Abstract
Circadian rhythms regulate diverse physiological processes including homeostatic functions of steroid hormones and their receptors. Perturbations of these rhythms are associated with pathogenic conditions, such as depression, diabetes, and cancer. Androgens play an important role in both normal development and carcinogenesis of the prostate. In the present study, we investigated a potential role for the core clock factor, Per1, in the pathogenesis of prostate cancer. Serum-shocked synchronized prostate cancer cells displayed disrupted circadian rhythms compared to the normal prostate tissue. Using Oncomine to perform a meta-analysis of microarray expression studies, we found that Per1 is downregulated in human prostate cancer samples compared with normal prostates. Reporter assays demonstrated that Per1 inhibited transactivation of the androgen receptor (AR) both in 293T cells overexpressing the AR and in the prostate cancer cell line, LNCaP. Forced expression of Per1 in LNCaP cells, diminished the expression of known androgen-sensitive genes following stimulation with dihydrotestosterone (DHT). We showed that Per1 physically interacted with AR; and in addition, we found that Per1 itself is regulated by androgens in prostate cancer cells. Overexpression of Per1 in prostate cancer cells resulted in significant growth inhibition and apoptosis. Our results support the emerging role of circadian genes as key players in malignant transformation. Further elucidating the connections between clock genes and the AR pathway could benefit the development of new therapeutic strategies for prostate cancer, as well as, provide insights into chronotherapy as a way to optimize current therapies.
Keywords: circadian rhythms, Per1, androgen receptor, prostate cancer
Introduction
Circadian rhythms reflect an internal timing system as exemplified by sleep/wake cycles. In mammals under the control of an intrinsic master clock, a multitude of physiological processes and behavioral patterns are directed by circadian rhythms (1). The mammalian circadian system is composed of three components: the input pathways, the central pacemaker and the output pathways. The input pathways transmit environmental signals to the central pacemaker which coordinates the external signals with the central endogenous rhythm of the body (2). Of the environmental signals, light is the most powerful circadian synchronizer. Light signals are captured by the retina and transmitted by the retino-hypothalamic tract to the suprachiasmatic nucleus (SCN) (3, 4). The SCN, located in the anterior hypothalamus acts as a master pacemaker generating neural and hormonal signals to peripheral clocks throughout the body (5, 6).
In the SCN, circadian rhythms are generated by a set of core clock genes including period (Per1-3), Casein kinase Iε (CKIε), Clock, Bmal1 and cryptochrome (Cry1-2) (2). Clock and Bmal1, two basic helix–loop–helix heterodimer transcriptional activators are positive regulators that bind E-boxes in the promoter of various genes including Per and Cry. Per and Cry proteins form oligomers that move from the cytoplasm to the nucleus where they interfere with Clock/Bmal1 activity, thereby forming the major negative circadian feedback loop. Further adding to the complexity, Clock/Bmal1 heterodimers induce the expression of the nuclear orphan receptor Rev-erbα, resulting in the repression of the transcription of Bmal1 through direct binding to a REV-ERB response element in the Bmal1 promoter (7, 8). In addition, posttranslational modification aid in regulating the circadian clock (9). Similar to the SCN, the molecular clockwork in peripheral cells is composed of autoregulatory transcription-translation feedback loops orchestrated by the clock genes (10, 11). The peripheral oscillators synchronized by the central clock, control the expression of downstream clock-controlled genes in a tissue-specific manner (12, 13).
Circadian rhythms influence many physiological processes and pathological conditions including cancer (14, 15). Epidemiological studies have shown that disruption of normal circadian rhythm may increase the risk of developing various cancer types such as breast, prostate, colorectal and endometrial cancers (16-21). Additionally, Per1 and Per2 have been reported to be deregulated in several human cancers (2). Furthermore, overexpression of either Per1 or Per2 in cancer cells inhibits their growth. Thus, Per genes may act as tumor suppressors.
Prostate cancer (PCa) is the most frequently diagnosed and the second leading cause of cancer mortality in males in the U.S.A (22). Androgens are necessary for male sexual differentiation and development, as well as the maintenance of sexual organs in the adult. In addition, androgens contribute to the development and progression of age-associated pathologies including benign prostatic hyperplasia and PCa. Androgen binds and activates the androgen receptor (AR) which acts as a transcription factor. Gene expression profiling studies show that approximately 1.5–4.3% of genes are either directly or indirectly regulated by androgens in PCa cells (23). These AR-regulated genes have a notable impact on diverse cellular processes. The aim of this study was to investigate a possible association between the core clock gene, Per1 and the AR signaling pathway in PCa.
Materials and Methods
Animals
Male BALB/c mice of 10 weeks of age were purchased from Vital River Laboratory Animal Technology Co. Ltd. (Beijing, China). Mice were housed in a temperature-controlled room under a 12 hrs-light/-dark cycle, given free access to water, and fed ad libitum on a standard chow. All animal care and procedures were in accordance with the institutional Animal Care and Use Committee guidelines. After being housed for 2-3 weeks, mice were sacrificed by cervical dislocation at 4 hrs intervals over 24 hrs with one mouse at each time point for each group. These experiments were done 3 times. Prostate tissue was dissected, rapidly frozen in liquid nitrogen, homogenized in TRIZOL reagent (Invitrogen) and stored at −80 °C until RNA extraction. Total RNA for Real-time PCR was extracted according to the manufacturer's protocol (Invitrogen).
Reagents
Dihydrotestosterone (DHT) was from Sigma-Aldrich (St. Louis, MO). Per1 (N-20), AR (N-20), PSA (C-19), myc (9E10), and PARP (H-250) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA); β-actin antibody was from Sigma-Aldrich (St. Louis, MO). Control immunoglobulin G (normal rabbit immunoglobulin G) (catalog no. sc-2027) was purchased from Santa Cruz Biotechnology.
Oncomine
Oncomine is a web-based data-mining platform www.oncomine.org (24). We compared Per1 expression in prostate cancers and normal prostate tissues with p<0.05 in 9 studies (Table S1).
Cell culture and transfection
LNCaP and 22Rv1 human prostate cancer cells were grown in RPMI 1640 (Invitrogen) with 10% fetal bovine serum and 100 U/ml penicillin/streptomycin. 293T human embryonic kidney cell line, as well as PC3 and DU145 human prostate cancer cell lines were cultured in Dulbecco's modified Eagle medium (Invitrogen) with 10% fetal bovine serum and 100 U/ml penicillin/streptomycin. Transient transfection assays of the 293T cells were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For transfection of LNCaP, 22Rv1 and DU145 cells, TransIT®-Prostate Transfection Kit (Mirus, Madison, WI) was used following the manufacturer's protocol. PC3 cells were transfected using Nucleofector Technology (Amaxa Biosystems, Cologne, Germany) with the Cell Line Nucleofector Kit-V according to the manufacturer's instructions.
Plasmids and luciferase reporter assay
Myc-tagged mPer1 was a generous gift of Dr. David M. Virshup as described (25). ARE4-E4Lux, which has the multimerized four consensus AREs from the PSA promoter cloned upstream of the luciferase gene in the pGL3 vector (Promega, Chicago, IL) and PSA P/E-Luc, which has a 564-bp fragment of the PSA promoter with a 2.4-kb enhancer sequence (−5322 to −2925) cloned upstream of luciferase were described previously (26). mPer1-7.2k luc was a kind gift of Dr. Hitoshi Okamura as described (27). Luciferase activity was measured with Dual-luc reporter 1000 assay system (Promega) and was normalized by Renilla values. All transfection experiments were carried out in triplicate wells and repeated separately at least three times.
Serum shock
Serum shock of prostate cancer cells was performed as described previously (28). PC3, DU145, LNCaP, 22Rv1 and NIH 3T3 cells were grown to 100% confluency. At Time point 0 (unstimulated cells), cells were incubated with 50% horse serum (Gibco) in culture medium and replaced with serum-free culture medium after 2 hrs of incubation. The cells were harvested at the indicated times and subjected to quantitative Real-time reverse transcription-PCR (RT-PCR) analysis.
RNAi
Small interfering RNA (siRNA) directed against Per1 (5′-CGCUCGCCCUGGCCAAUAAdTdT-3′ and 5′-UUAUUGGCCAGGGCGAGCGdGdG-3′) (synthesized by Qiagen) (29) and nonspecific control siRNA (Qiagen) were transfected into 293T using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Reverse transcription and quantitative Real-time PCR
Total RNA from cultured cells was extracted using Trizol reagent (Invitrogen). Two micrograms RNA were processed directly to cDNA by reverse transcription with Superscript III following the manufacturer's instructions (Invitrogen). PCR primers for each gene were designed using Real-time PCR Primer Design (https://www.genscript.com/ssl-bin/app/primer); sequences used in this study were shown in Table S2. We used SYBR®Premix Ex Taq™ (Perfect Real Time) (TAKARA BIO INC., Japan) for PSA, Nkx 3.1, B2M, cry1, Bmal1, Rev-erbα, Dbp and AR quantitative Real-time PCR with Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems) according to the manufacturer's instructions. The specificity of PCR products was checked on agarose gel. TaqMan probes were used in Quantitative Real-time PCR for Per1 and 18S rRNA as follow: 18S rRNA probe 5′-AGCAGGCGCGCAAATTACCC-3′, Per1 probe 5′-TCTACATTTCGGAGCAGGCAGCCG-3′. These were purchased from Applied Biosystems (Foster City, CA, USA) and were labeled with the reporter dye FAM in the 5′ end and the quencher dye BHQ in the 3′ end. Expression levels of 18S rRNA were used as an endogenous reference.
Western blotting and immunoprecipitation
Cell lysates were prepared using the lysis buffer [50 mmol/L Tris-HCl (pH 7.4), 150 mmol/L NaCl, 0.5% NP40]. For Western blotting, total cellular protein was separated on 4%–15% Ready-to-use SDS-PAGE gel (Bio-Rad) and transferred to nitrocellulose membranes which were then incubated with the primary antibodies. After incubation with appropriate secondary antibodies, the immunoblots were developed using SuperSignal Western blotting kits (Pierce Biotechnology, Rockford, IL) and exposed to X-ray film according to the manufacturer's protocol. Western blots were stripped between hybridizations with stripping buffer [10 mmol/L Tris-HCl (pH 2.3), 150 mmol/L NaCl]. For immunoprecipitation, 500 μg total cell lysates were immunoprecipitated with 3 μg of indicated antibodies at 4 °C, overnight. Immunocomplexes were captured with 20 μL of protein A/G gel slurry (Santa Cruz, CA), and the samples were allowed to mix at 4 °C for 2 hrs. The beads were washed three times
Nuclear extracts and electrophoretic mobility shift analyses (EMSA)
LNCaP cells were kept in RPMI without serum for 24 hrs and then treated either with or without DHT (10 nM) for another 24 hrs. Nucleoplasmic proteins were extracted using CelLytic™ NuCLEAR™ Extraction Kit (Sigma-Aldrich, St. Louis, MO). The nuclear extract (10 μg) was incubated at a final volume of 20 μL containing DNA-binding buffer [10 mmol/L Tris-HCl (pH, 7.5), 150 mmol/L NaCl, 1 mmol/L EDTA, 1 mmol/L DTT, 1 mmol/L phenylmethylsulfonyl fluoride, 20% v/v glycerol] and 1.5 μg of poly(deoxyiosinic-deoxycytidic acid) (Roche) for 10 min at room temperature and then incubated for 20 min at room temperature (30) with double-stranded 32P-labeled (specific activity of about 300,000 cpm) Per1-ARE oligonucleotide (5′-CAAGAGAACATGATGTTCCCAAG-3′) and Per1-ARE mutant oligonucleotide (5′-CAAGAAAAAATGATGTTCCCAAG-3′). In cold competition experiments, unlabeled competitor Per1-ARE oligos (100×)were incubated with nuclear extracts for 15 min on ice before the addition of the 32P-labeled probe.
Chromatin immunoprecipitation
LNCaP or 22Rv1 cells were kept in RPMI without serum for 24 hrs, treated with 10 nmol/L of DHT for 3 hrs, paraformaldehyde cross-linked and sonicated. Chromatin immunoprecipitation (ChIP) assays were done using EZChIP kit according to the manufacturer (Millipore, Billerica, MA). The following Per1 promoter-specific primers were used: 5′-GCAGATGGGAGTCCTGAAAA-3′ and 5′-GAGGCTGGAGAGACTGGAGA-3′. Primers for the PSA gene region ARE III were used as positive control (31).
Analysis of apoptosis and cell death
Cells were stained with propidium iodide and Annexin V-FITC (BD Biosciences). Briefly, after overexpression of either Per1 or empty vector, cells were selected in G418 (800 μg/ml for LNCaP, 500 μg/ml for PC3 and DU145) for over 1 week, resuspended in PBS and incubated with propidium iodide and Annexin V-FITC according to the manufacturer's instructions. Cells were analyzed by a Becton Dickinson FACScan flow cytometer using CellQuest software (Becton Dickinson).
Results
Rhythmic expression of clock genes and AR in normal prostate
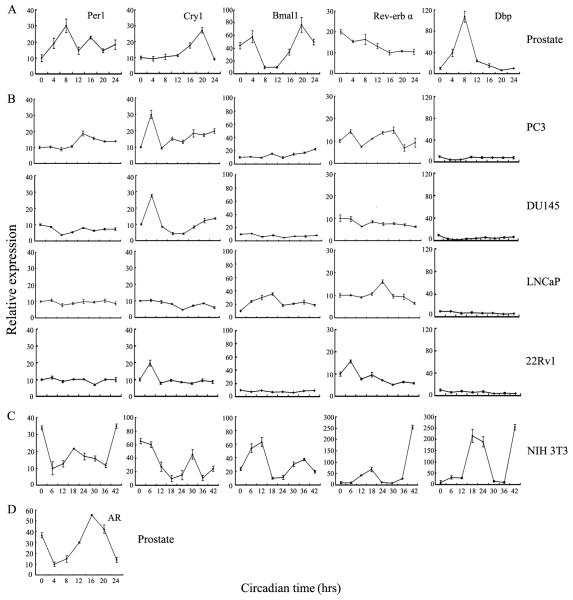
To begin to understand the relationship between circadian rhythm in normal and cancer prostate cells, we surveyed the expression of major oscillator genes, Per1, Cry1, Bmal1, and Rev-erbα, in normal prostate tissues by quantitative real time PCR (qPCR, Fig. 1A). Rhythmic expression of Per1, Cry1, Bmal1, and Rev-erbα, as well as Dbp [a PAR leucine zipper transcription factor known to be a clock controlled gene (32)] was noted. This result is in agreement with a recent study showing that core clock genes, Per1, Per2 and Bmal1, are rhythmically expressed in mouse prostate (33).
Figure 1.
Expression of clock genes in prostate tissue and prostate cancer cell lines. A, mice were entrained to 12 hrs-light/-dark cycles. Prostate tissue was collected at the indicated circadian times. Light was turned on and off at Zeitgeber 0 and 12, respectively. Levels of the indicated genes were quantified by Real-time PCR. B and C, PC3, DU145, LNCaP and 22Rv1 PCa cells and NIH 3T3 normal fibroblasts were synchronized by serum-shock. Samples were collected every 6 hrs for a total of 42 hrs. Expression of clock genes and Dbp were quantified by Real-time PCR. Results are means ± SD of three measurements. D, mRNA levels of AR expression in prostate tissue at the indicated circadian times were quantified by Real-time PCR. Data represent means ± SD of triplicate samples.
Rhythmic expression of core circadian genes can be induced in cultured cells by exposure to high concentration of serum (28, 29). To see whether core clock genes are rhythmically expressed in human PCa cell lines, we examined their expression in PC3, DU145, LNCaP and 22Rv1 cells (Fig. 1B). qPCR showed that the pattern of the clock gene expression was disrupted in PCa cell lines (Fig. 1B). Serum shocked NIH 3T3 cells were used as positive control, and as expected (34), showed rhythmic expression of the clock genes (Fig. 1C). Additionally, we found that AR mRNA levels oscillate in prostate tissue of mice entrained to 12 hrs-light/-dark cycles (Fig. 1D).
Downregulation of Per1 in PCa tissue
We performed in silico analysis of Per1 expression in human normal prostate, benign prostatic hyperplasia, prostate carcinoma and metastatic prostate cancer using microarray expression studies published in Oncomine (24). Per1 expression was significantly lower in PCa compared with normal prostate in either five (p<0.01) or nine (p<0.05) microarray expression studies (Fig. 2A and Table S1). Similar analysis of Per2 demonstrated that Per2 was also significantly downregulated in either four (p<0.01) or six (p<0.05) microarray expression studies (data not shown).
Figure 2.
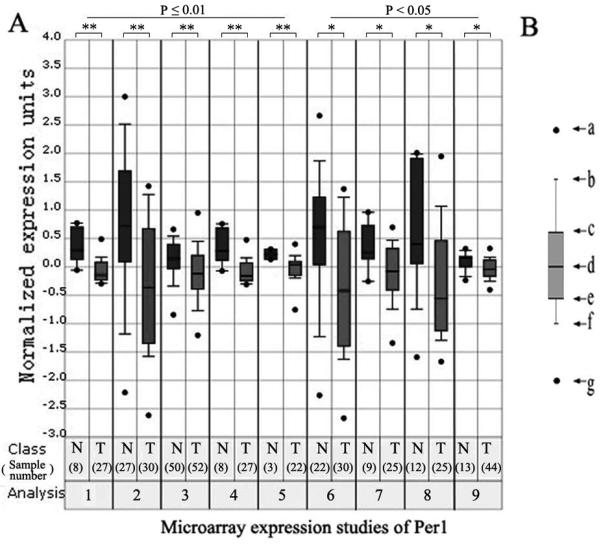
Downregulation of Per1 in prostate tumors (T) compared to normal prostate tissue (N). A, Data analysis were performed with the ONCOMINE 3.0 database (24) using standard settings (Table S1). B, Explanation of box plot. The dots in the box plots are the maximum and minimum values (a and g, respectively); the whiskers are the 90th and 10th percentile values (b and f, respectively); the horizontal lines in the box denote the 75th, 50th, and 25th percentile values (c, d and e, respectively).
Per1 inhibits the transcriptional activity of AR
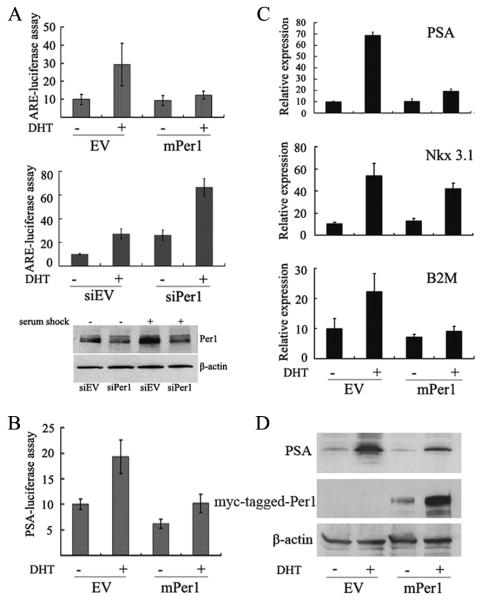
To examine whether Per1 affects the AR transcriptional activity, we performed reporter assays. 293T cells were cotransfected with promoter-reporter vector containing androgen response elements (ARE4-E4Luc), AR expression vector, and either the Per1 or empty vector, and treated with DHT. Results showed that Per1 inhibited DHT-induced luciferase activity (Fig. 3A, upper panel). Similarly, Per1 reduced luciferase activity in LNCaP cells (data not shown). In contrast, silencing of Per1 by siRNA increased the reporter activity about 3-fold in cultures either with or without DHT treatment (Fig. 3A, middle panel). Silencing of Per1 was confirmed by Western blotting (Fig. 3A, lower panel). Per1 also decreased AR-mediated stimulation of the reporter vector PSA P/E-Luc in LNCaP cells (Fig. 3B).
Figure 3.
Per1 suppresses the transcriptional activity of AR. A, upper panel: 293T cells were cotransfected with AR expression vector, ARE-luciferase reporter and either empty vector (EV) or Per1 (mPer1) expression vector. Luciferase activity was measured either with or without exposure of cells to DHT (10 nM, 24 hrs); middle panel, 293T cells were cotransfected with ARE-luciferase construct and either with control siRNA (siEV) or Per1 siRNA (siPer1). Effectiveness of Per1 siRNA is shown by Western blot (lower panel). B, LNCaP cells were cotransfected with PSA-luciferase construct and either empty vector or mPer1. For panels A and B, luciferase activity was assayed with either DHT (10 nM, 24 hrs) or diluent control. Results represent the fold-increase of luciferase activity compared to untreated control cells. Shown are means ± SD of triplicate samples. C and D, LNCaP cells were transfected with either empty vector or mPer1, and either untreated or treated with DHT. C. Real-time PCR analysis. Data represent means ± SD of triplicate samples. D. Western blot analysis.
Using the AR-positive PCa cell line, LNCaP, we measured the affect of Per1 on endogenous expression of known AR target genes, PSA, NKX3.1 and B2M. LNCaP cells were transfected with either Per1 or control vector and cultured ± DHT. qPCR analysis showed that while expression of Per1 alone had little effect on these genes, it strongly inhibited DHT-mediated induction of PSA and B2M (3.5- to 2-fold, respectively) and modestly depressed NKX3.1 (1.2-fold) expression compared to control cells (Fig. 3C). Per1 had a parallel effect on the protein level of PSA (Fig. 3D).
Per1 interacts with AR
To determine whether Per1 interacts with AR, 293T cells were cotransfected with myc-tagged mPer1 and either AR or empty vector. Immunoprecipitation experiments showed that Per1 is associated with AR (Fig. 4), and that DHT treatment does not affect the interaction.
Figure 4.
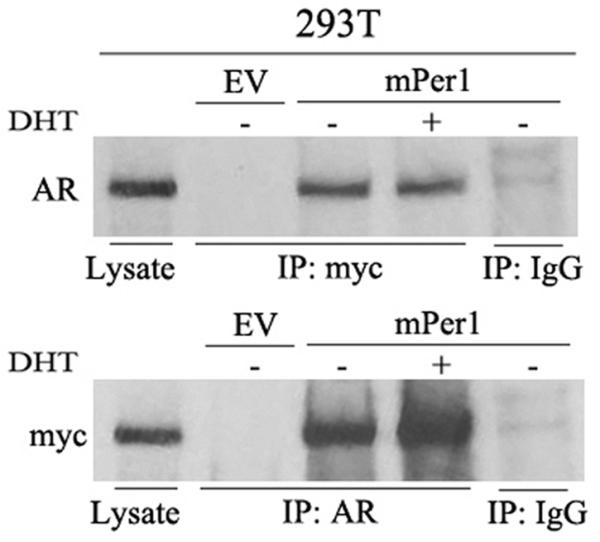
Per1 interacts with AR. 293T cells were co-transfected with AR and either EV or myc-tagged-mPer1 (upper), or the cells were co-transfected with myc-tagged-mPer1 and either EV or AR (lower). Cells were either untreated or treated with DHT (10 nM, 4 hrs). Lysates were immunoprecipitated (IP) and probed with the indicated antibodies. Cell lysates loaded directly were used to control for molecular size. IP with IgG was used as negative control.
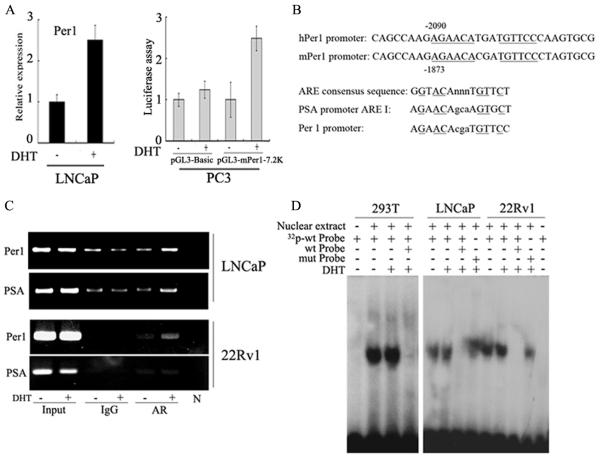
Per1 is induced by activated AR
We hypothesized that activated AR may stimulate transcription of Per1 as a feed-back pathway. Indeed, Per1 mRNA levels increased in LNCaP cells stimulated with DHT (Fig. 5A, left panel), showing that the human Per1 gene is DHT-inducible in prostate epithelial cells. Similarly, reporter assays using a mPer1-luciferase vector in PC3 cells, demonstrated that activated AR induces Per1 expression (Fig. 5A, Right panel). Sequence analysis showed that the Per1 promoter has a potential ARE binding site at −2090 bp from the start site of Per1 transcription; and that these and the surrounding nucleotides are conserved between the human and mouse, suggesting their functional significance (Fig. 5B). The highest percentage of homology to the ARE consensus sequence is the sequence AGAACAtgaTGTTCC. Compared with the ARE consensus GGTACAnnnTGTTCT (35), these sequences are identical in 6 of the most essential positions (positions 2, 3, and 5 in each half-site, underlined). Chromatin immunoprecipitation (Chip) assays using LNCaP and 22Rv1 cells detected the presence of AR at the same promoter region of Per1 following DHT treatment (Fig. 5C), demonstrating that endogenous AR binds to the Per1 promoter. Furthermore, electrophoretic mobility shift analyses demonstrated protein-DNA binding between the ARE element from the Per1 promoter and nuclear extracts from 293T overexpressing AR, as well as nuclear lysates from LNCaP and 22Rv1 PCa cells (Fig. 5D). Specificity of binding was confirmed by competition with excess unlabeled ARE oligonucleotides, which successfully competed for the binding, whereas a mutated Per1 ARE probe was not an effective competitor (Fig. 5D).
Figure 5.
Per1 is regulated by AR. A. Left panel: Real-time PCR analysis of Per1 expression. Data represent means ± SD of triplicate samples. Right panel: Reporter assay with Per1-luciferase construct. Cells were either untreated or treated with DHT (10 nM, 24 hrs). Data represent means ± SD of triplicate samples. B. Upper: Sequence alignment of the murine and human Per1 promoters. Potential ARE is underlined. Lower: ARE I in the PSA promoter and the putative ARE in the Per1 promoter are compared. C. ChIP assays. LNCaP and 22RV1 cells were either untreated or treated with DHT (10 nM, 3 hrs). D. EMSA. 293T transfected with AR, LNCaP and 22RV1 cells were either untreated or treated with DHT (10 nM, 24 hrs). Nuclear proteins were incubated with either wild type or mutant labeled probes of the putative ARE region from Per1 promoter. Cold competition assay was done using 100 × unlabeled wild type probe.
Per1 inhibits growth of PCa cells
To evaluate the effect of Per1 on proliferation of PCa cells, we transfected PC3, DU145 and LNCaP cells with either Per1-Neo expression vector or Neo control vector, and selected the cells with G418. Per1 induced profound growth inhibition in all 3 cell lines (Fig. 6A). In addition, apoptosis was measured by Annexin-V/PI staining. Compared to vector controls, Per1 transfected LNCaP, PC-3 and DU145 cells had 4.6-, 1.5- and 7.0-fold increased level of apoptosis, respectively (Fig. 6B).
Figure 6.
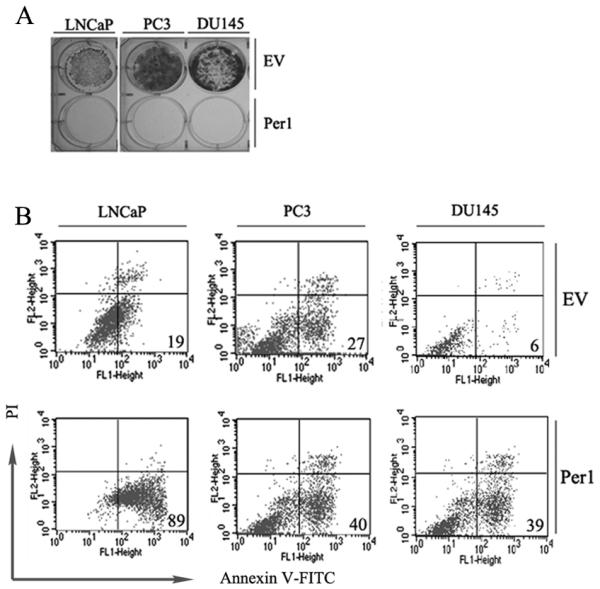
Per1 inhibits growth and induces apoptosis in PCa cell lines. LNCaP, PC3 and DU145 cells were transfected with either empty vector (EV) or Per1 expression vector followed by: A. Colony formation assay; B. Annexin-V/propidium iodide staining. The percentage of early apoptotic cells is shown. Results are representative from 3 independent experiments.
Discussion
Despite tremendous efforts to improve our understanding of PCa, its etiology remains largely unknown and it continues to be a major health problem. Older age, family history of the disease and race are well-established risk factors for the disease. Recent epidemiological data show that rotating shift workers have a higher risk of PCa compared with only day- or only night-shift workers (18, 19).
Oscillation of clock genes was found in various tissues including liver, skeletal muscle, white and brown adipose tissue (36). Also mammalian fibroblasts in vitro have functional clocks after their synchronization by either serum-shock (37) or other stimuli. On the other hand, deregulated circadian rhythm has been associated with accelerated growth of malignant tumors (38). We found a clear circadian expression of the core clock genes, Per1, Cry1, Bmal1 and Rev-erbα, in mouse prostate tissue. This result is in agreement with a recent report showing a similar expression profile of Per1 and Bmal1 in the prostate (33), as well as previously circadian patterns observed in the liver and muscle (36). In contrast, the rhythmic expression of these core clock genes was disrupted in PCa cell lines. Interestingly, genetic variants of clock genes have been associated with risk of development of PCa in a population-based study (39).
The SCN synchronizes the peripheral oscillators by producing diffusible neural and hormonal molecules, as well as directly targeting other regions of the brain, allowing animals to adapt their feeding, activity and metabolism to predictable daily changes in the environment (2). Studies show circadian rhythms in lipid and glucose metabolism coupled with rhythmic expression of several nuclear receptors in metabolically active tissues (36). A testosterone circadian rhythm has been reported which is usually absent in elderly men (40). Recently it was demonstrated that the mRNA levels of 28 of 49 murine nuclear receptors oscillate in metabolic tissues (36). In those metabolic tissues examined, AR did not show circadian expression. Remarkably, we found that AR mRNA levels oscillate in prostate tissue, suggesting that AR might link peripheral clocks to systemic hormonal regulation.
Using the Oncomine database, we found a number of studies in which Per1 levels were significantly downregulated in PCa samples compared with normal prostate tissue. Downregulation of Per1 and other clock genes was previously reported in a variety of cancers including acute myeloid leukemia (41), as well as breast (42), lung (43), endometrial (44) and pancreatic (45) cancers. We showed that forced expression of Per1 in PCa cells inhibited AR transcriptional activity, including DHT stimulation of PSA. Conversely, silencing of Per1 expression using siRNA was associated with increased transcriptional activity of AR. Also, Per1 physically bound to AR. Thus, Per1 appears to act as a negative regulator of AR in PCa cells. In addition, we showed that activated AR increased the transcription of Per1 associated with the recruitment of AR to the ARE of the Per1 promoter.
Taken together, our results suggest that activated AR stimulates Per1, which in turn attenuates AR activity, thereby helping to maintain hormonal homeostasis. Furthermore, our data support the hypothesis that disruption of circadian function may contribute to prostate tumorigenesis. Further elucidating the circadian-AR connections could pave the way for the development of novel therapeutic approaches.
Supplementary Material
Acknowledgements
Grant support: NIH grant, H. and C. Koeffler funds, the Inger Foundation and the China Scholarship Council.
We thank Dr. David M. Virshup for the Myc-tagged mPer1 vector and Dr. Hitoshi Okamura for the mPer1-7.2k luciferase reporter. We also want to thank Chunzhang Yang for the help with the Real-time PCR analysis.
References
- 1.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 2.Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–361. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 3.Gooley JJ, Lu J, Chou TC, Scammell TE, Saper CB. Melanopsin in cells of origin of the retinohypothalamic tract. Nat Neurosci. 2001;4:1165. doi: 10.1038/nn768. [DOI] [PubMed] [Google Scholar]
- 4.Ruby NF, Brennan TJ, Xie X, et al. Role of melanopsin in circadian responses to light. Science. 2002;298:2211–2213. doi: 10.1126/science.1076701. [DOI] [PubMed] [Google Scholar]
- 5.Liu C, Weaver DR, Strogatz SH, Reppert SM. Cellular construction of a circadian clock: period determination in the suprachiasmatic nuclei. Cell. 1997;91:855–860. doi: 10.1016/s0092-8674(00)80473-0. [DOI] [PubMed] [Google Scholar]
- 6.Yamazaki S, Numano R, Abe M, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 7.Albrecht U, Eichele G. The mammalian circadian clock. Curr Opin Genet Dev. 2003;13:271–277. doi: 10.1016/s0959-437x(03)00055-8. [DOI] [PubMed] [Google Scholar]
- 8.Brown SA, Ripperger J, Kadener S, et al. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science. 2005;308:693–696. doi: 10.1126/science.1107373. [DOI] [PubMed] [Google Scholar]
- 9.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 10.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 11.Shearman LP, Sriram S, Weaver DR, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 12.Panda S, Antoch MP, Miller BH, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 13.Miller BH, McDearmon EL, Panda S, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci U S A. 2007;104:3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 15.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 16.Hansen J. Increased breast cancer risk among women who work predominantly at night. Epidemiology. 2001;12:74–77. doi: 10.1097/00001648-200101000-00013. [DOI] [PubMed] [Google Scholar]
- 17.Davis S, Mirick DK, Stevens RG. Night shift work, light at night, and risk of breast cancer. J Natl Cancer Inst. 2001;93:1557–1562. doi: 10.1093/jnci/93.20.1557. [DOI] [PubMed] [Google Scholar]
- 18.Kubo T, Ozasa K, Mikami K, et al. Prospective cohort study of the risk of prostate cancer among rotating-shift workers: findings from the Japan collaborative cohort study. Am J Epidemiol. 2006;164:549–555. doi: 10.1093/aje/kwj232. [DOI] [PubMed] [Google Scholar]
- 19.Conlon M, Lightfoot N, Kreiger N. Rotating shift work and risk of prostate cancer. Epidemiology. 2007;18:182–183. doi: 10.1097/01.ede.0000249519.33978.31. [DOI] [PubMed] [Google Scholar]
- 20.Schernhammer ES, Laden F, Speizer FE, et al. Night-shift work and risk of colorectal cancer in the nurses' health study. J Natl Cancer Inst. 2003;95:825–828. doi: 10.1093/jnci/95.11.825. [DOI] [PubMed] [Google Scholar]
- 21.Viswanathan AN, Hankinson SE, Schernhammer ES. Night shift work and the risk of endometrial cancer. Cancer Res. 2007;67:10618–10622. doi: 10.1158/0008-5472.CAN-07-2485. [DOI] [PubMed] [Google Scholar]
- 22.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer Statistics, 2009. CA Cancer J Clin. 2009 doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 23.Dehm SM, Tindall DJ. Molecular regulation of androgen action in prostate cancer. J Cell Biochem. 2006;99:333–344. doi: 10.1002/jcb.20794. [DOI] [PubMed] [Google Scholar]
- 24.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vielhaber E, Eide E, Rivers A, Gao ZH, Virshup DM. Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon. Mol Cell Biol. 2000;20:4888–4899. doi: 10.1128/mcb.20.13.4888-4899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hisatake JI, Ikezoe T, Carey M, Holden S, Tomoyasu S, Koeffler HP. Down-Regulation of prostate-specific antigen expression by ligands for peroxisome proliferator-activated receptor gamma in human prostate cancer. Cancer Res. 2000;60:5494–5498. [PubMed] [Google Scholar]
- 27.Yamaguchi S, Mitsui S, Miyake S, et al. The 5' upstream region of mPer1 gene contains two promoters and is responsible for circadian oscillation. Curr Biol. 2000;10:873–876. doi: 10.1016/s0960-9822(00)00602-3. [DOI] [PubMed] [Google Scholar]
- 28.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 29.Cai W, Rambaud J, Teboul M, et al. Expression levels of estrogen receptor beta are modulated by components of the molecular clock. Mol Cell Biol. 2008;28:784–793. doi: 10.1128/MCB.00233-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zoubeidi A, Zardan A, Beraldi E, et al. Cooperative interactions between androgen receptor (AR) and heat-shock protein 27 facilitate AR transcriptional activity. Cancer Res. 2007;67:10455–10465. doi: 10.1158/0008-5472.CAN-07-2057. [DOI] [PubMed] [Google Scholar]
- 31.Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol Cell. 2002;9:601–610. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 32.Lopez-Molina L, Conquet F, Dubois-Dauphin M, Schibler U. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. Embo J. 1997;16:6762–6771. doi: 10.1093/emboj/16.22.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bebas P, Goodall CP, Majewska M, Neumann A, Giebultowicz JM, Chappell PE. Circadian clock and output genes are rhythmically expressed in extratesticular ducts and accessory organs of mice. Faseb J. 2009;23:523–533. doi: 10.1096/fj.08-113191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marquez S, Crespo P, Carlini V, et al. The metabolism of phospholipids oscillates rhythmically in cultures of fibroblasts and is regulated by the clock protein PERIOD 1. Faseb J. 2004;18:519–521. doi: 10.1096/fj.03-0417fje. [DOI] [PubMed] [Google Scholar]
- 35.Cleutjens KB, van Eekelen CC, van der Korput HA, Brinkmann AO, Trapman J. Two androgen response regions cooperate in steroid hormone regulated activity of the prostate-specific antigen promoter. J Biol Chem. 1996;271:6379–6388. doi: 10.1074/jbc.271.11.6379. [DOI] [PubMed] [Google Scholar]
- 36.Yang X, Downes M, Yu RT, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 37.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 38.Filipski E, King VM, Li X, et al. Host circadian clock as a control point in tumor progression. J Natl Cancer Inst. 2002;94:690–697. doi: 10.1093/jnci/94.9.690. [DOI] [PubMed] [Google Scholar]
- 39.Chu LW, Zhu Y, Yu K, et al. Variants in circadian genes and prostate cancer risk: a population-based study in China. Prostate Cancer Prostatic Dis. 2008;11:342–348. doi: 10.1038/sj.pcan.4501024. [DOI] [PubMed] [Google Scholar]
- 40.Seidman SN. Androgens and the aging male. Psychopharmacol Bull. 2007;40:205–218. [PubMed] [Google Scholar]
- 41.Gery S, Gombart AF, Yi WS, Koeffler C, Hofmann WK, Koeffler HP. Transcription profiling of C/EBP targets identifies Per2 as a gene implicated in myeloid leukemia. Blood. 2005;106:2827–2836. doi: 10.1182/blood-2005-01-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ, Chang JG. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26:1241–1246. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- 43.Gery S, Komatsu N, Kawamata N, et al. Epigenetic silencing of the candidate tumor suppressor gene Per1 in non-small cell lung cancer. Clin Cancer Res. 2007;13:1399–1404. doi: 10.1158/1078-0432.CCR-06-1730. [DOI] [PubMed] [Google Scholar]
- 44.Yeh KT, Yang MY, Liu TC, et al. Abnormal expression of period 1 (PER1) in endometrial carcinoma. J Pathol. 2005;206:111–120. doi: 10.1002/path.1756. [DOI] [PubMed] [Google Scholar]
- 45.Pogue-Geile KL, Lyons-Weiler J, Whitcomb DC. Molecular overlap of fly circadian rhythms and human pancreatic cancer. Cancer Lett. 2006;243:55–57. doi: 10.1016/j.canlet.2005.11.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.