Abstract
Adenovirus small early region 1a (e1a) protein drives cells into S phase by binding RB family proteins and the closely related histone acetyl transferases p300 and CBP. The interaction with RB proteins displaces them from DNA-bound E2F transcription factors, reversing their repression of cell cycle genes. However, it has been unclear how the e1a interaction with p300 and CBP promotes passage through the cell cycle. We show that this interaction causes a threefold reduction in total cellular histone H3 lysine 18 acetylation (H3K18ac). CBP and p300 are required for acetylation at this site because their knockdown causes specific hypoacetylation at H3K18. SV40 T antigen also induces H3K18 hypoacetylation. Because global hypoacetylation at this site is observed in prostate carcinomas with poor prognosis, this suggests that processes resulting in global H3K18 hypoacetylation may be linked to oncogenic transformation.
The adenovirus small e1a protein drives contact-inhibited primary cells through the cell cycle dependent on three conserved regions (CRs) in e1a (fig. S1) (1). e1a is not a DNA binding protein, but it binds several other proteins. e1a CR2 binds the retinoblastoma protein (RB) and its paralogs p107 (RBL1) and p130 (RBL2) with high affinity and, together with a lower affinity binding region within CR1, displaces RB proteins from E2F family transcription factors (heterodimers of E2F1 through E2F5 with DP1 or DP2) (1–3). Release of RB family proteins and their associated repressing chromatin modifying activities (4) results in de-repression of cell cycle genes (1). Although the N terminus of e1a is not as extensively conserved among primate adenoviruses as CR1 and CR2, it is nonetheless well conserved among these viruses (5) and is required to drive cells into the cell cycle (1). The N terminus and residues in CR1 that are required for e1a transforming activity bind to several cellular proteins involved in the regulation of chromatin structure, including p300 and CBP, PCAF, GCN5, and p400 (1). However, the interaction with p300/CBP is the most important for transformation (5) and inhibits p300/CBP-dependent co-activator activity in transient trans-fection assays (6). It is not understood how the e1a interaction with p300/CBP contributes to cell cycling.
In studies potentially related to this question, Seligson et al. (7) reported that the risk of tumor recurrence in patients with low-grade prostate cancer (tumors with well-formed glandular structures) is related to the global cellular levels of H3K18 acetylation and H3K4 methylation observed by the intensity of nuclear staining with antibodies specific for these histone modifications. Global H3K18 hypoacetylation plus H3K4 hypomethylation strongly correlate with increased risk of tumor recurrence (7). Because the e1a interaction with p300/CBP is required for transformation by e1a, and because all the p300 and CBP in a nuclear extract of adenoviros type 5 (Ad5)–transformed 293 cells co-elutes with e1a on a gel filtration column in a complex of ∼ 600 kD (8), we asked whether e1a induces a global decrease in histone acetylation and whether such an activity might contribute to e1a's transforming activity.
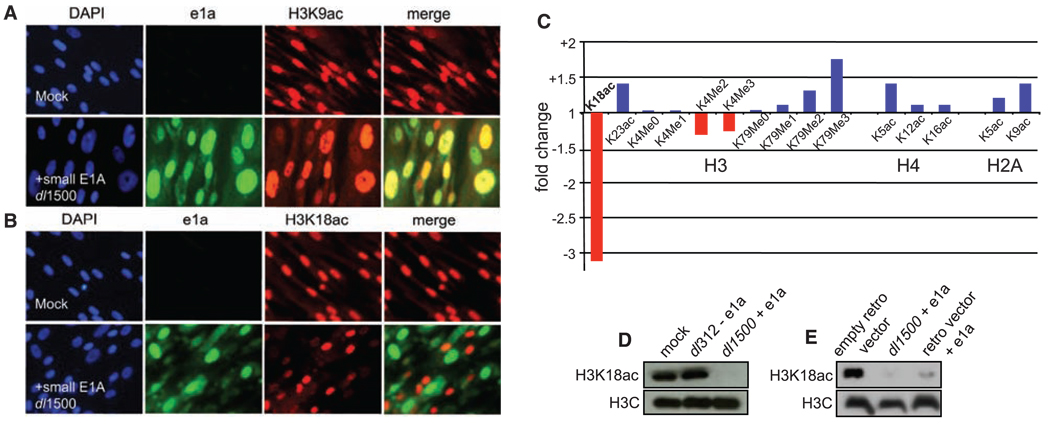
To address these questions, we infected human primary IMR90 embryo lung fibroblasts with Ad5 dl1500 that expresses only e1a (9) or Ad5 dl312 with a deletion of the E1A region (10). At 24 hours postinfection (p.i.), cells were immu-nostained with highly specific antibodies for acetylated lysines on histones H3 (K9, K14, K18, K23, and K27) and H4 (K5, K8, K12, and K16) (11), di- and trimethylation at H3K4, and dimethylation at H3K9. Nuclei containing e1a had very low H3K18ac (Fig. 1B), with no detectable changes in H3K9ac (Fig. 1A) or any of the other 10 modifications examined (fig. S2). The immunofluorescence data were confirmed by mass spectrometry, which also revealed slight H3K4 hypomethylation (Fig. 1C, Supplemental fig. S3, and table S1), and Western blot of acid-extracted histones (Fig. 1D and fig. S4). Similar results were observed in HeLa cells (table S2 and figs. S3 and S5) and after retrovirus vector expression of e1a (Fig. 1e), ruling out the possible influence of other adeno-virus proteins expressed at a very low level in the absence of large E1A. Thus, e1a induces hypo-acetylation of H3K18 and to a lesser extent hypomethylation of H3K4, similar to chromatin changes observed in primary prostate cancers with poor prognosis (7). Global H3K18 hypoacetyl-ation requires e1a's interaction with p300/CBP because hypoacetylation was not induced by e1a mutants Δ4–25, R2G, and I5G, which interfere with p300/CBP binding, but was induced by H7A, which does not (5), and by e1a deletion mutants in its high-affinity binding site for RB proteins Δ111–123 (ΔCR2) (3), CtBP (Δ233–237) (11), and p400 (Δ25–36) (12) (Supplemental figs. S6 and S7). The same e1a N-terminal mutations that prevented e1a-induced H3K18 hypoacetylation (Δ4–25, R2G, and I5G) were unable to induce S phase in quiescent human cells (13) or transformation of rodent cells (5).
Fig. 1.
e1a induces hypoacetylation of H3K18. Contact inhibited human primary IMR90 fibroblasts were mock-infected or infected with Ad5 dl1500 (e1a only). 24 hours p.i., cells were fixed and immunostained with antibodies to e1a (green), either (A) H3K9ac (red) or (B) K3K18ac (red), and 4´,6´-diamidino-2-phenylindole (DAPI) to stain nuclei. Merges of green and red signals are also shown. (C) Fold change in histone modifications after dl1500-infection of IMR90 cells determined by mass spectrometry (fig. S2). Western blots of histones from (D) IMR90 cells infected with dl312 (no e1a) or dl1500 (e1a only) and (E) HeLa cells transduced with the indicated retrovirus vectors or dl1500-infected.
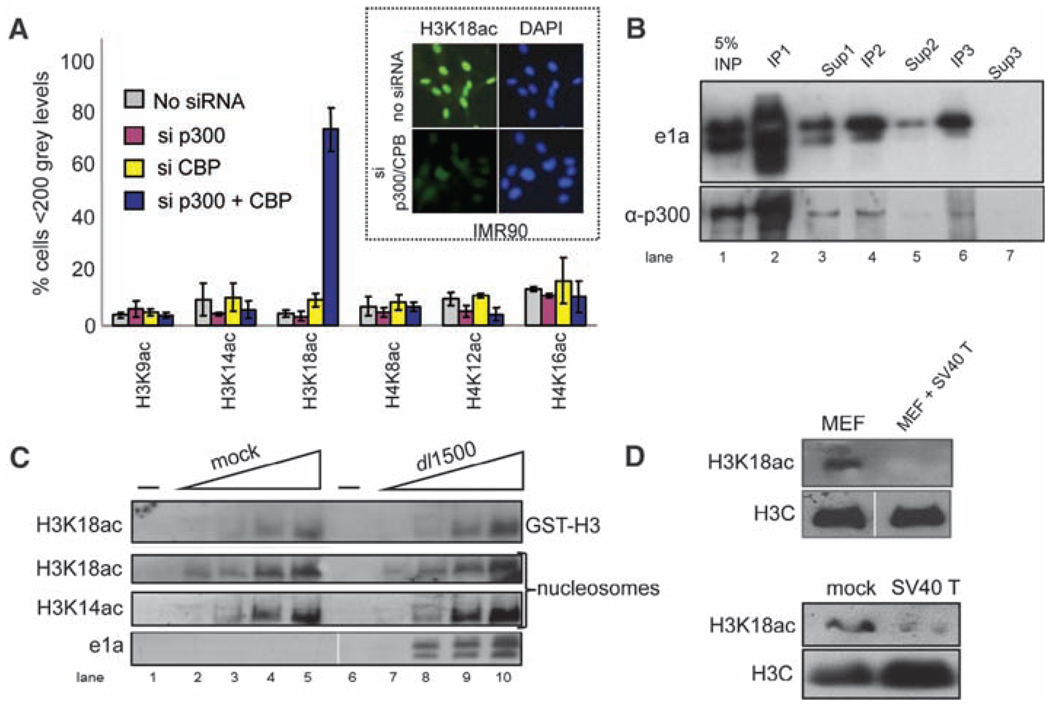
In vitro, CBP and p300 acetylate several histone lysines, including H3K18 (14). However, small interfering RNA (siRNA) knockdown of both CBP and p300 (but not individually) resulted in specific H3K18 hypoacetylation in vivo in IMR90 and HeLa cells (Fig. 2A and fig. S8). Dual knockdown did not affect five other sites of histone modification assayed. Consequently, p300/CBP, and possibly PCAF which associates with them (15), are the major histone acetyltransferases (HATs) required for maintenance of global H3K18ac in vivo. e1a-induced H3K18 hypoacetylation was not due to inhibition of p300/CBP HAT enzymatic activities generally, or specifically for H3K18, even though they were quantitatively bound by e1a when isolated from infected cells (Fig. 2, B and C).
Fig. 2.
CBP or p300 is required for acetylating H3K18. (A) Computational image analysis of immunofluorescent micrographs after IMR90 siRNA transfections against p300 and CBP (18) (fig. S7). The fraction T SD of nonstaining nuclei is plotted, n = 4. (Inset) Typical micrographs. (B) Nuclear extract from dl1500-infected HeLa cells 24 hours p.i. was subjected to three successive immunoprecipitations (IP1 to IP3) with equal amounts of monoclonal antibody (mAb) M73 against e1a. One-tenth of the total IP and supernatant (Sup1 to Sup3) after each IP were subjected to Western blotting. Differences in post-translational modifications caused e1a to appear as a doublet. (C) Twofold increasing amounts of p300/CBP IPed from mock- or dl1500-infected HeLa cells were incubated with glutathione S-transferase (GST)–H3 N terminus or hypoacetylated nucleosomes plus acetyl-coenzyme A (18), and products were assayed by Western blotting with anti-H3K18ac or anti-H3K14ac. (D) Histones from MEFs transformed with a retrovirus vector expressing SV40 large T antigen or the parental MEFs (top) and from IMR90 fibroblasts mock-infected or infected with an Ad expression vector for SV40 large T antigen 24 hours p.i. (bottom) were subjected to Western blotting.
Simian virus 40 (SV40) large T antigen, like e1a, is a DNA tumor virus transforming protein that stimulates cell cycling through interactions with RB family proteins (16) as well as other cellular proteins, possibly including p300/CBP (16). Consequently, we examined H3K18ac in IMR90 cells expressing SV40 large T antigen and primary murine embryo fibroblasts (MEFs) transformed with an SV40 T antigen–expressing ret-rovirus vector (17) (fig. S9). Western blotting revealed decreased H3K18ac in both cell types expressing SV40 large T antigen (Fig. 2D). Induction of global H3K18 hypoacetylation by both e1a and SV40 large T antigen suggests that hypo-acetylation at this site may be a general consequence of DNA tumor virus oncoprotein activity.
The accompanying report (13) shows that e1a causes a massive, genomewide relocalization of p300, CBP, and PCAF, targeting them to the promoter regions of a limited, but biologically related, set of genes that promote cell growth and division, causing H3K18 hyperacetylation in their promoter regions and strong induction. e1a also causes the redistribution of RB family proteins to promoter regions of genes involved in antiviral responses and differentiation, inducing their hypo-acetylation at H3K18 and repression (13). This redistribution of p300/CBP/PCAF and RB family members may explain the observed global H3K18 hypoacetylation by restricting these HATs to the ∼one-third of all genes involved in cell growth and replication that are activated by e1a (13).
Supplementary Material
Footnotes
Information about obtaining reprints of this article or about obtaining permission to reproduce this article in whole or in part can be found at: http://www.sciencemag.org/about/permissions.dtl
Supporting Online Material
www.sciencemag.org/cgi/content/full/321/5892/1084/DC1
Materials and Methods
Figs. S1 to S9
Tables S1 and S2
References and Notes
References and Notes
- 1.Berk AJ. Oncogene. 2005;24:7673. doi: 10.1038/sj.onc.1209040. [DOI] [PubMed] [Google Scholar]
- 2.Nevins JR. Curr. Opin. Genet. Dev. 1994;4:130. doi: 10.1016/0959-437x(94)90101-5. [DOI] [PubMed] [Google Scholar]
- 3.Liu X, Marmorstein R. Genes Dev. 2007;21:2711. doi: 10.1101/gad.1590607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blais A, Dynlacht BD. Curr. Opin. Cell Biol. 2007;19:658. doi: 10.1016/j.ceb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasti M, Grand RJ, Mymryk JS, Gallimore PH, Turnell AS. J. Virol. 2005;79:5594. doi: 10.1128/JVI.79.9.5594-5605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rochette-Egly C, Fromental C, Chambon P. Genes Dev. 1990;4:137. doi: 10.1101/gad.4.1.137. [DOI] [PubMed] [Google Scholar]
- 7.Seligson DB, et al. Nature. 2005;435:1262. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 8.Wang G, Berk AJ. J. Virol. 2002;76:9186. doi: 10.1128/JVI.76.18.9186-9193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montell C, Courtois G, Eng C, Berk A. Cell. 1984;36:951. doi: 10.1016/0092-8674(84)90045-x. [DOI] [PubMed] [Google Scholar]
- 10.Jones N, Shenk T. Proc. Natl. Acad. Sci. U.S.A. 1979;76:3665. doi: 10.1073/pnas.76.8.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chinnadurai G. Mol. Cell. 2002;9:213. doi: 10.1016/s1097-2765(02)00443-4. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs M, et al. Cell. 2001;106:297. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 13.Ferrari R, et al. Science. 2008;321:1086. doi: 10.1126/science.1155546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kouzarides T. Cell. 2007;128:693. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. Nature. 1996;382:319. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 16.Ahuja D, Saenz-Robles MT, Pipas JM. Oncogene. 2005;24:7729. doi: 10.1038/sj.onc.1209046. [DOI] [PubMed] [Google Scholar]
- 17.O'Hare MJ, et al. Proc. Natl. Acad. Sci. U.S.A. 2001;98:646. [Google Scholar]
- 18.Materials and methods are available on Science Online.
- 19.Supported by NIH grant CA25235 to A.J.B. and a Beckman Young Investigator Award to S.K.K. We thank C. Eng for technical assistance.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.