Abstract
Endosomal sorting complexes required for transport (ESCRT-0, I, II, III) are selectively recruited to cellular membranes to exert their function in diverse processes such as multivesicular body biogenesis, enveloped virus budding and cytokinesis. ESCRT-III is composed of members of the CHMP protein family- cytosolic proteins which are targeted to membranes via yet unknown signals. Membrane targeting is thought to result in a membrane-associated protein network that presumably acts at a late budding step. Here we provide structural evidence based on small angle X-ray scattering data that ESCRT-III CHMP3 can adopt two conformations in solution; a closed globular form that most likely represents the cytosolic conformation and an open extended conformation that might represent the activated form of CHMP3. Both the closed and the open conformation of CHMP3 interact with AMSH with high affinity. Although the C-terminal region of CHMP3 is required for AMSH interaction a peptide thereof reveals only weak binding to AMSH suggesting that other regions of CHMP3 contribute to the high affinity interaction. Thus AMSH including its MIT domain interacts differently with ESCRT-III CHMP3 than reported for the Vps4 MIT domain CHMP protein interactions.
Keywords: CHMP3, ESCRT-III, AMSH, Budding, Autoinhibition
Introduction
Multivesicular biogenesis, cell division cytokinesis and enveloped virus budding are seemingly unrelated processes, which involve a topologically identical membrane-remodeling event namely budding away from the cytosol. These normal cellular and pathological membrane remodeling processes are orchestrated by either the entire or a sub-set of the class E vacuolar protein sorting machinery that was first implicated in receptor down regulation via multivesicular body (MVB) formation 1; 2; 3; 4; 5. Receptor down regulation and MVB biogenesis is largely catalyzed by ESCRT-0, I, II and III (Endosomal Sorting Complexes Required for Transport) complexes which are sequentially recruited to the endosomal membrane and exert their action in protein sorting and vesicle budding 5; 6 with the assistance of a variety of cytosolic accessory proteins and protein complexes. ESCRT complexes 0, I and -II contain ubiquitin binding domains that recognize ubiquitinated cargo and have been thus implicated in receptor sorting into vesicles that bud off into the endosomal lumen 3. Although ESCRT-III is uncoupled from direct ubiquitin recognition, ESCRT-III family members recruit deubiquitinating enzymes AMSH 7; 8; 9; 10; 11and UBPY 12. The deubiquitin isopeptidase activity of both UBPY (Doa4) as well as AMSH, precede cargo incorporation into vesicles, thereby ensuring a constant pool of free ubiquitin and early disengagement of cargo from the MVB pathway 13. Although AMSH depletion does not impair EGFR degradation 14; 15, abrogation of its recruitment to the endosome by ESCRT-III CHMP3 impairs EGFR degradation 11. Furthermore, over-expression of a catalytically inactive form of AMSH exerts a dominant negative effect on HIV-1 budding 9.
ESCRT-III is composed 6 and 11 members in yeast 16 and mammals respectively, which have been named CHMP (CHarged MVB Protein) proteins 17; 18. CHMP proteins form binary subcomplexes 16; 17; 18 and have been suggested to polymerize into a protein lattice on membranes, which acts at a late budding step 17; 19; 20; 21. CHMP proteins are targets for the AAA type ATPase Vps4 whose MIT domain interacts with a C-terminal peptide region of CHMP1 and 2 22 and yeast Vps2 (CHMP2) and Vps24 (CHMP3)23. Recruitment of Vps4 in turn is required for efficient disassembly and recycling of ESCRT complexes from membranes 16; 24; 25; catalytic inactive Vps4 thus acts as a dominant negative inhibitor of retroviral budding 17; 19. CHMP family members recruit accessory ESCRT factors such as Alix via CHMP4 17; 19; 26, AMSH via CHMP3 9; 10 and UBPY via CHMP7 12. Furthermore they communicate with ESCRT-I via a Vps28-CHMP6/Vps20 interaction 27; 28 and ESCRT-II via a Vps25-Vps20/CHMP6 interaction 27; 29.
A hallmark of the ESCRT-III family members is a biased distribution of charged amino acids, with two thirds of the N-terminus carrying a basic charge and the remaining C-terminal region being acidic. The crystal structure of CHMP3 revealed a flat helical arrangement with two distinct and mutually exclusive CHMP-CHMP interaction surfaces and a large basic surface required for membrane targeting; both membrane targeting and CHMP-CHMP interactions were shown to be required for HIV-1 budding 21. The crystal structure lacks part of the C-terminal acidic region (residues 184 to 222) including the Vps4 and AMSH MIT domain interaction sites 9; 23; the structure was suggested to represent the activated conformation of CHMP3, since removal of the acidic C-terminus leads to membrane targeting in vivo while full length CHMP3 is found predominantly in the cytosol when expressed in the absence of any additional signals 21. The C-terminal region has been also suggested to exert a control function keeping CHMP proteins in a cytosolic state which is consistent with the activation hypothesis 9; 30. A potential release of the autoinhibition in CHMP3 was indirectly attributed to its interaction with AMSH, since it turned full length CHMP3 into a potent HIV-1 budding inhibitor 9.
Here we present structural evidence for a closed and open conformation of full length CHMP3, which confirms the activated state represented by the CHMP3 crystal structure. Both conformations do not reveal any changes in their secondary structure content or in their overall thermal stability, indicating that a first step in activation entails detachment of the C-terminal module from the N-terminal core. Both conformations reveal high affinity binding to AMSH in vitro as measured by isothermal titration calorimetry. This suggests that the AMSH MIT domain interaction with CHMP3 differs substantially from the low affinity Vps4 MIT domain interaction 22; 23.
Results
Characterization of CHMP3 conformations
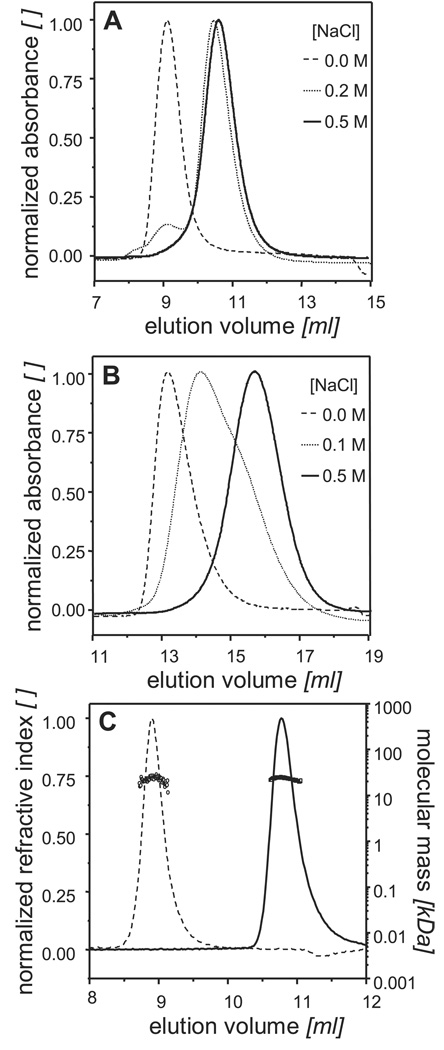
While performing size exclusion chromatography (SEC) on a Superdex™ 75 column with 200HBS as the running buffer, purified CHMP3 showed two distinct peaks at elution volumes 9.1 ml and 10.8 ml (Figure 1A). It had been previously hypothesized that the C-terminal acidic tail of CHMP proteins may interact with its N-terminal basic region via electrostatic interactions. We, therefore, examined the effect of ionic strength on the polydispersity of CHMP3 by SEC. CHMP3 eluted as a single symmetric peak in 0HBS and 500HBS at 9.1 and 10.8 ml respectively (Figure 1A), revealing a distinct effect of the ionic strength on the elution profile. This salt dependent change in elution profile was further checked on a Superdex™ 200 column with three different NaCl concentrations namely 0, 100 and 500 mM. While in 100HBS CHMP3 eluted as a broad peak, with 0HBS and 500HBS as the running buffers a single symmetric peak at 13.1 ml and 15.7 ml respectively, was observed (Figure 1B). Furthermore separation of the high-salt form in 0HBS produced a single peak at 13.1 ml and separation of the low-salt form in 500HBS revealed a peak at 15.7 ml (data not shown). Collectively these findings suggested that the two forms of CHMP3 were in a dynamic equilibrium with one another and, depending upon the ionic strength of the medium, one form could be exclusively converted into the other in a reversible fashion thus providing preparative means for isolating a pure CHMP3 form at a time.
Figure 1.
Enrichment of CHMP3 conformers: Analytical SEC recorded by measuring, absorbance at 280 nm (A) on a Superdex™ 75 10/300 GL column with the running buffers 0HBS ( ), 200HBS (
), 200HBS ( ) and, 500HBS (----); (B) on a Superdex™ 200 10/300 GL column with the running buffers 0HBS (
) and, 500HBS (----); (B) on a Superdex™ 200 10/300 GL column with the running buffers 0HBS ( ), 100HBS (
), 100HBS ( ) and 500HBS (----). A small change in NaCl concentration from 100HBS to 200HBS produces a large change in the elution profiles. (C) SEC in combination with RI and MALLS performed on a Shodex Protein KW-804 column reveals that CHMP3 in 0HBS(----) and 500HBS (
) and 500HBS (----). A small change in NaCl concentration from 100HBS to 200HBS produces a large change in the elution profiles. (C) SEC in combination with RI and MALLS performed on a Shodex Protein KW-804 column reveals that CHMP3 in 0HBS(----) and 500HBS ( ) elutes with the observed Mw of 23.3 ± 2 kDa (○) and 24.1 ± 0.2 kDa (□) respectively, which is in excellent agreement with the calculated molecular mass of 24.5 kDa for monomeric CHMP3.
) elutes with the observed Mw of 23.3 ± 2 kDa (○) and 24.1 ± 0.2 kDa (□) respectively, which is in excellent agreement with the calculated molecular mass of 24.5 kDa for monomeric CHMP3.
In order to determine the oligomeric state of these two CHMP3 forms analytical SEC in combination of multi-angle laser light scattering (MALLS) was performed (Figure 1C). As expected, depending upon the ionic strength of the running buffer CHMP3 eluted as a single symmetric peak at two different elution volumes. With 500HBS as the running buffer, CHMP3 eluted at 10.8 ml with the observed average molecular mass (Mw) of 24.1 ± 0.2 kDa which is in excellent agreement with the calculated molecular mass of 24.5 kDa for the monomeric CHMP3. In 0HBS, CHMP3 eluted at 8.9 ml, with a Mw of 23.3 ± 2 kDa implying that the change in elution-volume/hydrodynamic-radius did not result from CHMP3 oligomerization but from a conformational change instead.
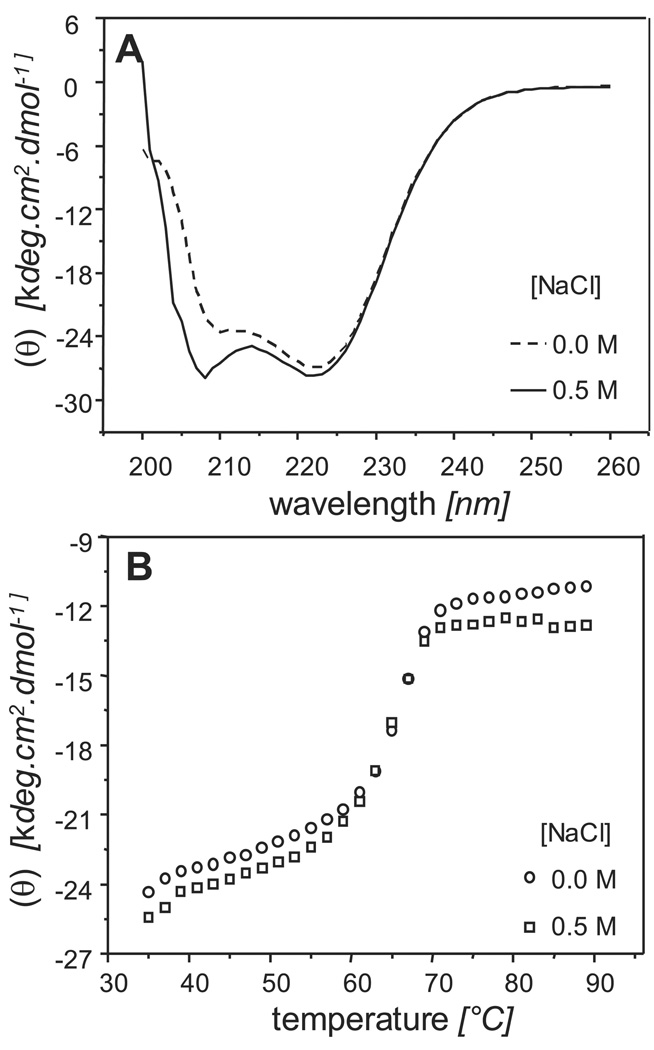
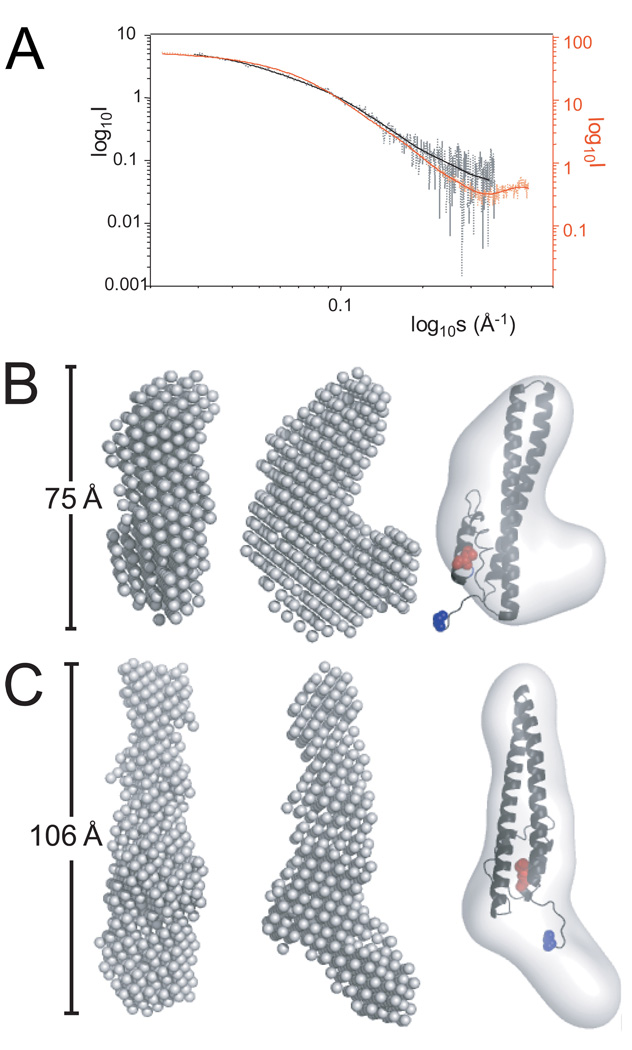
Circular dichroism (CD) analysis on both CHMP3 conformers showed no substantial difference in the secondary structure content as the observed spectra in 0HBS and 500HBS (Figure 2A) were very similar with a small difference at around 208 nm which most likely resulted from a difference in NaCl concentration. Two minima observed at 208 nm and 222 nm are consistent with the α-helical fold of CHMP3 21. Thermal unfolding showed thermal transitions at ~ 65°C for both 0HBS as well as 500HBS buffer conditions (Figure 2B), indicating that high salt had no effect on the stability of the helical structure content. Structural models of CHMP3 conformations based on small angel X-ray scattering analyses. Both CHMP3 conformers were analyzed by small angle X-ray scattering, which produced interpretable scattering intensity patterns (Figure 3A). Maximal protein dimensions (Dmax) of approximately 8.5 Ǻ and 11 Ǻ were found in 0HBS and 500HBS respectively by the distance distribution function p(r) computed by a Fourier transformation of the scattering intensity. The shapes of the CHMP3 conformers were determined ab initio from the scattering data using the program DAMMIN 31. 10 independent models of CHMP3 in 0HBS and 500HBS fitted the corresponding experimental data with the discrepancy χ of 1.66 to 1.98 and 1.58 to 1.62, respectively (Figure 3A). The ab initio modeling of CHMP3 in 0HBS showed a compact ~ 75Å long elongated structure (Figure 3B). The overall shape is consistent with the crystal structure of a C-terminally truncated form of CHMP3 and well accommodates the 70 Å long helical hairpin of the four helical bundle core structure 21 as can be seen from the manual fit of the structure into the molecular envelope of the SAXS model (Figure 3B). The volume of the model (~35 %) not accounted for by the crystal structure (136 residues out of 222) appears to wrap around the central body of the structure (Figure 3B). In contrast, in the model of high salt CHMP3 this part extends from the main body and generates an elongated ~ 106 Å long structure (Figure 3C). The core CHMP3 structure composed of the four helical bundle can be fitted manually into the elongated molecular envelope (Figure 3C). Thus the data indicates that the CHMP3 model obtained under no salt conditions represents the “closed” conformation of CHMP3 where the C-terminal acidic tail interacts with the larger N-terminal basic core; the model obtained under high salt conditions represents the “open” conformation where the C-terminal acidic tail is displaced and extends from the N-terminal basic core.
Figure 2.
Circular dichroism shows that both the “low-“ and the “high-salt” conformations of CHMP3 have a: (A) similar α-helical content as shown by the CD spectra in 0HBS (---) and 500HBS ( ); (B) same thermal unfolding transition at ~ 65° as was measured by recording ellipticity at 222 nm in 0HBS (○) and 500HBS (□) as a function of temperature.
); (B) same thermal unfolding transition at ~ 65° as was measured by recording ellipticity at 222 nm in 0HBS (○) and 500HBS (□) as a function of temperature.
Figure 3.
Small angle X-ray scattering (SAXS) analysis of CHMP3. (A) SAXS profiles of CHMP3 in 0HBS (dotted black line) and 500HBS (dotted red line). Log of scattered intensity is shown vs. log of s (s = 4πsin(θ)/λ, 2θ = scattering angle and λ = wavelength), log10s scale is chosen for aiding the visualization of the differences between the scattering curves at high as well as low s values. SAXS profiles calculated from the the ab initio models are shown as solid black and red lines. (B) The ab initio calculated model of CHMP3 in 0HBS buffer conditions reveals an elongated L-shaped molecule. Two orientations rotated by 90 °are shown (left two panels). The core of the CHMP3 crystal structure (grey ribbon) composed of an elongated four helical bundle was fitted manually into the molecular envelope calculated from the SAXS model (right panel). (C) The calculated model of CHMP3 in 0HBS buffer conditions reveals an elongated 106 Å long molecule. Two orientations rotated by 90° are shown (left two panels). The core of the CHMP3 crystal structure (grey ribbon) was fitted into the elongated structure (right panel). The N-terminal residue is indicated in blue and the C-terminla residue in red.
AMSH-CHMP3 interaction
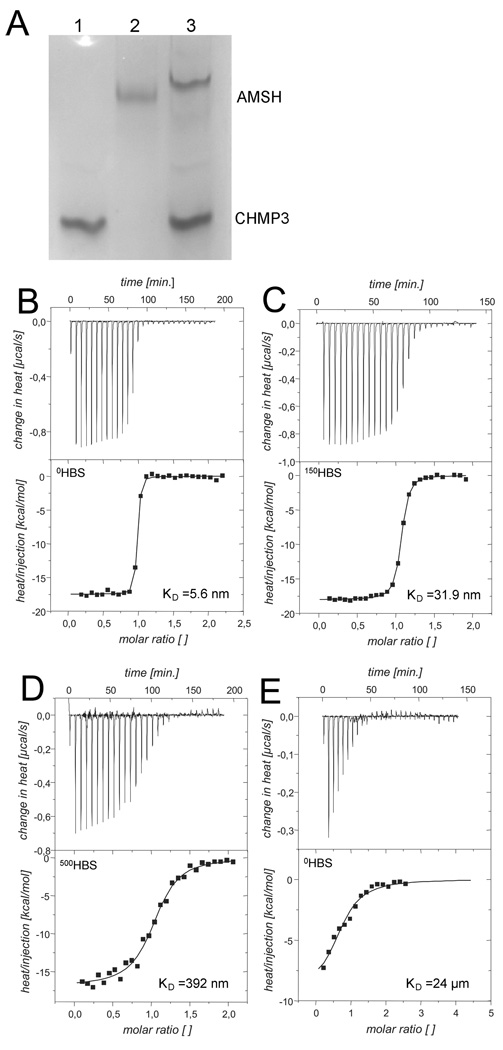
Having established that the two forms of CHMP3 were structurally different from each other, we analyzed their interaction with the deubiquitin isopeptidase AMSH which requires the C-terminal region of CHMP3 for interaction 9; 10. Consistent with the proposed interaction we detect binding of AMSH(1–206) to CHMP3(9–222) by gel filtration chromatography (data not shown) and native gel electrophoresis; both CHMP3(9–222) and AMSH(1–206) migrate as distinct bands on a native gel (Figure 4A, lanes 1 and 2 respectively); a complex formed by the two proteins results in a new band migrating slower than AMSH(1–206) alone (Figure 4A, lane 3). In order to better understand the interaction and the functional properties of the salt induced conformational change in CHMP3 we assessed AMSH binding affinity to both the closed and the open conformation of CHMP3 by using isothermal titration calorimetry (ITC). Table 1 summarizes the enthalpy ΔH°, interaction stoichiometry n, and equilibrium dissociation constant KD, as obtained by fitting to the binding isotherm a 1:n interaction model, which assumes a single type of binding sites. We measured a dissociation constant (KD) of 5.6 nM for the “closed” conformer in 0HBS (Figure 4B), 31.9 nM at physiological 150HBS buffer conditions (Figure 4C) and a KD of 392 nM for the open form in 500HBS (Figure 4D), thus a ~70 fold difference in affinity. No substantial difference in enthalpy was observed as we determined a ΔH° of −16.9 kcal/mol and −17.5 kcal/mol in 500HBS and 0HBS respectively. Since it has been proposed that the C-terminal tail of CHMP3 is necessary for the interaction with AMSH, we titrated the C-terminal CHMP3 peptide (residues 196–222) with AMSH in 0HBS and 500HBS, while no interpretable isotherm was produced with 500 mM NaCl, a KD of 24 µM (Figure 4E) was determined with 0 mM NaCl which is ~4500 times larger as compared to the CHMP3(9–222) interaction; this result thus establishes that AMSH must form additional contacts with CHMP3. Since the C-terminal region carries a substantial negative charge, the NaCl concentration dependent difference in KD suggests that the electrostatic interactions may play a role in CHMP3 AMSH interaction. Very similar ΔH° of −17.5, −18.0 and −16.9 kcal/mol were observed in 0HBS, 150HBS and 500HBS respectively thus demonstrating that the interaction enthalpy was independent of the ionic strength. Trials to determine whether CHMP3 binds only to the AMSH MIT domain could not be performed since we were unable to obtain a soluble monodisperse form of the AMSH MIT domain.
Figure 4.
AMSH(1–206) intercation with CHMP3. (A) Native gel electrophoresis of CHMP3(9–222), lane 1; AMSH(1–206), lane 2; AMSH(1–206) in complex with CHMP3(9–222), lane 3;. Isothermal titration calorimetry (ITC) binding curves for the AMSH (1–206) – CHMP3 (9–222) interaction in (B) 0HBS; (C) 150HBS; (D) 500HBS; and (E) AMSH(1–206) titration curve with the CHMP3 peptide (residues 196 to 222) in 0HBS.
Table 1.
Isothermal titration calorimetry of the CHMP3-AMSH interaction
| CHMP3 (9–222) | CHMP3(196–222) | |||
|---|---|---|---|---|
| 0HBS | 150HBS | 500HBS | 0HBS | |
| n | 0.950 | 1.04 | 1.04 | 0.706 |
| KD [nM] | 5.6 | 31.9 | 392 | 23690 |
| ΔH0 [kcal/mol] | −17.5 | −18.0 | −16.9 | −12.5 |
| [AMSH] (cell) [µM] | 15 | 16 | 15 | 7 |
| [CHMP3] (syringe) [µM] | 160 | 150 | 150 | 150 |
Discussion
ESCRT-III proteins are predominantly cytosolic proteins that are specifically targeted to cellular membranes upon yet unknown signals. Removal of the C-terminal residues 151 to 222 of CHMP3 induced efficient membrane targeting in vivo indicating that the presence of the C-terminus prevents membrane interaction 21. Furthermore, CHMP3 residues 151 to 222 were used to pull down the core structure of CHM3 composed of residues 1 to 150 9, demonstrating an interaction between the N-terminal core and the C-terminal region. The models of CHMP3 in its closed and open conformation based on the SAXS analysis reveal an extension of the C-terminal region in the open form that wraps around the N-terminal core in the closed form. Both structures are in good agreement with the four helical bundle core of CHMP3. The SAXS models confirm further the previous suggestion that the crystal structure represents the “activated” form of CHMP3, since the C-terminal region containing residues 140 to 183 is detached from the core four helical bundle structure and involved in intermolecular interactions that are important for CHMP protein polymer formation in vivo 21. The fact that ionic strength can change the conformation of CHMP3 reveals that monomeric CHMP3 is in a metastable conformation. The removal and reattachment of the C-terminal region towards the core is reversible and has no apparent influence on the secondary structure content and overall thermostability of CHMP3. CHMP3 contains a low activation barrier since it exists already as a mixture of both closed and open forms at physiological salt conditions in vitro. Including the complete N-terminus of wild type CHMP3 (conserved residues 1– 8) did not change the presence of two peaks observed by SEC under 0HBS and 500HBS buffer conditions and produced similar peaks as those obtained for CHMP3(9–222) SEC analysis; thus the conserved N-terminus of CHMP proteins does not seem to contribute to the stability of the closed conformation of CHMP3 in vitro.
The open CHMP3 conformation under high ionic strength conditions does not polymerize in vitro most likely due to the presence of high salt, since polar interactions have been previously implicated in CHMP polymer formation 21. It is yet unclear which signals will actually target CHMP proteins to cellular membranes. One possibility is that their interaction partners confer membrane recruitment or vice versa; CHMP4 via the interaction with Alix or Bro1 17; 19; 26, CHMP3 via AMSH 9; 10, CHMP7 via UBPY 12, Vps20/CHMP6 via ESCRT-I Vps28. 27; 28 and/or ESCRT-II 27; 29. Any of these interactions may lead to activation of individual CHMP proteins. Furthermore, heteromeric CHMP-CHMP protein interactions contribute to membrane targeting since mutations in CHMP3 dimer interface which abrogate dimerization in vitro also abolish membrane targeting of CHMP3 in vivo 21.
The structure of the human Vps4b MIT domain in complex with a peptide derived from CHMP1b revealed three conserved leucine residues (Leu187, Leu190 and Leu194; corresponding to CHMP3 Met213, Leu217, Leu220) binding to a hydrophobic pocket on the MIT domain; the interaction is further stabilized by salt bridges (Glu184, Arg190, Arg 195; corresponding to CHMP3 Arg216, Arg221)23. The interaction motif is the same as the one described for the yeast MIT-Vps2 complex, namely Asp/Glu)xxLeuxxArgLeuxxLeu(lys/Arg) 22 which is not exactly conserved within the C-terminus of CHMP3 (210-LeuxxMetxxArgLeuxxLeuArg-221). However, mutational analysis of the CHMP3-AMSH interaction implicated CHMP3 residues Arg216 and Leu217, both important for Vps4 interaction, in the AMSH MIT domain interaction 9. This implies that AMSH recognizes the same CHMP3 region as Vps4A MIT. The binding studies of 17 amino acid long peptides corresponding to the extreme C-terminus of CHMP1A, CHMP1B, CHMP2A and CHM2B revealed variable KD’s between ~4 and 200µM; a similar binding affinity (~28 µM) was determined for the yeast Vps2p C-terminal peptide (residues 183–232) binding to Vps4p MIT 22. Although, the C-terminal CHMP3 peptide shows a similar affinity for AMSH interaction, full length CHMP3 interacts much tighter with AMSH revealing nanomolar dissociation constants (KD of 5.6 nM), thus indicating that other regions of CHMP3 contribute substantially to the AMSH interaction. Although the C-terminal binding motif may be crucial for recruitment of CHMP3 to AMSH 9 other regions of CHMP3 seem to confer tight association with AMSH. Since we could not express a soluble form of the AMSH MIT domain alone, we don’t know whether CHMP3 interacts only with the MIT domain or also with other regions of AMSH. Interestingly, the structure Vps4p MIT in complex with Vps2 (residues 183 to 232) reveals a second MIT interaction site formed by two short additional helical segments; however, the binding affinity of this longer Vps2p peptide is similar to those determined for short mammalian CHMP peptides, namely 28 µM 22. Nevertheless, the structure may indicate how the C-termini of full-length CHMP proteins might interact with MIT domains via a larger surface leading to substantially higher binding affinity.
We show that AMSH binds to CHMP3 in 0HBS (closed conformation) and 500HBS (open conformation) with a dissociation constant of 5.6 nM and 392 nM respectively, implying that the closed conformer binds tighter to AMSH as opposed to the open conformer There are however two reasons why the AMSH CHMP3 interaction may induce CHMP3 opening as suggested 9. First, the low activation barrier of CHMP3 favors stabilization of an open conformation by AMSH. Second, the high affinity interaction requires more than the C-terminal CHMP3 peptide region; this implies indirectly that the high affinity interaction is mediated via one distinct CHMP3 conformation. Thus the interaction occurs most likely via the open conformation, which suggests that AMSH may indeed open a closed CHMP3 conformation; the observed difference in affinity may simply result from a difference in NaCl concentration as charged residues have been implicated in the interaction 9 On the other hand mutagenesis analyses showed that CHMP3 residues implicated in AMSH binding (Arg216, Leu217) do not affect the interaction of the CHMP3 C-terminus with its N-terminal core 9, indicating that AMSH interaction does not compete with the CHMP3 C-terminus binding to its own core. The high affinity CHMP3-AMSH interaction may explain why AMSH overexpression renders full length CHMP3 dominant negative with respect to HIV-1 budding 9 since it may sequester CHMP3 in a state which inhibits its function during budding. Since CHMP3 within a functional ESCRT-III complex seems to act late in budding, the presence of a high affinity CHMP3–AMSH interaction would ensure efficient deubiquitination before budding is completed. In principle, CHMP3 could interact with Vps4 via a similar region; this however raises the question how a high affinity CHMP3-AMSH interaction switches to a low affinity CHMP3-Vps4 interaction, since Vps4 will not be able to compete off AMSH when bound to CHMP3. Alternatively, CHMP3 may not be a target for Vps4 and the high affinity interaction ensures stable complex formation coupled to continuous deubiquitination guaranteeing a free pool of ubiquitin. This is furthermore consistent with the proposal that deubiquitination precedes cargo incorporation into vesicles 13.
Materials and methods
Proteins expression and purification
CHMP3 cDNA encoding amino acid 9–222 was expressed and purified as described 21. Briefly, N-terminally His-tagged CHMP3 was expressed in E. coli Bl21 codon+ cells (Invitrogen) and purified by immobilized metal ion chromatography (IMAC). The His-tag was cleaved off by using TEV protease and CHMP3 was separated from uncleaved CHMP3 and TEV protease by another round of IMAC. A final purification step included size exclusion chromatography (SEC) in a buffer containing 20 mM HEPES, pH 7.6, 150 mM NaCl (150HBS). Further exchanges in buffers containing 20 mM HEPES, pH 7.6 and variable ionic strength ranging from 0, 100, 150, 200 and 500 (0HBS, 100HBS, 150HBS, 200HBS and 500HBS respectively) were done by SEC as indicated. The cDNA corresponding to AMSH (accession code 095630) residues 1–206 (AMSH(1–206)) was cloned into a pBADM-41 vector (N-terminal MBP fusion) and expressed in BL21 (DE3) codon+ cells (Invitrogen). Cells were lysed in HEPES buffer (50 mM HEPES, pH 8.0, 100 mM NaCl) and AMSH(1–206) was purified on an amylose column in the same buffer. For further purification the protein was bound on a HiTrapQ column (GE Healthcare) in HEPES buffer (50mM HEPES pH 8.0, 50mM NaCl) and eluted with a linear gradient of NaCl. Finally, a buffer exchange in 0HBS, 100HBS, 150HBS and 500HBS was done by SEC on a Superdex™ 200 10/300 GL (GE Healthcare)
Size exclusion chromatography and multi-angle laser light scattering
SEC was performed with a Shodex Protein KW-804 HPLC column (300 mm × 8.0 mm). The column was equilibrated in 0HBS and 500HBS; the runs were perform at 20°C with a flow rate of 0.8 ml/min. On-line detection was performed by multi-angle laser light scattering (MALLS) using a DAWN-EOS detector (Wyatt Technology Corp., Santa Barbara, CA) equipped with a laser emitting at 690 nm and by refractive index measurement using a RI2000 detector (Schambeck SFD). Light scattering intensities were measured at different angles relative to the incident beam, and analysis of the data was performed with the ASTRA software (Wyatt Technology Corp., Santa Barbara, CA). The excess light scattering intensity at angle θ (Rθ) is related to molecular mass of solute particle according to Zimm’s formalism of the Rayleigh–Debye–Gans model for a diluted polymer solution 32. For small (gyration radius smaller than 100 nm), non-interacting particles Rθ is given by
| (Eq. 1) |
where Rθ is the measured excess Rayleigh ratio, C is the protein concentration (g/ml) as measured by on-line refractive index detector, M is the molar mass (g/mol), and K* is an optical constant given by the following equation:
| (Eq. 2) |
where NA is Avogadro’s number, n0 is the refractive index of the solvent at the incident radiation wavelength (1.33 for a diluted aqueous buffer), dn/dc (ml/g) is the specific refractive index increment of the solute (0.185 ml/g), and λ is the wavelength of the incident light in void (690 nm). Within the elution peak, the chromatogram is divided in slices, and for each slice, MALLS and refractive index measurements are used to calculate the molecular weight. Weight-averaged (Mw) molecular weights are obtained from the molecular weight distribution across the elution peak.
Circular Dichroism
Circular dichroism (CD) measurements were carried out on a Jasco J-810 (JascoLtd, Tokyo, Japan) spectropolarimeter equipped with a temperature control. Thermal unfolding of CHMP3 (3µM) in buffer 0HBS and 500HBS was monitored by recording the ellipticity at 220 nm as a function of temperature.
Small Angle Scattering Data Collection and Analysis
The synchrotron radiation X-ray scattering data were collected following standard procedures on the X33 SAXS beam line (Deutsches Elektronensynchrotron, DESY and EMBL, Hamburg) 33; 34 A MAR345 image plate with online readout (MarResearch, Germany) was used at a sample - detector distance of 2.7 m covering the range of momentum transfer 0.012 < s < 0.45 Å−1 (s = 4π sin(θ)/λ, where θ is the scattering angle and λ = 0.15 nm is the X-ray wavelength). The s-axis was calibrated by the scattering pattern of Silver-behenate salt (d-spacing 5.84 nm). Scattering data from CHMP3 protein solutions with concentrations 7.2 and 2.8 mg/ml in 0HBS and 4.2, 2.3 and 0.9 mg/ml in 500HBS were measured to check for particle-particle interactions. Repetitive measurements of 180 sec at 15 °C of the same protein solution were performed in order to check for radiation damage. Stable intensities especially at low angles indicated that no protein aggregation took place during the exposure times. The data were normalized to the intensity of the incident beam; the scattering of the buffer was subtracted and the difference curves were scaled for concentration. All the data processing steps were performed using the program package PRIMUS 35. The forward scattering I(0) and the radius of gyration Rg were evaluated using the Guinier approximation 36 assuming that at very small angles (s < 1.3/Rg) the intensity is represented by I(s) = I(0) exp(−(sRg)2/3). These parameters were also computed from the entire scattering patterns using the indirect transform package GNOM (Svergun, 1992), which also provide the distance distribution function p(r) of the particle as defined:
| (Eq. 3) |
The molecular mass of both CHMP3 conformers was calculated by comparison with the forward scattering from the reference solution of bovine serum albumin (BSA). From this procedure a relative calibration factor for the molecular mass (MM) can be calculated using the known molecular mass of BSA (66kDa) and the concentration of the reference solution.
Ab inito shape modeling of CHMP3
Low-resolution models of both CHMP3 conformers were built by the program DAMMIN 31, which represents the protein as an assembly of dummy atoms inside a search volume defined by a sphere of the diameter Dmax. Starting from a random model, DAMMIN employs simulated annealing to build a scattering equivalent model fitting the experimental data Iexp(s) to minimize discrepancy:
| (Eq. 4) |
where N is the number of experimental points, c a scaling factor and Icalc(sj) and σ(sj) are the calculated intensity and the experimental error at the momentum transfer sj, respectively. 10 independent DAMMIN bead models were calculated for both CHMP3 conformers and final models were obtained by superposition of the independent shape reconstructions using the program packages DAMAVER 37 and SUBCOMP 38.
Isothermal titration calorimetry
Calorimetric measurements were carried out using a VP-ITC instrument with a cell volume of 1.4569 ml (MicroCal, LLC). CHMP3 and AMSH were exchanged in the same batch of a given buffer (0HBS, 150HBS, 500HBS) by SEC. The same batch of the buffer was used for further dilutions and for dissolving the CHMP3 peptide (containing residues 196–222). Interaction constants characterizing the AMSH and CHMP3 protein/ peptide interaction were determined by a direct titration in 0HBS, 150HBS, and 500HBS at 25°C. The concentrations used are summarized in Table 1. The solution in the cell was stirred at 286 rpm to ensure rapid mixing. The interaction isotherms were analyzed using the Origin software package supplied by MicroCal. An interaction model assuming n independent and equivalent binding sites was applied, and the stoichiometry n, change in enthalpy ΔH0 and binding constant KD were iteratively fitted.
Acknowledgements
This work was supported by Deutsche Forschungsgemeinschaft SPP-1175 (W.W.), the ANRS (W.W.), the University Joseph Fourier (W.W.), the CNRS (W.W.) and the NIH (H. G.). S. Lata was supported by EMBO and HFSP post doctoral fellowships.
Abbreviations
- ESCRT
endosomal complex required for transport
- MVB
multivesicular body
- CHMP
charged MVB protein
- SAXS
small angle x-ray scattering
- MALLS
multi angle laser-light scattering
References
- 1.Morita E, Sundquist WI. Retrovirus budding. Annual Review of Cell and Developmental Biology. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 2.Hurley JH, Emr SD. The ESCRT Complexes: Structure and Mechanism of a Membrane-Trafficking Network. Annual Review of Biophysics and Biomolecular Structure. 2006;35 doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams RL, Urbe S. The emerging shape of the ESCRT machinery. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 4.Carlton JG, Martin-Serrano J. Parallels Between Cytokinesis and Retroviral Budding: A Role for the ESCRT Machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 5.Saksena S, Sun J, Chu T, Emr SD. ESCRTing proteins in the endocytic pathway. Trends in Biochemical Sciences. 2007;32:561–573. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nature Reviews Molecular Cell Biology. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 7.Tsang HT, Connell JW, Brown SE, Thompson A, Reid E, Sanderson CM. A systematic analysis of human CHMP protein interactions: Additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics. 2006;88(3):333–346. doi: 10.1016/j.ygeno.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 8.McCullough J, Row PE, Lorenzo O, Doherty M, Beynon R, Clague MJ, Urbe S. Activation of the endosome-associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body-sorting machinery. Current Biology. 2006;16:160–165. doi: 10.1016/j.cub.2005.11.073. [DOI] [PubMed] [Google Scholar]
- 9.Zamborlini A, Usami Y, Radoshitzky SR, Popova E, Palu G, Gottlinger H. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc. Natl. Acad. Sci. 2006;103:19140–19145. doi: 10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agromayor M, Martin-Serrano J. Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo. J Biol Chem. 2006;281(32):23083–23091. doi: 10.1074/jbc.M513803200. [DOI] [PubMed] [Google Scholar]
- 11.Ma YM, Boucrot E, Villen J, Affar EB, Gygi SP, Gottlinger HG, Kirchhausen T. Targeting of AMSH to endosomes is required for EGF receptor degradation. J. Biol. Chem. 2007;282(13):9805–9812. doi: 10.1074/jbc.M611635200. [DOI] [PubMed] [Google Scholar]
- 12.Row PE, Liu H, Hayes S, Welchman R, Charalabous P, Hofmann K, Clague MJ, Sanderson CM, Urbe S. The MIT domain of UBPY constitutes a CHMP binding and endosomal localisation signal required for efficient EGF receptor degradation. J. Biol. Chem. 2007;282(42):30929–30937. doi: 10.1074/jbc.M704009200. [DOI] [PubMed] [Google Scholar]
- 13.Clague MJ, Urbe S. Endocytosis: the DUB version. Trends Cell Biol. 2006;16:551–559. doi: 10.1016/j.tcb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Bowers K, Piper SC, Edeling MA, Gray SR, Owen DJ, Lehner PJ, Luzio JP. Degradation of endocytosed epidermal growth factor and virally ubiquitinated major histocompatibility complex class I is independent of mammalian ESCRTII. J Biol Chem. 2006;281:5094–5105. doi: 10.1074/jbc.M508632200. [DOI] [PubMed] [Google Scholar]
- 15.McCullough J, Clague MJ, Urbe S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol. 2004;166:487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. ESCRT-III: An endosome-associated heterooligomeric protein complex required for MVB sorting. Developmental Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 17.von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, Scott A, Krausslich HG, Kaplan J, Morham SG, Sundquist WI. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 18.Horii M, Shibata H, Kobayashi R, Katoh K, Yorikawa C, Yasuda J, Maki M. CHMP7, a novel ESCRT-III-related protein, associates with CHMP4b and functions in the endosomal sorting pathway. Biochem J. 2006;400:23–32. doi: 10.1042/BJ20060897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strack B, Calistri A, Popova E, Gottlinger H. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 20.Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci U S A. 2003;100:12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muziol T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Gottlinger H, Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, Emr SD, Williams RL. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 23.Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. ESCRT-III recognition by VPS4 ATPases. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 24.Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshimori T, Yamagata F, Yamamoto A, Mizushima N, Kabeya Y, Nara A, Miwako I, Ohashi M, Ohsumi M, Ohsumi Y. The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol Biol Cell. 2000;11:747–763. doi: 10.1091/mbc.11.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katoh K, Shibata H, Suzuki H, Nara A, Ishidoh K, Kominami E, Yoshimori T, Maki M. The ALG-2-interacting protein Alix associates with CHMP4b, a human homologue of yeast Snf7 that is involved in multivesicular body sorting. J Biol Chem. 2003;278:39104–39113. doi: 10.1074/jbc.M301604200. [DOI] [PubMed] [Google Scholar]
- 27.Bowers K, Lottridge J, Helliwell SB, Goldthwaite LM, Luzio JP, Stevens TH. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic. 2004;5:194–210. doi: 10.1111/j.1600-0854.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- 28.Pineda-Molina E, Belrhali H, Piefer AJ, Akula I, Bates P, Weissenhorn W. The crystal structure of the C-terminal domain of Vps28 reveals a conserved surface required for Vps20 recruitment. Traffic. 2006;7:1007–1016. doi: 10.1111/j.1600-0854.2006.00440.x. [DOI] [PubMed] [Google Scholar]
- 29.Teo H, Perisic O, Gonzalez B, Williams RL. ESCRT-II, an Endosome-Associated Complex Required for Protein Sorting: Crystal Structure and Interactions with ESCRT-III and Membranes. Dev Cell. 2004;7:559–569. doi: 10.1016/j.devcel.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Shim S, Kimpler LA, Hanson PI. Structure/Function Analysis of Four Core ESCRT-III Proteins Reveals Common Regulatory Role for Extreme C-Terminal Domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 31.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wyatt PJ. Submicrometer particle sizing by multiangle light scattering following fractionation. J. Coll. Interf. Sci. 1998;197:9–20. doi: 10.1006/jcis.1997.5215. [DOI] [PubMed] [Google Scholar]
- 33.Koch MHJ, Bordas J. X-ray diffraction and scattering on disordered systems using synchrotron radiation. Nucl. Instrum. Methods. 1983;208:461–469. [Google Scholar]
- 34.Roessle MW, Klaering R, Ristau U, Robrahn B, Jahn D, Gehrmann T, Konarev PV, Round A, Fiedler S, Hermes S, Svergun DI. Upgrade of the Small Angle X-ray scattering Beamline X33 at the EMBL Hamburg. J. Appl. Crystallogr. 2007 in press. [Google Scholar]
- 35.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS- a Windows-PC based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 36.Guinier A. La diffraction des rayons X aux tres petits angles; application a l'etude de phenomenes ultramicroscopiques. Ann Phys (Paris) 1939;12:161–237. [Google Scholar]
- 37.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 2001;34:33–41. [Google Scholar]