Abstract
Recent advances have revealed that modification of chromatin structure is an important determinant of cell fate and function. DNA methylation and covalent modifications of histone tails contribute to changes in chromatin architectures, either enhancing or repressing gene expression. Another mechanism underlying the modification of chromatin structure relies on the activity of the SWI/SNF-related ATP-dependent chromatin remodeling complexes that control the accessibility of DNA sequences to transcription factors. There is increasing evidence that one particular family of ATP-dependent chromatin remodeling complexes based on the alternative DNA-dependent ATPases, Brg1 and Brm, plays essential roles during neural development. This remodeling complex has dedicated functions at different stages of neural development that appear to arise by combinatorial assembly of its subunits.
Introduction
Vertebrate genomes contain about 30 genes encoding ATP-dependent chromatin remodeling enzymes that are often subunits of large polymorphic complexes resembling the yeast SWI/SNF complex [1]. Recent studies have revealed that one family of complexes based on the Brg1 and Brm ATPases (BAF complexes) has particularly critical dosage-dependent roles in the development of the nervous system [2–4]. Exchanges of subunits within BAF complexes accompany the transitions from pluripotent, to multipotent stem cells and finally to post mitotic neurons, and appear to be critical for these transitions [5,6].
ATP-dependent chromatin remodeling and the development of the invertebrate nervous system
As summarized in Table 1, screens for genes involved in neurogenesis or morphogenesis of dendrites of neurons in invertebrate systems identified subunits of BAF complexes to be important for various aspects of neural development. Recently, Parrish et al. performed an RNAi screening to identify transcription factors that influenced the dendrite formation of class I da neurons in the peripheral nervous system (PNS) in Drosophila embryos [7]. Close examination of defects in dendrite morphogenesis identified brahma (a Brg1/Brm homolog) and its associated factors, Bap55 and Bap60 (BAF53b and BAF60 in vertebrates, respectively) to display misrouting phenotypes when their functions were reduced [7]. RNAi against Bap55 and Snr1 (BAF47) also resulted in reduced arborization and lateral branching phenotypes, implying the role of BAF complex in dendritic formation of neurons. Additionally, reducing the function of moira (BAF170) and SAYP (a likely homologue of BAF45a [1]) caused abnormal commissures and loss of neurons in the PNS [8]. In C. elegans, loss-of-function of most of the components of BAF complex lead to embryonic lethality [9], which makes the analysis of neural phenotypes in later developmental stages difficult. In addition, RNAi-based screening for neuronal phenotypes in growing larval stages has been difficult owing to the inefficiency of RNAi in neurons in C. elegans. Recently, a RNAi-hypersensitive mutant was isolated, which allowed genome-wide RNAi-screening to identify genes important for axon-guidance in C. elegans [10]. Screening genes in chromosomes I and III, Schmitz et al. identified ZK1128.5 (tag-246, a BAF60 homolog) whose reduced function resulted in axon-guidance defects. In a separate study, Sawa et al. conducted mutagenesis screening to identify genes important for organogenesis of the phasmid socket that arises from the neurogenic asymmetric division of precursor-like T blast cells in C. elegans. Loss-of-function mutations in two genes, psa-1 and psa-4 (BAF155 and Brg1, respectively) lead to phasmid socket absent (Psa) phenotype resulting from a failure of T cells to generate neural cells [11]. psa-1 and psa-4 mutations genetically synergized to enhance the Psa phenotype, suggesting that they act in the same complex, and RNAi against other subunits, R07E5.3 (a BAF47 homolog), tag-246 and C18E3.2 (homologs of BAF60) all resulted in the Psa phenotype, further supporting the role BAF complex in the neurogenic asymmetric divisions of T cells. Although it remains unknown whether the involvement of BAF complex is evolutionarily conserved, it is noteworthy that the expression of Emx2, a homeobox gene proposed to be involved in asymmetric division during neurogenesis [12] and Foxg1, the winged helix/forkhead transcription factor that controls the timing of neurogenesis [13], are regulated by Brg1 in neural progenitors [3].
Table 1.
Summary of components of BAF complex and the phenotypes associated with their reduced function in invertebrates.
| Mammals | Drosophila | Neural phenotype (Drosophila) |
C. elegans | Neural phenotype (C. elegans) |
|---|---|---|---|---|
| Brg1/Brm | brahma | Misrouting defect in dendrite morphogenesis |
psa-4 | Psa phenotype (defect in symmetric division T blast cells) |
| BAF45a/b/c/d | SAYP | Lack ventral commissures | C28H8.9 | N/A |
| BAF47 | Snr1 | Increased primary branch extension; reduced lateral branching defect in dendrites |
R07E5.3 | Psa phenotype |
| RNAi: 100% embryonic lethality → zygotic | ||||
| RNAi: Psa phenotype | ||||
| BAF53a/b | Bap55 | Reduced arborization, misrouting defect in dendrite formation |
ZK616.4 | N/A |
| BAF57 | Dalao/Bap111 | N/A | Y71H2AM.17 | N/A |
| BAF60a/b/c | Bap60 | Misrouting defect in dendrites |
tag-246 | RNAi: Psa phenotype, axon-guidance defect |
| C18E3.2 | RNAi: 50% embryonic lethality → escapers have Psa phenotype |
|||
| BAF155/SRG3 | moira | Abnormal commissures and loss of neurons in the peripheral nervous system |
psa-1 | Psa phenotype |
| BAF170 | moira | Same as above | psa-1 | Same as above |
| BAF180 |
Polybromo/ Bap180 |
N/A | pbrm-1 | N/A |
| BAF200 | Bap170 | N/A | C08B11.3 | N/A |
| BAF250a/b | Osa | N/A |
psa-10/let- 526 |
Psa phenotype (Uchida, M and Sawa, H, personal communication) |
Genetics and biochemical features of BAF complexes in mammals
In mammals, the composition of BAF complexes is polymorphic with subunits encoded by homologous gene families, members of which assume mutually exclusive occupancy in the complex [14–16]. The core ATPase subunit is encoded by Brg1 and Brm in mammals. Although genome wide mapping in neurons has not been done, studies in embryonic stem (ES) cells indicate that there are about 10,000 BAF binding sites per genome and about half of these occur near genes most commonly near the transcription start sites [5]. Whereas inactivation of Brm did not lead to any obvious neural phenotype [17], Brg1 appears to be important for various aspects in neural development. Mice deficient with Brg1 die in pre-or peri-implantation stage, and heterozygous mutants display defects in neural tube formation when developed into later embryonic stages [3,18]. Similarly, mice homozygous with the null mutation of BAF155 (Srg3 or mSWI3) die during peri-implantation stage, and heterozygous mutants develop also with neural tube defects [19]. Srg3/BAF155 was proposed to protect BAF complexes from proteasomal degradation and affect the nuclear localization of the complex [20], suggesting the requirement of functional BAF complexes during neural tube development. Recent studies suggest that the neural tube defect seen with Brg1 or BAF155 mutations may arise from improper maintenance and differentiation of neural progenitors. Conditional inactivation of Brg1 using Cre recombinase specifically expressed in neural progenitors leads to exencephaly which typically results from neural tube defects in developing embryos [3]. Whereas Brg1 inactivation in Xenopus lead to expansion of neural progenitors [21], the role of Brg1 in higher vertebrates appears to be the opposite. Brg1 inactivation interfered with self-renewal of neural progenitors and eventual reduction in neural progenitor population [3,22]. The reduced proliferation with Brg1 deletion is specific for neural progenitors, as the absence of Brg1 in fibroblasts did not lead to defects in survival and proliferation [18]. The inferred specific function of BAF complex in the self-renewal of neural progenitors suggests that the neural tube defect may be due to the failure of proper maintenance and differentiation of neural progenitors. While this hypothesis remains to be examined more thoroughly, recent identification of Vangl1 and Vangl2 that were associated with congenital abnormalities of neural tube formation in humans [23], may provide another explanation for the association between the reduced function of BAF complex and neural tube defects. Vangl1 is expressed in developing neural tubes in mouse embryos, and mice with loss-of-function for Vangl1 and Vangl2 displayed defects in neural tube closure [24]. Our recent data suggest that Brg1 occupies the promoter of Vangl1 at least in ES cells [5]. This observation raises the possibility that Brg1 may control the expression of Vangl1 in neural cells and may explain the gene dosage-dependence on Brg1 and Vangl1 in neural tube phenotypes. More detailed analysis remains necessary to determine the possible functional relationship between BAF complex and Vangl1 expression.
Physical interactors with BAF complex
Numerous studies identified proteins that interact with BAF complexes, and in this review, we focus on a number of the interacting proteins important for neural development. The variety of interactions appears to arise from the different subunit compositions of the complexes such that the chromatin remodeler is tailored to the needs of a specific cell type (Figure 1). Neural restrictive silencing factor (NRSF or REST) is a zinc finger domain transcription factor that binds to its target sites (RE1) and represses its target genes by recruiting its co-repressors (CoREST, MeCP2 and Sin3A) [25–28]. NRSF/REST expression is restricted to non-neuronal cells, and the reduced activity of NRSF/REST in neurons allows the expression of neuronal genes [25–27]. BAF57, BAF170 and Brg1 were shown to form a larger complex with NRSF/REST and its co-repressors [29]. Moreover, the repressor activity of NRSF/REST on its target genes required a functional BAF complex; Brg1 was recruited to RE1 sites of NRSF/REST target genes mediated by the interaction between the bromo domain of Brg1 and NRSF/REST. Inhibiting Brg1 activity resulted in increased expression of NRSF/REST target genes, implying a synergistic relationship between BAF complex and NRSF/REST in controlling the expression of neuronal genes [29]. Similar results were shown in a separate study where abnormal expression of neuronal genes is elevated in lung carcinoma cells when Brg1 was inhibited, and suppressed by exogenous expression of Brg1 [30]. Brg1 also enhanced deacetylation of histone H4 around the binding sites of RE-1, suggesting the mechanistic relationship between BAF complex, NRSF/REST and the chromatin architecture to inhibit NRSF/REST target genes [30]. Although these studies were performed using immortalized and carcinoma cell lines and need further confirmation in animal models, they strongly suggest an important role of BAF complex in controlling neuronal gene expression. Interestingly, depletion of Brg1 in neural progenitors is associated with increased expression of several neuronal genes including Calbindin1 and Down syndrome cell adhesion molecule (Dscam) ([3], unpublished data), also predicted to be targets of NRSF/REST [31]. Dscam was recently shown to be important for neurite morphogenesis [32] and axon guidance [33]. Furthermore, overexpression of human MeCP2 in neurons, the eye and wing veins lead to impaired motor function, glassy external eye and extra vein phenotypes, respectively in Drosophila[34]. These phenotypes were suppressed by the reduced function of Osa (BAF250 homolog in Drosophila), supporting the role of BAF complex for NRSF/REST activity [34].
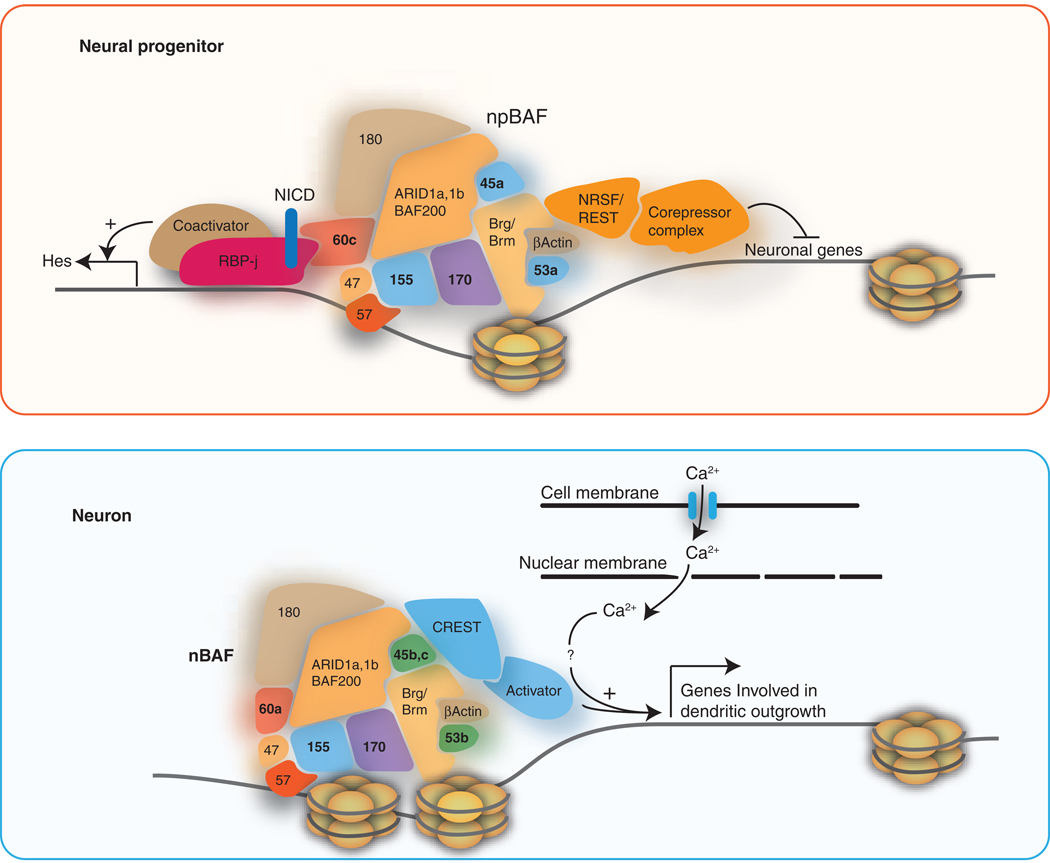
Figure 1.
A model of BAF complex activity in neural progenitors and post-mitotic neurons based on previously identified interactors. Subunits of BAF complexes contain motifs known to bind to histones, including bromo-, chromo- and PHD domains. npBAF likely activates Hes1 and Hes5 downstream of Notch signaling through BAF60c and in turn inhibit differentiation and promote proliferation in progenitors. Through the interaction with NRSF/REST, npBAF would facilitate the inhibition of neuronal genes (shown in red enclosure). It is not currently known whether the subunit switching of BAF complex during differentiation would interfere with the interaction with NRSF/REST. Neuron-specific nBAF complex has a specific function dedicated to the neuronal function. BAF53b was shown to promote activity-dependent dendritic outgrowth of neurons mediated by CREST (shown in blue enclosure). One of the genes regulated by nBAF-CREST complex to promote the dendritic outgrowth is Ephexin1 [4].
Geminin, a key regulator of cell cycle progression, was also found to physically interact with Brg1 in HEK293 cell extracts, and the synergistic genetic interaction between Brg1 and Geminin was suggested by the enhancement of phenotypes caused by Brg1 inhibition with overexpressing Geminin in Drosophila [35]. Brg1 mediates the transactivation of neuronal genes by basic Helix-loop-helix (bHLH) transcription factors Neurogenins and NeuroD [21]. Geminin was shown to interfere with the Brg1-mediated activation of neuronal genes by competing with Brg1 to bind bHLH factors [35]. Morever, loss of Brg1 function or Geminin overexpression interfered with NeuroD-mediated neuronal differentiation of P19 cells [21,35]. Overexpression of Geminin also enhanced Sox2 expression, which was abolished by dominant negative form of Brm in chick embryos, indicating a role of BAF complexes in regulating Sox2 [36]. As Sox2 is essential for the maintenance of neural progenitor cells [37], the depletion of neural progenitor population seen with reduced Brg1 function [3] may be related to the consequent dysregulation of Sox2 expression. In ES cells, Brg1 is part of an ES cell-specific complex, esBAF, which binds to the Sox2 gene and is essential for Sox2 expression [5,6].
Activity dependent neuroprotective protein (ADNP), whose haplodeficiency also leads to defects in neural tube formation in mice [38], was shown to interact with BAF complexes [39]. Using anti-GFP antibody to purify proteins that interact with ADNP-GFP fusion protein in HEK293 cell lines, Brg1, BAF250a and BAF170 were recovered as interactors with ADNP. Similar results were separately shown where BAF complex purification using Brg1 antibody recovered ADNP in P0 mouse brain extract ([3], unpublished data) and in ES cells [6]. Interestingly, inhibiting ADNP activity lead to reduced neurite numbers in P19 cell-derived neurons [40], a phenotype similar to that seen when BAF complex activity is compromised in hippocampal neurons [4].
BAF subunit switching during neural development
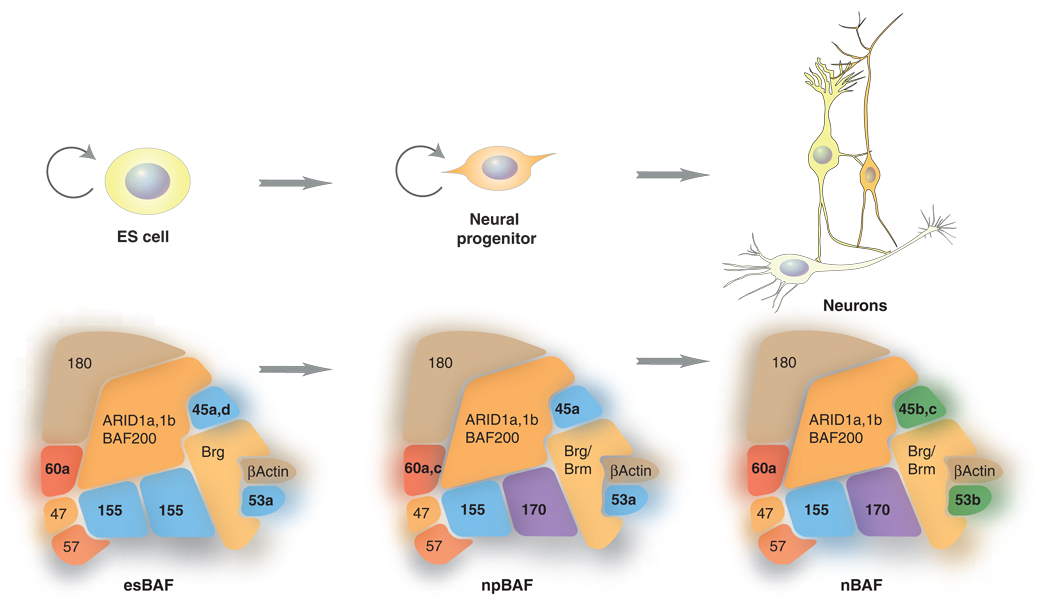
In mammals, the composition of BAF complexes is highly polymorphic due to multiple gene families that encode the subunits. The resulting combinatorial assembly of BAF complexes plays an essential role during neural development. Two subunits, BAF45a and BAF53a are assembled into BAF complexes in neural progenitors (designated as npBAF complex). During neuronal differentiation, the expression of BAF45a and BAF53a diminishes and their places are replaced by homologous members, BAF45b and BAF53b (nBAF) specifically expressed in post-mitotic neurons [3]. The npBAF complex is dedicated to maintain self-renewal capacity of neural progenitors evidenced by the reduced proliferation of neural progenitors with BAF45a or BAF53a inhibition and increased proliferation with BAF45a overexpression [3]. Expressing BAF45b in progenitors did not have an effect on proliferation indicating that the progenitor-specific function of npBAF was governed by the BAF subunits dedicated to neural progenitors [3]. A recent report proposed that BAF60c (which is essential for heart morphogenesis [41]) is also an npBAF subunit [42]. BAF60c expression was enriched in neural progenitors and absent in differentiated neurons in the developing brain and neural tubes of mouse embryos, and overexpression of BAF60c enhanced proliferation of progenitors as with BAF45a and BAF53a [42]. BAF60c enhances Notch signaling as BAF60c stabilizes the interaction between activated Notch (NICD) and its DNA-binding partner, RBP-j [43]. The NICD-RBP-j-dependent transcription for Nodal and Hes1 required functional Baf60c and Brg1 function [43]. Notch signaling inhibits neuronal differentiation and maintains neural progenitors by inducing expression of the basic Helix-loop-helix (bHLH) factors Hes1 and Hes5 that in turn repress expression of proneural genes and Notch ligand genes [44–46]. Furthermore, inhibiting RBP-j results in precocious differentiation of progenitors, thus sustained activation of Notch signaling is critical for keeping the proliferative state of progenitors [47]. Microarray analysis of genes affected by depletion of Brg1 in neural progenitors identified components of Notch signaling pathway to be reduced [3], further suggesting the involvement of npBAF in maintaining neural progenitors by enhancing Notch signaling pathway. The expression of Rhomboid, a target of Notch signaling in Drosophila gut tissue [48], was decreased with reduced function of Brg1 in progenitors [3]. Meanwhile, Jagged 1, one of the ligands for Notch important for self-renewal of neural stem cells [49], was increased with reduced function of npBAF. Jagged 1 is expressed in the subventricular zone of postnatal brain lining the ventricle juxtaposed to Notch-expressing neural stem cells and colocalizes with the non-neuronal GFAP-positive cells [49], suggesting that npBAF functions to minimize the expression of Jagged 1 in neural progenitors. Interestingly, mutagenesis screening for modifying phenotypes caused by overexpressing dominant-negative form of brahma maps to mutations of genes in Notch signaling pathway in Drosophila [50].
As npBAF function is specialized in promoting self-renewal of neural stem cells, nBAF appears to be dedicated to neuron-specific functions. Null mutation of BAF53b lead to defect in activity-dependent dendritic outgrowth of neurons [4]. BAF53a overexpression did not rescue this lethal phenotype indicating that combinatorial assembly of these complexes produces specific biological functions. During the purification of endogenous nBAF complexes peptides were found from CREST, which promotes activity-dependent dendritic formation: i) CREST and Brg1 both control dendritic formation upon stimulation of neurons, ii) CREST and nBAF complex physically interact, and iii) they synergistically target ephexin1 that promotes dendritic formation. Ephexin1 expression is reduced by either reducing CREST or BAF53b activity [4]. A recent study also found Brg1 to interact with CREST, and to repress the activation of c-fos by Brg1-dependent recruitment of HDAC and Rb [51]. Upon stimulation and Ca2+ influx, calcineurin-dependent dephosphorylation of Rb releases HDAC, allowing acetylation of H4 histone subunit and the subsequent activation of target genes by CREST/CREB-mediated recruitment of CBP, a transcriptional coactivator [51]. However, the observation that loss-of-function of BAF53b leads to decreased expression of Ephexin1 [4], another target of CREST, implies that the simplistic view of Brg1’s function as a general repressor of activity-dependent program is likely to be more complicated. In addition, c-fos transcription was not found to be changed in Brg- or BAF53b-mutant mice [3,4]. Future directions should be geared towards elucidating how nBAF complexes can be both activators and repressors. Interestingly, in C. elegans, loss-of-function mutations in lin-35 (a homolog of Rb) genetically synergize with psa-1 (BAF155) to augment the defect in asymmetric division of T cells [52], suggesting the conservation of the functional relationship between BAF complexes and Rb.
Pluirpotent ES cells contain another specialized form of BAF complexes termed esBAF, distinguished by the presence of a BAF155 homodimer (but not BAF170) and Brg1 (but not Brm). esBAF complexes are an essential component of the core pluripotency transcriptional circuit and appear to be tailored to interact with ES cell specific transcription factors such as Oct4, Dppa2, Dppa4 and Sox2 thereby providing robustness and stability to an undifferentiated state [5,6]. Thus, the three separate transitions from pluripotent stem cell, to neural stem cell to post-mitotic neuron are accompanied by changes in subunit composition (figure 2). While more studies are needed, the changes in subunit composition appear to be critical to each transition. One possible meaning of these subunit exchanges could be that the complexes must be tailored to interact with the ambient set of transcription factors at each stage of neural determination and later neural function. Additional genetic and biochemical studies will be essential to test this speculation.
Figure 2.
Schematic diagrams depicting subunit compositions specific for ES cells (esBAF), neural progenitors (npBAF) and post-mitotic neurons (nBAF). The arrows indicate the self-renewal capacity of ES cells and neural progenitors.
Conclusion
ATP-dependent chromatin remodeling has generally been considered to play a permissive role in development. However, recent evidence is suggesting a far more complex and programmatic function in the development of the nervous system of both invertebrates and vertebrates. The combinatorial assembly of the complexes makes genetic analysis complicated, yet current evidence indicates that combinatorial assembly underlies refinement and specificity of their functions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest have been highlighted as:
* of special interest
** of outstanding interest
- 1. Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. This review introduces the idea that chromatin regulatory complexes assume biological specificity in the way that letters assume meaning in words: by combinatorial assembly.
- 2.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 3. Lessard J, Wu J, Ranish J, Wan M, Winslow M, Staahl B, Wu H, Aebersold R, Graef I, Crabtree G. An Essential Switch in Subunit Composition of a Chromatin Remodeling Complex during Neural Development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. This study offers the first demonstration of switching of subunit composition of BAF complexes during neural development, and focuses on the role of neural progenitor-specific BAF complexes.
- 4. Wu J, Lessard J, Olave I, Qiu Z, Ghosh A, Graef I, Crabtree G. Regulation of Dendritic Development by Neuron-Specific Chromatin Remodeling Complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. Using null mutation of BAF53b, this paper reveals a specific role of BAF53b in the dendritic outgrowth of neurons and demonstrates that the neuron-specific role of BAF53b is not interchangable with BAF53a.
- 5. Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An Embryonic Stem Cell Chromatin Remodeling Complex, esBAF is an Essential Component of the Core Pluripotency Transcriptional Network. PNAS. doi: 10.1073/pnas.0812888106. in press. Using Chip-seq technique and microarray data, this study identified the genomic occupancy of Brg1 in ES cells and revealed the essential function of BAF complex in the transcriptional network that keeps the pluripotency of ES cells.
- 6. Ho L, Ronan JL, Wu J, Staahl B, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An Embryonic Stem Cell Chromatin Remodeling Complex, esBAF is Essential for Embryonic Stem Cell Self-Renewal and Pluripotency. PNAS. doi: 10.1073/pnas.0812889106. in press. By purifying BAF complex using an antibody against Brg1 in ES cells and thorough biochemical analysis, this work identified an ES cell-specific assembly of BAF complex and the essential role of the esBAF complex in the pluripotency and self-renewal of ES cells.
- 7. Parrish JZ, Kim MD, Jan LY, Jan YN. Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes & Development. 2006;20:820–835. doi: 10.1101/gad.1391006. This RNAi-based approach designed with careful analysis of dendritic phenotypes identified transcription factors associated with morphogenesis of sensory neuron dendrites in Drosophila. Notably, this screening identified most of the components of BAF complex to be important for the dendritic formation of neurons.
- 8.Ivanov AI, Rovescalli AC, Pozzi P, Yoo S, Mozer B, Li HP, Yu SH, Higashida H, Guo V, Spencer M, et al. Genes required for Drosophila nervous system development identified by RNA interference. Proc Natl Acad Sci U S A. 2004;101:16216–16221. doi: 10.1073/pnas.0407188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 10. Schmitz C, Kinge P, Hutter H. Axon guidance genes identified in a large-scale RNAi screen using the RNAi-hypersensitive Caenorhabditis elegans strain nre-1(hd20) lin-15b(hd126) Proc Natl Acad Sci U S A. 2007;104:834–839. doi: 10.1073/pnas.0510527104. Using an RNAi-hypersensitive strain of C. elegans, this study identified a BAF60 homolog to be important for proper axon guidance. This work provides the first evidence supporting the role of BAF complex in axon guidance in C. elegans.
- 11.Sawa H, Kouike H, Okano H. Components of the SWI/SNF complex are required for asymmetric cell division in C. elegans. Molecular Cell. 2000;6:617–624. doi: 10.1016/s1097-2765(00)00060-5. [DOI] [PubMed] [Google Scholar]
- 12.Gangemi RM, Daga A, Marubbi D, Rosatto N, Capra MC, Corte G. Emx2 in adult neural precursor cells. Mech Dev. 2001;109:323–329. doi: 10.1016/s0925-4773(01)00546-9. [DOI] [PubMed] [Google Scholar]
- 13.Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- 14.Olave I, Reck-Peterson S, Crabtree G. NUCLEAR ACTIN AND ACTIN-RELATED PROTEINS IN CHROMATIN REMODELING. Annu. Rev. Biochem. 2002;71:755–781. doi: 10.1146/annurev.biochem.71.110601.135507. [DOI] [PubMed] [Google Scholar]
- 15.Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 17.Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 19.Kim JK, Huh SO, Choi H, Lee KS, Shin D, Lee C, Nam JS, Kim H, Chung H, Lee HW, et al. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol Cell Biol. 2001;21:7787–7795. doi: 10.1128/MCB.21.22.7787-7795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sohn DH, Lee KY, Lee C, Oh J, Chung H, Jeon SH, Seong RH. SRG3 interacts directly with the major components of the SWI/SNF chromatin remodeling complex and protects them from proteasomal degradation. J Biol Chem. 2007;282:10614–10624. doi: 10.1074/jbc.M610563200. [DOI] [PubMed] [Google Scholar]
- 21.Seo S, Richardson GA, Kroll KL. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development. 2005;132:105–115. doi: 10.1242/dev.01548. [DOI] [PubMed] [Google Scholar]
- 22.Matsumoto S, Banine F, Struve J, Xing R, Adams C, Liu Y, Metzger D, Chambon P, Rao M, Sherman L. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Developmental Biology. 2006;289:372–383. doi: 10.1016/j.ydbio.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 23.Kibar Z, Torban E, McDearmid JR, Reynolds A, Berghout J, Mathieu M, Kirillova I, De Marco P, Merello E, Hayes JM, et al. Mutations in VANGL1 associated with neural-tube defects. N Engl J Med. 2007;356:1432–1437. doi: 10.1056/NEJMoa060651. [DOI] [PubMed] [Google Scholar]
- 24.Torban E, Patenaude AM, Leclerc S, Rakowiecki S, Gauthier S, Andelfinger G, Epstein DJ, Gros P. Genetic interaction between members of the Vangl family causes neural tube defects in mice. Proc Natl Acad Sci U S A. 2008;105:3449–3454. doi: 10.1073/pnas.0712126105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 26.Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- 27.Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 28.Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, et al. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- 29.Ooi L, Belyaev ND, Miyake K, Wood IC, Buckley NJ. BRG1 chromatin remodeling activity is required for efficient chromatin binding by repressor element 1-silencing transcription factor (REST) and facilitates REST-mediated repression. J Biol Chem. 2006;281:38974–38980. doi: 10.1074/jbc.M605370200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe H, Mizutani T, Haraguchi T, Yamamichi N, Minoguchi S, Yamamichi-Nishina M, Mori N, Kameda T, Sugiyama T, Iba H. SWI/SNF complex is essential for NRSF-mediated suppression of neuronal genes in human nonsmall cell lung carcinoma cell lines. Oncogene. 2006;25:470–479. doi: 10.1038/sj.onc.1209068. [DOI] [PubMed] [Google Scholar]
- 31.Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Gottgens B, Buckley NJ. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci U S A. 2004;101:10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuerst PG, Koizumi A, Masland RH, Burgess RW. Neurite arborization and mosaic spacing in the mouse retina require DSCAM. Nature. 2008;451:470–474. doi: 10.1038/nature06514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ly A, Nikolaev A, Suresh G, Zheng Y, Tessier-Lavigne M, Stein E. DSCAM is a netrin receptor that collaborates with DCC in mediating turning responses to netrin-1. Cell. 2008;133:1241–1254. doi: 10.1016/j.cell.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cukier HN, Perez AM, Collins AL, Zhou Z, Zoghbi HY, Botas J. Genetic modifiers of MeCP2 function in Drosophila. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000179. e1000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seo S, Herr A, Lim JW, Richardson GA, Richardson H, Kroll KL. Geminin regulates neuronal differentiation by antagonizing Brg1 activity. Genes & Development. 2005;19:1723–1734. doi: 10.1101/gad.1319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papanayotou C, Mey A, Birot AM, Saka Y, Boast S, Smith JC, Samarut J, Stern CD. A mechanism regulating the onset of Sox2 expression in the embryonic neural plate. Plos Biol. 2008;6:e2. doi: 10.1371/journal.pbio.0060002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 38.Pinhasov A, Mandel S, Torchinsky A, Giladi E, Pittel Z, Goldsweig AM, Servoss SJ, Brenneman DE, Gozes I. Activity-dependent neuroprotective protein: a novel gene essential for brain formation. Brain Res Dev Brain Res. 2003;144:83–90. doi: 10.1016/s0165-3806(03)00162-7. [DOI] [PubMed] [Google Scholar]
- 39.Mandel S, Gozes I. Activity-dependent neuroprotective protein constitutes anovel element in the SWI/SNF chromatin remodeling complex. J Biol Chem. 2007;282:34448–34456. doi: 10.1074/jbc.M704756200. [DOI] [PubMed] [Google Scholar]
- 40.Mandel S, Spivak-Pohis I, Gozes I. ADNP differential nucleus/cytoplasm localization in neurons suggests multiple roles in neuronal differentiation and maintenance. J Mol Neurosci. 2008;35:127–141. doi: 10.1007/s12031-007-9013-y. [DOI] [PubMed] [Google Scholar]
- 41.Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 42. Lamba DA, Hayes S, Karl MO, Reh T. Baf60c is a component of the neural progenitor-specific BAF complex in developing retina. Dev Dyn. 2008;237:3016–3023. doi: 10.1002/dvdy.21697. According to the expression analysis in the developing neural tube and retina of mouse embryos, BAF60c appears to be a member of npBAF complex. This study also identified a role of BAF60c in promoting the proliferation of neural progenitors.
- 43.Takeuchi JK, Lickert H, Bisgrove BW, Sun X, Yamamoto M, Chawengsaksophak K, Hamada H, Yost HJ, Rossant J, Bruneau BG. Baf60c is a nuclear Notch signaling component required for the establishment of left-right asymmetry. Proc Natl Acad Sci USA. 2007;104:846–851. doi: 10.1073/pnas.0608118104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 45.Gaiano N, Fishell G. The role of notch in promoting glial and neural stem cell fates. Annu Rev Neurosci. 2002;25:471–490. doi: 10.1146/annurev.neuro.25.030702.130823. [DOI] [PubMed] [Google Scholar]
- 46.Greenwald I. LIN-12/Notch signaling: lessons from worms and flies. Genes Dev. 1998;12:1751–1762. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- 47.Zhu X, Zhang J, Tollkuhn J, Ohsawa R, Bresnick EH, Guillemot F, Kageyama R, Rosenfeld MG. Sustained Notch signaling in progenitors is required for sequential emergence of distinct cell lineages during organogenesis. Genes Dev. 2006;20:2739–2753. doi: 10.1101/gad.1444706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fuss B, Hoch M. Notch signaling controls cell fate specification along the dorsoventral axis of the Drosophila gut. Curr Biol. 2002;12:171–179. doi: 10.1016/s0960-9822(02)00653-x. [DOI] [PubMed] [Google Scholar]
- 49.Nyfeler Y, Kirch RD, Mantei N, Leone DP, Radtke F, Suter U, Taylor V. Jagged1 signals in the postnatal subventricular zone are required for neural stem cell self-renewal. EMBO J. 2005;24:3504–3515. doi: 10.1038/sj.emboj.7600816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Armstrong JA, Sperling AS, Deuring R, Manning L, Moseley SL, Papoulas O, Piatek CI, Doe CQ, Tamkun JW. Genetic screens for enhancers of brahma reveal functional interactions between the BRM chromatin-remodeling complex and the delta-notch signal transduction pathway in Drosophila. Genetics. 2005;170:1761–1774. doi: 10.1534/genetics.105.041327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Qiu Z, Ghosh A. A calcium-dependent switch in a CREST-BRG1 complex regulates activity-dependent gene expression. Neuron. 2008;60:775–787. doi: 10.1016/j.neuron.2008.09.040. This paper provides detailed analyses of mechanistic roles of CREST and Brg1 in activity-dependent regulation of c-fos and NR2B.
- 52.Cui M, Fay DS, Han M. lin-35/Rb cooperates with the SWI/SNF complex to control Caenorhabditis elegans larval development. Genetics. 2004;167:1177–1185. doi: 10.1534/genetics.103.024554. [DOI] [PMC free article] [PubMed] [Google Scholar]