentiation of lens fibers. Proc Natl Acad Sci U S A. 1974 Dec;71(12):5073–5077. doi: 10.1073/pnas.71.12.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M. V., Barrio L. C., Bargiello T. A., Spray D. C., Hertzberg E., Sáez J. C. Gap junctions: new tools, new answers, new questions. Neuron. 1991 Mar;6(3):305–320. doi: 10.1016/0896-6273(91)90241-q. [DOI] [PubMed] [Google Scholar]
- Beyer E. C., Kistler J., Paul D. L., Goodenough D. A. Antisera directed against connexin43 peptides react with a 43-kD protein localized to gap junctions in myocardium and other tissues. J Cell Biol. 1989 Feb;108(2):595–605. doi: 10.1083/jcb.108.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer E. C., Paul D. L., Goodenough D. A. Connexin family of gap junction proteins. J Membr Biol. 1990 Jul;116(3):187–194. doi: 10.1007/BF01868459. [DOI] [PubMed] [Google Scholar]
- Beyer E. C., Paul D. L., Goodenough D. A. Connexin43: a protein from rat heart homologous to a gap junction protein from liver. J Cell Biol. 1987 Dec;105(6 Pt 1):2621–2629. doi: 10.1083/jcb.105.6.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Dahl G., Miller T., Paul D., Voellmy R., Werner R. Expression of functional cell-cell channels from cloned rat liver gap junction complementary DNA. Science. 1987 Jun 5;236(4806):1290–1293. doi: 10.1126/science.3035715. [DOI] [PubMed] [Google Scholar]
- Ebihara L., Beyer E. C., Swenson K. I., Paul D. L., Goodenough D. A. Cloning and expression of a Xenopus embryonic gap junction protein. Science. 1989 Mar 3;243(4895):1194–1195. doi: 10.1126/science.2466337. [DOI] [PubMed] [Google Scholar]
- Emptage N. J., Duncan G., Croghan P. C. Internal acidification modulates membrane and junctional resistance in the isolated lens of the frog Rana pipiens. Exp Eye Res. 1992 Jan;54(1):33–39. doi: 10.1016/0014-4835(92)90066-2. [DOI] [PubMed] [Google Scholar]
- Goodenough D. A., Dick J. S., 2nd, Lyons J. E. Lens metabolic cooperation: a study of mouse lens transport and permeability visualized with freeze-substitution autoradiography and electron microscopy. J Cell Biol. 1980 Aug;86(2):576–589. doi: 10.1083/jcb.86.2.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough D. A. Lens gap junctions: a structural hypothesis for nonregulated low-resistance intercellular pathways. Invest Ophthalmol Vis Sci. 1979 Nov;18(11):1104–1122. [PubMed] [Google Scholar]
- Goodenough D. A., Paul D. L., Jesaitis L. Topological distribution of two connexin32 antigenic sites in intact and split rodent hepatocyte gap junctions. J Cell Biol. 1988 Nov;107(5):1817–1824. doi: 10.1083/jcb.107.5.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C. R., Harfst E., Gourdie R. G., Severs N. J. Analysis of the rat liver gap junction protein: clarification of anomalies in its molecular size. Proc R Soc Lond B Biol Sci. 1988 Mar 22;233(1271):165–174. doi: 10.1098/rspb.1988.0016. [DOI] [PubMed] [Google Scholar]
- Gruijters W. T., Kistler J., Bullivant S., Goodenough D. A. Immunolocalization of MP70 in lens fiber 16-17-nm intercellular junctions. J Cell Biol. 1987 Mar;104(3):565–572. doi: 10.1083/jcb.104.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haefliger J. A., Bruzzone R., Jenkins N. A., Gilbert D. J., Copeland N. G., Paul D. L. Four novel members of the connexin family of gap junction proteins. Molecular cloning, expression, and chromosome mapping. J Biol Chem. 1992 Jan 25;267(3):2057–2064. [PubMed] [Google Scholar]
- Helms C., Graham M. Y., Dutchik J. E., Olson M. V. A new method for purifying lambda DNA from phage lysates. DNA. 1985 Feb;4(1):39–49. doi: 10.1089/dna.1985.4.39. [DOI] [PubMed] [Google Scholar]
- Kistler J., Bullivant S. Protein processing in lens intercellular junctions: cleavage of MP70 to MP38. Invest Ophthalmol Vis Sci. 1987 Oct;28(10):1687–1692. [PubMed] [Google Scholar]
- Kistler J., Christie D., Bullivant S. Homologies between gap junction proteins in lens, heart and liver. Nature. 1988 Feb 25;331(6158):721–723. doi: 10.1038/331721a0. [DOI] [PubMed] [Google Scholar]
- Kistler J., Kirkland B., Bullivant S. Identification of a 70,000-D protein in lens membrane junctional domains. J Cell Biol. 1985 Jul;101(1):28–35. doi: 10.1083/jcb.101.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984 Sep 25;12(18):7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird D. W., Puranam K. L., Revel J. P. Turnover and phosphorylation dynamics of connexin43 gap junction protein in cultured cardiac myocytes. Biochem J. 1991 Jan 1;273(Pt 1):67–72. doi: 10.1042/bj2730067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias R. T., Riquelme G., Rae J. L. Cell to cell communication and pH in the frog lens. J Gen Physiol. 1991 Dec;98(6):1085–1103. doi: 10.1085/jgp.98.6.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T. M., Goodenough D. A. Evidence for two physiologically distinct gap junctions expressed by the chick lens epithelial cell. J Cell Biol. 1986 Jan;102(1):194–199. doi: 10.1083/jcb.102.1.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T., Dahl G., Werner R. Structure of a gap junction gene: rat connexin-32. Biosci Rep. 1988 Oct;8(5):455–464. doi: 10.1007/BF01121644. [DOI] [PubMed] [Google Scholar]
- Musil L. S., Beyer E. C., Goodenough D. A. Expression of the gap junction protein connexin43 in embryonic chick lens: molecular cloning, ultrastructural localization, and post-translational phosphorylation. J Membr Biol. 1990 Jun;116(2):163–175. doi: 10.1007/BF01868674. [DOI] [PubMed] [Google Scholar]
- Paul D. L., Ebihara L., Takemoto L. J., Swenson K. I., Goodenough D. A. Connexin46, a novel lens gap junction protein, induces voltage-gated currents in nonjunctional plasma membrane of Xenopus oocytes. J Cell Biol. 1991 Nov;115(4):1077–1089. doi: 10.1083/jcb.115.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D. L. Molecular cloning of cDNA for rat liver gap junction protein. J Cell Biol. 1986 Jul;103(1):123–134. doi: 10.1083/jcb.103.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peracchia C. Calcium effects on gap junction structure and cell coupling. Nature. 1978 Feb 16;271(5646):669–671. doi: 10.1038/271669a0. [DOI] [PubMed] [Google Scholar]
- Rae J. L., Kuszak J. R. The electrical coupling of epithelium and fibers in the frog lens. Exp Eye Res. 1983 Mar;36(3):317–326. doi: 10.1016/0014-4835(83)90114-8. [DOI] [PubMed] [Google Scholar]
- Rae J. L. The electrophysiology of the crystalline lens. Curr Top Eye Res. 1979;1:37–90. [PubMed] [Google Scholar]
- Schuetze S. M., Goodenough D. A. Dye transfer between cells of the embryonic chick lens becomes less sensitive to CO2 treatment with development. J Cell Biol. 1982 Mar;92(3):694–705. doi: 10.1083/jcb.92.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson K. I., Jordan J. R., Beyer E. C., Paul D. L. Formation of gap junctions by expression of connexins in Xenopus oocyte pairs. Cell. 1989 Apr 7;57(1):145–155. doi: 10.1016/0092-8674(89)90180-3. [DOI] [PubMed] [Google Scholar]
- Zhang J. T., Nicholson B. J. Sequence and tissue distribution of a second protein of hepatic gap junctions, Cx26, as deduced from its cDNA. J Cell Biol. 1989 Dec;109(6 Pt 2):3391–3401. doi: 10.1083/jcb.109.6.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]