Abstract
Loss of PTEN and activation of PI3K are commonly observed in advanced prostate cancer. Inhibition of mTOR, a downstream target of PI3K signaling, results in cell cycle arrest and apoptosis in multiple in vitro and in vivo models of prostate cancer. However, single agent use of mTOR inhibition has limited clinical success and the identification of molecular events mitigating tumor response to mTOR inhibition remains a critical question. Here, using genetically engineered human prostate epithelial cells (PrEC), we demonstrate that MYC, a frequent target of genetic gain in prostate cancers, abrogates sensitivity to rapamycin by decreasing rapamycin-induced cytostasis and autophagy. Analysis of MYC and the mTOR pathway in human prostate tumors and PrECs demonstrated selective increased expression of 4EBP1 with gain in MYC copy number or forced MYC expression, respectively. We have also found that MYC binds to regulatory regions of the 4EBP1 gene. Suppression of 4EBP1 expression resulted in re-sensitization of MYC-expressing PrEC to rapamycin and increased autophagy. Taken together, our findings suggest that MYC expression abrogates sensitivity to rapamycin through increased expression of 4EBP1 and reduced autophagy.
Keywords: mTOR, rapamycin, MYC, autophagy, 4EBP, prostate cancer
Introduction
Prostate cancer is the second leading cause of death among men in the United States. Most prostate cancer deaths are due to advanced disease that is refractory to existing treatments. PTEN is a tumor suppressor gene that is often lost in advanced prostate cancer resulting in constitutive activation of the PI3K pathway. PI3K pathway subsequently activates multiple downstream targets including AKT1 and mTOR and promotes cell growth and inhibits apoptosis.
The importance of the PI3K-AKT1-mTOR pathway in tumor progression has been elegantly demonstrated in vivo using transgenic mouse models. Loss of PTEN or expression of activated AKT1 leads to malignant or pre-cancerous lesions in mouse prostate, respectively. Treating transgenic mice expressing myristoylated Akt1 under a probacin promoter with RAD001 (a Rapamycin analogue) for two weeks reverses the PIN phenotype, a precancerous lesion of the prostate (1). This dramatic impact of mTOR inhibition reversing the neoplastic phenotype is also observed in transgenic mice with selective prostate knock out of Pten (2). Thus, mTOR inhibition with rapamycin or rapamycin analogues is effective in PI3K-driven prostate cancer.
mTOR signaling is mediated primarily through two multi-protein complexes (TORC1 and TORC2) (reviewed in (3)). TORC1 includes mTOR, raptor, and mLST8 (also known as GβL) (4-6). When activated, TORC1 phosphorylates two primary down-stream effectors, eukaryotic initiation factor 4E (eIF-4E)-binding protein 1 (4E-BP1) and ribosomal protein S6 kinase 1 (S6K1) (7-9). Phosphorylation of 4E-BP1 results in release of eIF-4E and increased cap-dependent translation of a set of proteins involved in G1- to S-phase progression including cyclin D and MYC (10, 11). Phosphorylation of S6K1 results in the phosphorylation of the 40S ribosomal protein S6 and the enhancement of translation of mRNAs that possess a 5′-terminal oligopyrimidine tract (12, 13). The TORC2 complex is comprised of mTOR, rictor, and mLST8 and has been shown to phosphorylate AKT1 (14) and Paxillin (15) among others. While mTOR is activated in many human cancers, the relative biological importance of TORC1 and TORC2 activity in human cancers remains unknown.
The drug rapamycin binds to a cellular protein FKBP12, and this complex inhibits the TORC1 complex. Despite the dramatic effects observed after treatment of cells in tissue culture and transgenic animals with rapamycin and rapamycin-derivatives, limited clinical efficacy of single agent mTOR inhibition has been observed in advanced prostate cancer. Here, we used genetically engineered prostate epithelial cells and found that MYC expression profoundly reduces sensitivity to rapamycin. Importantly, MYC copy number gain is associated with increased 4EBP1 expression in human prostate tumors, MYC binds to regulatory regions of the 4EBP1 gene, and MYC increases 4EBP1 expression and decreases autophagy. Suppression of 4EBP1 re-sensitized cells to rapamycin and resulted in less autophagy. In fact, 4EBP1 expression limits autophagy induction by at least two inducers: rapamycin and tunicamycin. Thus, MYC over-expression is associated with increased 4EBP1 expression and decreased autophagy which may play a role in limiting the clinical effectiveness of single agent rapamycin or rapamycin analogues.
Materials and Methods
Cell lines and medium
Prostate cancer cell lines DU145, LNCaP and PC3 were obtained from American Type Culture Collection (ATCC). Cells were grown in RPMI medium (Cellgro) with 10% fetal bovine serum. This medium was supplemented with 10 mM HEPES, 1 mM Sodium Pyruvate, 2 mM L-Glutamine and Plasmocin at 1 μg/ml. Genetically engineered cell lines including PrEC, LHS, LHSR, LHSR-AR, BH10i-AR, and LHMB-AR were created and propagated as described previously (See Supplemental Table 1 for specific genetic constitution of each cell line) (16). The prostate epithelial cells were incubated at 37°C and 5% CO2 in PrEGM with defined supplements as recommended (Cambrex).
Retroviral Infection
Retroviruses were created as previously described (17) with minor modifications. 293 EBNA packaging cell lines were transfected with each of 3 plasmids containing 1) the Gag and Pol genes, 2) the VSV envelope protein gene, and 3) a pWZL retroviral vector containing the gene of interest using FuGENE 6 (3:1 volume (μL) to weight (μg) ratio). Packaging cell lines were transfected twice separated by 24 hours; virus was harvested 36-48 hours following the second transfection. Virus-containing media was centrifuged at 500 rpm for 5 min at 4° C, filtered (45 μm pore size (μStar)), mixed with 10% growth medium and 4 μg/ml polybrene, and placed upon cells. The retroviral supernatant was removed 24 hours later, cells were cultured another 24 hours in normal growth medium and selective media containing the appropriate agent (blasticidin or puromycin) was added. Lentiviral infections were performed following a similar protocol to the retroviral infections. Lentiviral packaging vectors (pLKO system) were obtained from Addgene (Trono laboratory). shRNA construct against 4EBP1 in lentiviral vector was from the RNAi consortium (Broad Institute).
Plasmids and Bacteria
All plasmids were propagated in DH5α Escherichia coli bacteria (Invitrogen) in Luria-Broth (LB) growth medium (Fisher Scientific) and grown at 37° C. The vector backbone pWZL was obtained as pWZL-MYC from Addgene (18). The EGFP insert was from pEGFP-C1 (BD biosciences). The short hairpin RNA construct against MYC, sh3-1 was a gift from Joseph Nevins at Duke University and is in pRETROSUPER (19).
Drug Sensitivity Assay
Rapamycin (Sigma Aldrich) and a PI3K inhibitor (BEZ235- Novartis) were used in prostate cancer cell lines (DU145, PC3, LNCaP) as well as in genetically engineered cell lines to determine the sensitivity of the cells to the two drugs using a propidium iodide-based cell cytotoxicity assay (20). Cells were plated at a density of 5000 cells/ well in 100 μl of tissue culture medium. Serial dilutions of the drug at varying concentrations were added in a volume of 100 μl the next day, one set of plates is taken out at this point as a time 0 control, and plates are incubated for 4-5 days after which they are removed from the incubator and frozen at -80° C. At least 12 hours after being frozen, the plates were thawed, 50 μl (stock 200 μg/ml) of propidium iodide was added to each well, and plates were incubated for one hour in dark. Fluorescent readings were measured with a FLUOstar OPTIMA (BMG labtechonologies) with excitation at 530 nm and emission at 590 nm wavelengths. Proliferation was normalized to the no drug control and plotted against increasing drug concentration using Microsoft Excel software. Inhibitory concentrations were obtained using GraphPad to obtain a nonlinear regression curve fit to calculate the EC50 (Prism Software).
Protein isolation and Western blotting
Cells were solubilized in RIPA lysis buffer (supplemented with mini cocktail EDTA free proteinase inhibitors – Roche). Adherent cells were washed in ice-cold PBS twice and harvested using a cell scraper. Cells were re-suspended in RIPA lysis buffer (supplemented with mini cocktail EDTA free proteinase inhibitors – Roche). When Rapamycin and BEZ235 were used to investigate the effect on protein phosphorylation, cells were starved for 48 hours in 0.2% serum containing medium (or PrGEM that has only 1/50th of the recommended supplements) and released in medium containing 10% serum for 30 minutes before protein isolation. Protein lysates (25 μg/sample) were separated on pre-cast gradient gels (BioRad) by electrophoresis and transferred to a nylon membrane (Immobilon-P). The membrane was blocked in 5% non-fat dry milk for one hour and hybridized to primary antibodies in TBS-T overnight at 4° C. Membrane was washed in TBS-T for 5 minutes three times and incubated in secondary antibody anti-mouse or anti-rabbit linked to Horse radish peroxidase (1:2000) for one hour. Following five washes of 5 minutes each, membrane was processed using ECL Western Blotting Reagents (Amersham, GE Healthcare) and exposed to film (Hyblot CL, Denville Scientific Inc.). The following primary antibodies were purchased from Cell Signaling Technology: LC3B (#2775), pS6 (#2211), Total S6 (#2217), pAKT1 (Ser473) (#9271), Total AKT1 (#9272), p4EBP1 (#9459), Total 4EBP1 (#9452), eIF4E (#9742), Caspase 3 (#9662). The antibodies against c-MYC (SC-42) and actin (SC-47778) were from Santa Cruz Biotechnology and the antibody against PARP was from Upstate Biotech (#06-557). The secondary antibodies were either HRP-linked anti-rabbit or anti-mouse both from Cell Signaling Technology.
Cell-cycle analysis
Cells were grown at low density on plates and rapamycin was added at different concentrations and exposed for different time periods. When harvesting cells, culture medium was saved and spun to preserve floating and loosely attached cells. Attached cells were detached using trypsin and all cells for each treatment were collected in a 1.5 ml micro centrifuge tube and spun at 2000 rpm for 2 minutes. Cells were washed in PBS, fixed in 70% ethanol, and re-suspending in ice-cold Nuclear Isolation Medium (NIM – 0.5% BSA, 0.1% NP-40 in PBS) containing 5 μg/ ml propidium Iodide and 100 μg/ml DNase-free RNase. Cell-cycle phases in 5000 – 10,000 cells were determined by flow cytometry at the Duke Flow Cytometry facility.
RNA expression and Tissue microarrays
De-identified local prostate cancer specimens were analyzed under an IRB approved protocol (Duke IRB 6679-04EX). PrEC cell lines and tumor specimens were processed for hybridization to Oligonucleotide microarrays (Affymetrix U133A) using standard methods (17, 21). Gene expression was determined from the CEL files using RMA with background correction and quantile normalization (22, 23). Expression of MYC, mTOR, 4EBP1, S6, S6K, and AKT were determined using gene specific probes on the U133A microarray.
Formalin-fixed, paraffin-embedded tissue for a subset of the samples for which frozen tissue was available for microarray analysis was used to create tissue microarrays. MYC copy number gain was determined using fluorescent in situ hybridization as previously described (24).
Non-parametric tests were used to determine statistical significance of differences between cell lines (Mann Whitney) or correlation between the expression of specific genes (Spearman's Rank Statistic).
Chromatin Immunoprecipitation assay
ChIP assay was performed as described previously (19). Briefly, 10% Formaldeyde solution was added to cells for 15 minutes to crosslink DNA-protein complexes. Isolated nuclear chromatin extracts were sonicated and incubated overnight at 4°C with antibodies from Santa Cruz Biotechnology (Santa Cruz, CA, USA) with either 5 μg of anti-c-MYC (N262) or 2.5 μg of normal rabbit IgG (SC-2707). This was followed by incubation with 20 ml of Protein A agarose beads (Roche) for 3-4 hours at 4°C. After extensive washing, DNA fragments were harvested by de-crosslinking the immunoprecipitates. Real-Time PCR utilizing SYBR Green master mix (Qiagen) and the ABI PRISM 7900 Sequence Detection System was used for quantitative analysis of the recovered DNA fragments from ChIP including regions of 4EBP1, ALB, and ODC1 (See Supplemental Table 2 for primer sequences). Relative occupancy of the MYC transcription factor at the different E-boxes were estimated by comparing its occupancy at the albumin promoter following methods described in Aparicio et al. (25). The ChIP assays were done at least three times for each cell line.
Autophagy Assays
Cells were grown in 10 cm plates in the presence of absence of rapamycin or tunicamycin for 24 hours and harvested for analysis. For Western blots, cell lysates were separated using PAGE (as described above) and hybridized to an antibody for LC3B type II (Cell Signaling Technology, Inc.) at 1:1000 to detect autophagosome formation. For flow cytometry, cells were exposed to 1 μg/ml acridine orange (Sigma Aldrich) for 20 minutes, trypsinized, and re-suspended in growth medium. Approximately 20,000 cells were analyzed using the PerCP-Cy5.5-A channel with a BD FACS Canto II (BD biosciences) flow cytometer. For fluorescent microscopy, cells were grown on tissue culture treated chamber slides (BD Falcon) and exposed to rapamycin or tunicamycin for 24 hours. The cells were briefly exposed to 1 μg/ml acridine orange (Sigma Aldrich) for 20 minutes and examined under a microscope without fixation. All images were acquired using an inverted Olympus IX71 microscope and Photometrics Cool Snap HQ camera connected to a Deltavision RT Restoration Imaging System (Applied Precision, Issaquah, WA). Images were acquired using the 20× Uplan SApo objective ((0.75 NA) and the Deltavision SoftWoRx Resolve 3D capture program and collected as a stack of 0.2 - 0.5 μm increments in the z axis (15-35 sections). Images were deconvolved using the conservative algorithm with 5 - 10 iterations. Deconvolved images were saved as separate z-sections or viewed as stacked (collapsed) images using the Quick Projection option. Images were then imported to Adobe Photoshop for presentation.
Results
Genetic determinants of prostate cell sensitivity to rapamycin
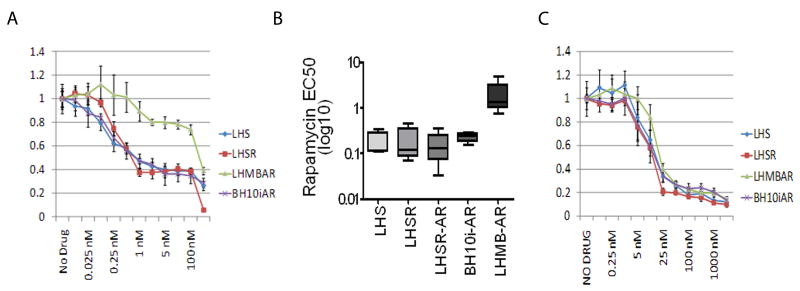
Prior studies (26) have shown that the PTEN status of established prostate cancer cell lines impacts sensitivity to rapamycin; PTEN wild-type cells (DU145) are less sensitive than PTEN-null cell lines (PC3 and LNCaP) (Suppl. Fig. 1a). However, the marked aneuploidy present in these established cancer cell lines (27-29) precludes refined analysis regarding specific interactions between genetic events and sensitivity to rapamycin. Therefore, genetically engineered immortalized and tumorigenic human prostate epithelial cells (PrEC) (16) were tested for rapamycin sensitivity. In these cell lines, neither activated H-Ras nor the androgen receptor (AR) significantly altered PrEC sensitivity to rapamycin (Fig. 1A, 1B, and Suppl. Fig. 1B). In contrast, the expression of MYC resulted in marked resistance to rapamycin (Fig. 1A and 1B). In multiple experiments, the EC50 for rapamycin was the same for cell lines with differential expression of RAS (LHS EC50 0.18 nM +/- 0.11 (mean +/- standard deviation) v LHSR EC50 0.21 nM +/- .16) or AR (LHSR EC50 0.21 nM +/- 0.16 v LHSR-AR EC50 0.16 nM +/- 0.14) whereas there was an approximately 10-fold difference in the EC50 between cell lines with differential expression of MYC (BH10i-AR EC50 = 0.23 nM +/- 0.060 vs. LHMB-AR EC50 = 2.13 nM +/- 1.963)(Mann Whitney exact p=0.0286). While MYC is known to be an important downstream target of mTOR-mediated regulation of cap-dependent translation (11), the ability of MYC to abrogate sensitivity to rapamycin has not been previously described.
Figure 1. Over-expression of the oncogene c-MYC in prostate cell lines confers resistance to the mTOR inhibitor Rapamycin but not to a PI3K inhibitor.
A) Normalized cell cytotoxicity (y-axis) for four genetically engineered prostate epithelial cell lines (PrECs) exposed to increasing concentrations of rapamycin (x-axis). Absorbance values are normalized to the no drug control individually for each cell line. B) The EC50 (Rapamycin) of the PrECs plotted on a log10 scale. C) Sensitivity of PrEC cell lines to a PI3K inhibitor. Cytotoxicity is normalized as in A.
Altering MYC levels in established prostate cancer cell lines mimics the effects seen in the PrEC
MYC expression also impacts rapamycin sensitivity in established prostate cell lines (PC3, DU145, and LNCaP). Consistent with our findings in PrEC, over-expression of MYC resulted in decreased sensitivity to rapamycin in LNCaP and DU145 (Fig. 2A and 2B). Increased expression of MYC in PC3 had minimal impact on rapamycin sensitivity and we note that baseline expression of MYC in PC3 is higher than DU145 and LNCaP (Fig 2A). RNAi mediated suppression of MYC increased rapamycin sensitivity in all three cell lines tested (Fig. 2C and 2D). Altering MYC in the established cell lines impacted the growth kinetics of each cell line independent of rapamycin exposure. To ensure that MYC activity rather than a change in cell growth altered rapamycin sensitivity, we tested a range of serum deprivation conditions to identify a concentration of serum that resulted in an equivalent reduction in cell growth as seen with MYC knock-down. PC3 cells growing in 2.5% serum had an equivalent reduction in cell growth as seen with MYC knock-down (∼70%) but were not more sensitive to rapamycin (data not shown). Overall, altering MYC expression in established prostate cancer cell lines consistently altered sensitivity to rapamycin albeit less dramatically as in the genetically engineered PrEC cell lines.
Figure 2. MYC impacts response to Rapamycin in Established Prostate Cancer Cell Lines.
A) Western blots for MYC (αMYC) and ACTIN (αACTIN) of antibiotic-resistant polyclonal cell lines DU145, PC3, and LNCaP after infection with a retrovirus construct containing GFP or MYC. B) Relative cell number after 4 days of growth in media containing 5 nM of rapamycin for DU145, PC3, and LNCaP cells expressing either GFP (grey bars) or MYC (white bars). Relative cell number is determined by the emission at 590 nm after staining with propidium iodide normalized to cells grown for 4 days without rapamycin. C) Western blot for MYC (αMYC) and ACTIN (αACTIN) of stable, polyclonal cell lines DU145, PC3, and LNCaP after infection with a retrovirus containing a scrambled shRNA or sh3.1 which targets MYC. D) Relative cell number after 4 days of growth in media containing 5 nM of rapamycin for DU145, PC3, and LNCaP cells infected with either a scrambled shRNA construct (“shØ”, grey bars) or sh3.1 targeting MYC (“shMYC”, white bars). Relative cell number is determined as above.
MYC alters rapamycin induced cytostasis and autophagy
In previous reports, mTOR inhibition with rapamycin has been found to cause cytostasis (1), apoptosis (1), and/or autophagy (30). In the PrEC cell lines, the impact of MYC appears to be primarily through decreased rapamycin-induced cytostasis and autophagy rather than apoptosis. In the BH10i-AR cells, rapamycin treatment results in a marked cytostasis with a decrease in the percentage of cells in the S and G2 phases of the cell cycle (Fig. 3A). Expression of MYC abrogates the rapamycin-induced cytostasis. The effect of MYC was not due to apoptosis as rapamycin failed to induce apoptosis in either BH10i-AR or LHMB-AR cell lines as detected by proteolytic cleavage of Caspase 3 (Suppl. Fig. 2A) or PARP (Suppl. Fig. 2B) although MYC expression did decrease the sub-G1 population of cells (the “M1” gate in Fig. 3A) with or without rapamycin. The specificity of MYC's observed impact on rapamycin sensitivity is supported by a failure of activated H-RAS in the PrEC to have a similar decrease in rapamycin induced cytostasis (Suppl. Fig. 2C).
Figure 3. Rapamycin sensitivity is mediated by alteration in cell-cycle progression and autophagy.
(A) LHMB-AR and BH10i-AR cells are grown in the presence or absence of Rapamycin for 24 hours and cell-cycle analysis was done by flow cytometry (described in materials and methods). (B) LC3B analysis by Western blotting for autophagy after exposure for 24 hours to 1 μM Rapamycin or 2.5 μM Tunicamycin. (C) Fluorescent images of cells grown for 24 hours at 20 nM Rapamycin and control cells without Rapamycin (as described in materials and methods). Autophagosomes are the red vesicles in the cytoplasmic region. Scale bars are 40 microns. (D) Autophagy analyzed using flow cytometry. Cells were grown in the presence of 20 nM rapamycin, 2.5 μM tunicamycin or no drug for 48 hours and stained with acridine orange. Each treated group for either cell line is statistically different from the untreated group (Mann Whitney test: p < 0.0001).
Cells detected to be in G1/G0 may simply be in a state of dormancy (e.g. G0) but they can exhibit alternative behaviors such as autophagy. In the PrEC, MYC expression decreased autophagy as measured by acridine orange staining, flow cytometry, and LC3B Western Blot (Fig. 3B) and limited the increase in autophagy following treatment with rapamycin (Fig. 3B, 3C, and 3D). Coincident exposure to bafilomycin A1 decreased baseline autophagy and eliminated rapamycin induced autophagy. MYC expression also limited autophagy caused by tunicamycin – a frequently used agent that reliably causes autophagy through inhibition of GlcNAc phosphotransferase and inhibition of glycoprotein synthesis - an mTOR-independent process. However, we did not see a difference in autophagy induction between the PrEC cell lines with and without MYC expression using Earle's balanced salt solution, N-Acetyl sphingosine, or Lithium Chloride (Suppl. Fig 2D).
Analysis of the expression of downstream mTOR components in vitro and in vivo in genetically engineered prostate epithelial cells and prostate tumors
After establishing an association between MYC activity and rapamycin sensitivity, we sought to investigate how MYC, a transcription factor, may impact the mTOR signaling cascade. To identify mTOR target proteins that may have altered expression due to MYC, we analyzed transcriptional microarray data derived from the PrEC cell lines and a set of prostate tumors for which we have microarray data and tissue microarrays.
First, we used gene expression profiles from the PrECs to determine if mTOR, S6K, 4EBP1, or AKT1 were differentially expressed in the presence or absence of MYC. In PrEC lines of different genetic constitutions, 4EBP1 was specifically over-expressed in cells expressing MYC and not altered by any other genetic manipulation (Fig. 4A). MYC, however, did not impact the expression of S6K, mTOR, or AKT1.
Figure 4. MYC expression leads to increased 4EBP1 in cell lines and tumor samples.
(A) The RNA expression levels of the different components of the mTOR pathway in the genetically engineered Prostate Epithelial Cell lines. The RNA levels are normalized to that expressed in the unmodified PrEC. (B) MYC amplification in prostate tumors from patients detected by Fluorescent In Situ Hybridization (FISH). Box plot of 4EBP1 expression for samples with gain of C-MYC (n=6) vs. those that are diploid (n = 15). In representative image of cells with C-MYC gain, C-MYC probe is in red and CEN8 probe in green. (C) Results of chromatin immunoprecipitation in LHMBAR and PC3 cell lines using non-specific rabbit polyclonal antibody (IgG, blue bars) or a MYC-specific antibody (MYC, red bars). Relative occupancy (y-axis) plotted for 3 regions containing an E-box adjacent to the 4EBP1 transcriptional start site (E-box 1-3) and ornithine decarboxylase (ODC). Locations of each E-box depicted in schematic of 4EBP1 gene. (D) Protein expression and phosphorylation status of mTOR effector proteins in prostate epithelial cells expressing hTERT, Large T, activated p110, and AR (“BH10iAR”) and this same cell line with the additional expression of MYC (“LHMBAR”). Cells were grown to log phase in normal medium and incubated for 48 hours in medium equivalent to serum-starved conditions and then transferred to normal medium for 30 min with or without Rapamycin.
Next, we tested if genetic gain of MYC in prostate cancer tumors was associated with increased expression of mTOR and/or target proteins. Using a set of 50 tumors for which both tissue microarrays and expression data were available, MYC copy number was definitively established in 21 specimens by FISH (Fig. 4B – left panel) and transcriptional microarray analysis was used to determine expression of specific mTOR targets. Here, while the number of samples with gain in MYC copy number was relatively small (n=6), 4EBP1 had increased expression in prostate tumors with increased MYC copy number compared to samples that were diploid for MYC (P=0.0091) (Fig. 4B – right panel). In addition, in a larger number of prostate cancer samples, MYC RNA expression significantly correlated with 4EBP expression (Spearman r = 0.453, p = 0.0391) while there was no correlation between MYC and mTOR (Spearman r = -.229, p = 0.316) (Suppl. Fig 3A).
Thus, in genetically engineered cell lines and localized prostate cancer specimens, there is a consistent correlation between MYC and increased expression of 4EBP1 that was not apparent with other target proteins of mTOR.
MYC binds to E-Box sequence adjacent to 4EBP1 gene
Given the correlation between MYC copy number gain and 4EBP1 expression, we performed chromatin immunoprecipitation to determine if MYC binds to regulatory regions of the 4EBP1 gene. One E-box sequence was identified immediately upstream of the 4EBP1 transcriptional start site and two additional E-box sequences were identified within the first intron of 4EBP1 (See schema in Fig. 4C). In LHMBAR, there was strong relative occupancy of E-box 1 and E-box 2 when compared to an unrelated albumin promoter region but no occupancy of E-box 3 (Fig. 4C). MYC occupancy of E-box 1 and E-box 2 was greater in LHMBAR than MYC occupancy of an E-box adjacent to the ornithine decarboxylase gene which is a well characterized direct MYC target (31). MYC also occupied E-box 1, E-box 2, and the E-box adjacent to ODC in the BH10iAR cell line, but at much lower relative occupancy (Suppl. Fig 3B). Binding of MYC to E-box 1 and E-box 2 was also observed in the established prostate cancer cell line PC3 although relative MYC occupancy was somewhat lower than at the ODC gene (Fig. 4C). Thus, MYC likely increases 4EBP1 expression through transcriptional regulation.
MYC expression increases expression of 4EBP1 protein
We next investigated MYC induced changes in the protein expression and phosphorylation status of mTOR target proteins in the genetically engineered prostate cell system. When the expression and phosphorylation of pS6, p4EBP1, and pAKT1 were assessed, we found a marked increase in total and phosphorylated 4EBP in the LHMB-AR cells whereas total and phosphorylated S6 levels did not change and AKT1 was only modestly increased (Fig. 4D). In the PrECs, rapamycin significantly decreases phosphorylation of S6 and increases phosphorylation of AKT1, but had limited impact on 4EBP phosphorylation in MYC expressing cells. This pattern appears to be specific to mTOR inhibition with rapamycin as inhibition of both PI3K and mTOR with BEZ235 inhibited phosphorylation of all three proteins (Suppl. Fig 4A and 4B). The increase in protein expression of 4EBP1 with MYC over-expression is also observed in PC3 cells, an established prostate cancer cell line (Suppl. Fig 4C). In addition, levels of eIF4E were consistent despite increased MYC expression and increased 4EBP1 expression (Suppl. Fig 4D) suggesting that along with increased total levels of 4EBP1 there was also an increase in the ratio of 4EBP1 to eIF4E.
4EBP1 suppression re-sensitizes MYC expressing PrEC to rapamycin and increases autophagy
The strong association between MYC and 4EBP1 expression suggests that MYC may confer increased resistance to rapamycin through increased 4EBP1 expression. While 4EBP1 does not co-immunoprecipitate with the TORC1 complex in the PrEC and established prostate cancer cell lines (data not shown), we remained interested in determining if decreasing 4EBP1 would alter rapamycin sensitivity in PrECs with high MYC expression. To test this, 4EBP1 was targeted with a shRNA in LHMB-AR PrECs. While suppression of 4EBP1 had no impact on LHMB-AR cell growth, the cells became significantly more sensitive to rapamycin (Fig 5A).
Figure 5. Decreased 4EBP1 leads to increased sensitivity to Rapamycin and increased autophagy.
(A) shRNA suppression of 4EBP1 increases sensitivity to rapamycin. Relative cell number at 4 days for each cell line is normalized to culture in the absence of rapamycin and plotted against increasing rapamycin concentration. (B) Decreasing 4EBP1 increases autophagy. Western blot for LC3B following 24-hour exposure to 1 μM Rapamycin or 2.5 μM Tunicamycin. (C) Accumulation of autophagosomes (red) during autophagy. Scale bar is 40 microns. (D) Autophagy analyzed using flow cytometry. Cells grown in the presence of 20 nM rapamycin, 2.5 μM tunicamycin or no drug for 48 hours were stained with acridine orange. Each treated group for either cell line is statistically different from the untreated group (Mann Whitney test: p < 0.0001).
As MYC decreased autophagy and limited the increase in autophagy resulting from rapamycin treatment, we determined if knock-down of 4EBP1 affected autophagy. Importantly, decreased 4EBP1 expression resulted in decreased baseline levels of autophagy based upon lipidation of LC3B and acridine orange fluorescence (Fig. 5B, 5C, and 5D). Indeed, whereas LHMB-AR cells expressing a lentivirus expressing a scrambled shRNA construct had baseline levels of autophagy similar to that of uninfected LHMB-AR, the LHMB-AR cells in which 4EBP1 expression was suppressed exhibited increased baseline levels of autophagy. Importantly, suppression of 4EBP1 also increased rapamycin- and tunicamycin-induced autophagy (Fig. 5D). Thus, 4EBP1 expression appears to have significant impact on autophagy, both baseline levels and selected inducers of autophagy including rapamycin.
Discussion
The PI3K pathway remains a promising target for therapeutic intervention in prostate cancer given the high frequency of PTEN loss. Indeed, recent findings from comprehensive cancer genome sequencing projects suggest that the PI3K pathway is frequently activated in breast cancer (32), pancreatic cancer (33), and glioblastoma (34). While mTOR inhibitors have demonstrated outstanding preclinical activity in prostate cancer models dependent on activation of the PI3K pathway, the limited clinical activity of mTOR inhibition observed in prostate cancer has been disappointing. While it has been shown that feedback loops through IGFR1 and mitogen activated kinases play a role in resistance to mTOR inhibitors, our work suggests that expression of MYC, a target of gain or amplification in 20 - 30 % of advanced prostate cancer (35-37), may also account for the limited activity of mTOR inhibition in prostate cancer.
While MYC has a myriad of transcriptional targets and profound effects on cell biology, our work identifies 4EBP1 as a novel target of MYC through which MYC activity impacts the response to rapamycin. 4EBP1 is believed to primarily control cap-dependent translation by binding and inactivating eIF4E; phosphorylation of 4EBP1 results in the release of eIF4E and subsequent activation of the eIF4G scaffolding protein (38). Thus, it is unclear how increased expression of phosphorylated 4EBP1 (p4EBP1) results in decreased rapamycin sensitivity. Recent work suggests that 4EBP1 may also have a positive impact as p4EBP1 can bind to and stabilize the TORC1 in vitro (39). Our work suggests that 4EBP1 may have as yet unrecognized actions that impact mTOR signaling and/or autophagy. Indeed, the impact of 4EBP1 on autophagy was significant as cell lines with 4EPB1 knock-down displayed increased baseline levels of autophagy, increased rapamycin induced autophagy, and increased tunicamycin-induced autophagy and the impact of 4EBP1 was abolished in the presence of the autophagy inhibitor bafilomycin A1. The consistency of these findings suggests that 4EBP1 may interact with specific proteins involved in autophagy; the subsequent discovery of how 4EBP1 impacts autophagy has the potential to provide new insight as to the role of 4EBP1 in cellular physiology and cancer.
Our discoveries of the significant impact of MYC on rapamycin, the impact of MYC on 4EBP1 expression, and the impact of 4EBP1 on autophagy suggest that these proteins and the process of autophagy are important to prostate tumor's response to mTOR inhibition. However, the results from established prostate cancer cell lines and 4EBP1 knock-down experiments suggest that MYC's impact on rapamycin sensitivity is not entirely explained by the increased expression of 4EBP1 and/or the modulation of autophagy. It is likely that MYC has additional roles that affect a cell's sensitivity to mTOR inhibition and we did note modest increase in pAKT with increased MYC expression suggesting that TORC2 activity may also be impacted. Fortunately, using a recently described dual PI3K and mTOR inhibitor, the impact of MYC on sensitivity is eliminated. Coincidently, BEZ235 dramatically reduces p4EBP and pAKT and increases autophagy and may prove to be a more effective, MYC-independent therapeutic strategy in advanced prostate cancer.
Our work provides a specific example of how confounding genetic events can impact the efficacy of a targeted therapy. As comprehensive genomic analyses for each of the major tumor types become available, work such as that presented in this paper will provide a means to understand how the co-occurrence of specific genetic events may impact a patient's response to a given targeted therapy. The use of genetically engineered cells can help identify specific modulating genetic events that are important to consider when choosing therapy for individuals with cancer. Our results suggest that the limited clinical efficacy of mTOR inhibition in prostate cancer may be partially attributed to the frequent genetic gain of MYC, increased expression of 4EBP, and decreased autophagy.
Supplementary Material
Acknowledgments
Phillip G. Febbo is a Damon Runyon-Lily Clinical Investigator and this work was supported by NCI123175 and the Prostate Cancer Foundation.
Footnotes
Conflicts of Interest: None
References
- 1.Majumder PK, Febbo PG, Bikoff R, et al. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat Med. 2004;10:594–601. doi: 10.1038/nm1052. [DOI] [PubMed] [Google Scholar]
- 2.Podsypanina K, Lee RT, Politis C, et al. An inhibitor of mTOR reduces neoplasia and normalizes p70/S6 kinase activity in Pten+/- mice. Proc Natl Acad Sci U S A. 2001;98:10320–5. doi: 10.1073/pnas.171060098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarbassov dos D, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–94. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 5.Xu G, Kwon G, Cruz WS, Marshall CA, McDaniel ML. Metabolic regulation by leucine of translation initiation through the mTOR-signaling pathway by pancreatic beta-cells. Diabetes. 2001;50:353–60. doi: 10.2337/diabetes.50.2.353. [DOI] [PubMed] [Google Scholar]
- 6.Dennis PB, Jaeschke A, Saitoh M, Fowler B, Kozma SC, Thomas G. Mammalian TOR: a homeostatic ATP sensor. Science. 2001;294:1102–5. doi: 10.1126/science.1063518. [DOI] [PubMed] [Google Scholar]
- 7.Kim DH, Sarbassov DD, Ali SM, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–75. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 8.Hara K, Maruki Y, Long X, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–89. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 9.Kim DH, Sarbassov DD, Ali SM, et al. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- 10.Hashemolhosseini S, Nagamine Y, Morley SJ, Desrivieres S, Mercep L, Ferrari S. Rapamycin inhibition of the G1 to S transition is mediated by effects on cyclin D1 mRNA and protein stability. J Biol Chem. 1998;273:14424–9. doi: 10.1074/jbc.273.23.14424. [DOI] [PubMed] [Google Scholar]
- 11.West MJ, Stoneley M, Willis AE. Translational induction of the c-myc oncogene via activation of the FRAP/TOR signalling pathway. Oncogene. 1998;17:769–80. doi: 10.1038/sj.onc.1201990. [DOI] [PubMed] [Google Scholar]
- 12.Jefferies HB, Reinhard C, Kozma SC, Thomas G. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc Natl Acad Sci U S A. 1994;91:4441–5. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jefferies HB, Fumagalli S, Dennis PB, Reinhard C, Pearson RB, Thomas G. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. Embo J. 1997;16:3693–704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarbassov DD, Ali SM, Kim DH, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 15.Jacinto E, Loewith R, Schmidt A, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–8. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 16.Berger R, Febbo PG, Majumder PK, et al. Androgen-Induced Differentiation and Tumorigenicity of Human Prostate Epithelial Cells. Cancer Res. 2004;64:8867–75. doi: 10.1158/0008-5472.CAN-04-2938. [DOI] [PubMed] [Google Scholar]
- 17.Febbo PG, Lowenberg M, Thorner AR, Brown M, Loda M, Golub TR. Androgen mediated regulation and functional implications of fkbp51 expression in prostate cancer. J Urol. 2005;173:1772–7. doi: 10.1097/01.ju.0000155845.44729.ba. [DOI] [PubMed] [Google Scholar]
- 18.Boehm JS, Hession MT, Bulmer SE, Hahn WC. Transformation of human and murine fibroblasts without viral oncoproteins. Mol Cell Biol. 2005;25:6464–74. doi: 10.1128/MCB.25.15.6464-6474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leung JY, Ehmann GL, Giangrande PH, Nevins JR. A role for Myc in facilitating transcription activation by E2F1. Oncogene. 2008;27:4172–9. doi: 10.1038/onc.2008.55. [DOI] [PubMed] [Google Scholar]
- 20.Skehan P. Cytotoxicity and Cell Growth Assays. In: Celis JE, editor. Cell Biology: A Laboratory Handbook. Second. San Diego: Acadmic Press; 1998. pp. 313–8. [Google Scholar]
- 21.Singh D, Febbo PG, Ross K, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–9. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 22.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 23.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reiter RE, Sato I, Thomas G, et al. Coamplification of prostate stem cell antigen (PSCA) and MYC in locally advanced prostate cancer. Genes Chromosomes Cancer. 2000;27:95–103. doi: 10.1002/(sici)1098-2264(200001)27:1<95::aid-gcc12>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 25.Current Protocols in Molecular Biology. John Wiley & sons; 2005. pp. 21.3.1–.3.33. [Google Scholar]
- 26.Neshat MS, Mellinghoff IK, Tran C, et al. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc Natl Acad Sci U S A. 2001;98:10314–9. doi: 10.1073/pnas.171076798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stone K, Mickey D, Wunderli H, Mickey G, Paulson D. Isolation of a human prostate carcinoma cell line (DU 145) Int J Cancer. 1978;21:274–81. doi: 10.1002/ijc.2910210305. [DOI] [PubMed] [Google Scholar]
- 28.Kaighn M, Narayan K, Ohnuki Y, Lechner J, Jones L. Establishment and characterization of a human prostate carcinoma cell line (PC-3) Invest Urol. 1979;17:16–23. [PubMed] [Google Scholar]
- 29.Horoszewicz JS, Leong SS, Kawinski E, et al. LNCaP model of human prostatic carcinoma. Cancer Res. 1983;43:1809–18. [PubMed] [Google Scholar]
- 30.Cao C, Subhawong T, Albert JM, et al. Inhibition of mammalian target of rapamycin or apoptotic pathway induces autophagy and radiosensitizes PTEN null prostate cancer cells. Cancer Res. 2006;66:10040–7. doi: 10.1158/0008-5472.CAN-06-0802. [DOI] [PubMed] [Google Scholar]
- 31.Bello-Fernandez C, Packham G, Cleveland JL. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci U S A. 1993;90:7804–8. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 33.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLendon R, Friedman A, Bigner D, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008 doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jenkins RB, Qian J, Lieber MM, Bostwick DG. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res. 1997;57:524–31. [PubMed] [Google Scholar]
- 36.Nupponen NN, Kakkola L, Koivisto P, Visakorpi T. Genetic alterations in hormone-refractory recurrent prostate carcinomas. Am J Pathol. 1998;153:141–8. doi: 10.1016/S0002-9440(10)65554-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mark HF, Samy M, Santoro K, Mark S, Feldman D. Fluorescent in situ hybridization study of c-myc oncogene copy number in prostate cancer. Exp Mol Pathol. 2000;68:65–9. doi: 10.1006/exmp.1999.2282. [DOI] [PubMed] [Google Scholar]
- 38.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–80. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 39.Wang L, Rhodes CJ, Lawrence JC., Jr Activation of mammalian target of rapamycin (mTOR) by insulin is associated with stimulation of 4EBP1 binding to dimeric mTOR complex 1. J Biol Chem. 2006;281:24293–303. doi: 10.1074/jbc.M603566200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.