Abstract
Despite advancements in knowledge from over a century of metastasis research, the genetic programs and molecular mechanisms required for cancer metastasis are still incompletely understood. Genes that specifically regulate the process of metastasis are useful tools to elucidate molecular mechanisms and may become markers and/or targets for anti-metastatic therapy. Recently, several non-coding regulatory RNA genes, microRNA (miRNA), were identified that play roles in various steps of metastasis, some without obvious roles in tumorigenesis. Understanding how these metastasis-associated miRNA, which we term metastamir, are involved in metastasis will help identify possible biomarkers or targets for the most lethal attribute of cancer - metastasis.
Keywords: microRNA, metastamir
Introduction
Cancer is currently the second leading cause of death in the United States and, if current trends continue, will soon be the leading killer. The major reasons for cancer deaths are complications arising from metastasis. Therefore, improved morbidity and mortality will require effective treatments targeting metastatic disease. Despite many advancements in knowledge from over a century of researching metastasis, the molecular mechanisms are still not completely understood. Several reports have described the involvement of a recently discovered class of non-coding regulatory RNA, termed microRNA (miRNA), in the regulation of cancer (oncomir; (1, 2)). More recently, a specialized family of miRNA, which we call metastamir, have been shown to have pro-and anti-metastatic effects.
miRNA were originally discovered because of their roles in controlling the timing of C. elegans larval development. Less than a decade later they were identified in plant and mammalian cells (see (3) for review. Typically, pri-miRNA are transcribed by RNA polymerase II before capping, polyadenylation and maturation of a hairpin loop structure by ribonuclease 3 (Drosha) into pre-miRNA. Following export into the cytoplasm, the pre-miRNA become associated with several ribonucleoproteins of the RISC (RNA-induced silencing complex), including Dicer and Argonaut family members from which a mature miRNA are formed. miRNA complement the 3′-UTR of mRNA in order to impair translation or alter message stability. Because of their small size, miRNA are predicted to be promiscuous and may have several hundred mRNA targets, meaning that a single miRNA can, by itself, impact the expression of hundreds of proteins (See (3) for review).
Metastasis involves multiple steps and multiple genes in which neoplastic cells dissociate from the primary tumor, enter body cavities or, more commonly, circulatory systems (lymphatics or blood vasculature), survive during transport until they arrest at discontiguous sites, exit the circulation, and proliferate at ectopic sites (colonization) in response to local growth factors (4). The process is extremely inefficient (of the ∼4 million cells entering the vascular compartment per gram of tumor per day, much less than 0.01% develop macroscopic masses elsewhere (4)). The inefficiency is perhaps because every step in the metastatic cascade is selective and rate-limiting (i.e., failure to complete any step precludes subsequent steps). Each step in metastasis requires coordinated temporal expression of genes and spacio-temporal expression of proteins.
Examination of mRNA expression patterns has yielded sometimes conflicting results related to roles in metastasis, prompting some to question even the existence of metastasis-regulatory genes. Yet, multiple labs, using several different human and rodent model systems, demonstrated the existence of gene products that affect metastasis without promoting or inhibiting tumorigenicity at orthotopic sites (5). So, while tumor formation is prerequisite to metastasis, tumorigenicity and metastasis are distinct phenotypes, the latter requiring genetic changes superimposed upon those needed to make the tumor. These considerations led us and others to predict the existence of metastamirs.
The invitation to write this mini-review was prompted by our discovery that the miR-146 family of miRNA could profoundly inhibit invasion and metastasis of MDA-MB-231 human breast carcinoma cells. In that report, we further showed that miR-146a/b was downstream of the BRMS1 metastasis suppressor and intermediate to BRMS1-regulated genes (6). Concurrently, we have shown that BRMS1 coordinately regulates entire families of metastamirs - up-regulating metastasis-suppressing miRNA and down-regulating metastasis-promoting miRNA (7).
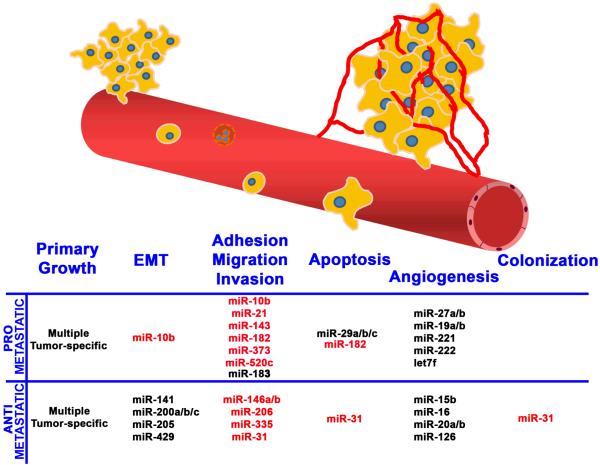
Those findings, coupled with an explosion of papers describing miRNA and metastasis-associated steps compelled us to expand the focus of this mini-review to consider the state of the field. To date, eleven miRNA have been shown to promote or inhibit metastasis in experimental models (Figure 1) and the number is likely to grow even further because more than 20 more have been shown to impact critical steps in the metastatic cascade, such as epithelial-mesenchymal transition (EMT), apoptosis, and angiogenesis (Figure 1). Furthermore, several clinical studies have identified correlations between miRNA expression and recurrence, development of metastases and/or survival (for a recent review, see (8)). Therefore, our goal is to focus on the evidence for metastamirs, the implications of their existence and some technical and theoretical considerations that emerge from their discovery.
Figure 1.
Critical steps in metastasis altered by metastamir. Pro- and anti-metastatic metastamir are listed with the steps in the metastatic cascade of which they affect. The metastamir that have been functionally tested for metastasis in vivo are highlighted in red.
Discovery of metastamir
In retrospect, it was self-fulfilling that miRNA regulating metastasis would be found because the process itself involves hundreds of genes. To date, metastamirs have typically been discovered using in vitro screens for steps in the metastatic cascade including cell growth, EMT, adhesion, migration, invasion, apoptosis and/or angiogenesis. Most commonly, metastamirs promoting cell migration and invasion have been described. Figure 1 shows a relatively current listing of metastamir impacting the cascade, but highlights (in red) those for which actual functional data have been collected for metastasis in vivo. The latter point is critical since it is not possible to study metastasis in vitro. Yet, at a conceptual level, it should be possible to design in vivo screens using miRNA or antagomir (miRNA antagonists) libraries to discover metastasis-promoting or metastasis-suppressing metastamir. Antagomir studies will depend upon yet-to-be-perfected technology to stably knock-down miRNA expression. Screening for metastamir in vivo would be cost-prohibitive in most laboratories; so, in vitro surrogates make economical sense.
Another interesting point is that the vast majority of metastamir have been identified in breast/mammary tumor cell lines. The reasons for this preponderance may be associated with funding levels or availability of robust metastasis models. Nonetheless, the number of metastamir identified in other tumor types is likely to expand in the near future. Until then, it is important not to summarily extrapolate function in breast tumors to all histiotypes.
Metastasis-suppressing metastamir
miR-335 and -206
The first suppressing metastamir was identified in Joan Massague’s lab by Tavazoie et al., who compared miRNA expression in metastatic variants derived from the human breast carcinoma cell line, MDA-MB-231 (9). They identified six miRNA with a low relative expression in the metastatic cells. Three of these, miR-335, -126, and -206, suppressed metastasis in vivo, however, miR-126 also inhibited cell proliferation and tumorigenesis. Therefore, we did not include miR-126 in our list of metastasis suppressors. Both miR-335 and -206 inhibited invasion and migration in vitro. miR-335 targets SOX4 (SRY-box containing transcription factor), PTPRN2 (receptor type tyrosine protein phosphatase), MERTK (c-Mer tyrosine kinase), and possibly TNC (tenascin C). Additionally, inhibition of SOX4 or TNC by shRNA inhibited invasion in vitro and metastasis in vivo. Their findings elegantly demonstrate how a single miRNA could impact several downstream pathways by arborizing signaling pathway components. There was also a clinical association of miR-335 expression with metastasis-free survival in a set of 20 primary breast tumor samples.
miR-146a/b
Several groups have shown a role for miR-146 in inflammation through regulation of NfkB (10). Although miR-146a and b are encoded on different chromosomes, their mature sequence differs by only two nucleotides at the 3′ region. So their mRNA targets are predicted to overlap significantly. Indeed, both miR-146a and b inhibit invasion and migration of breast cancer cells by down-regulating NFkB by targeting IRAK1 and TRAF6 (11). These studies were extended in vivo by demonstrating miR-146a and b suppressed metastasis that may involve targeting of EGF receptor (6) or ROCK1 (12), both of which are involved in promoting invasion and metastasis. miR-146a expression is inversely correlated with prostate cancer progression (12).
miR-31
Inhibition of any single step in metastasis results in metastasis suppression. Inhibition of multiple steps would therefore result in more robust inhibition of the metastatic process. miR-31 inhibits multiple steps of metastasis including invasion, anoikis, and colonization leading to a 95% reduction in lung metastasis in an orthotopic model of breast cancer (13). Clinically, miR-31 levels were lower in breast cancer patients with metastasis (n = 56 patients).
Metastasis promoting metastamir
miR-10b
Ma, Weinberg and colleagues were the first to discover a metastamir (14). They hypothesized that certain miRNA could regulate specific stages of tumor progression and found that miR-10b was highly expressed only in metastatic breast cancer cell lines compared to primary human mammary epithelial or spontaneously immortalized cells. After showing that miR-10b enhanced migration and invasion in vitro and metastasis in vivo, they identified a pathway where the pro-metastatic gene TWIST1 up-regulates miR-10b that targets HOXD10 leading to an increase in RHOC. Additionally, RTQ with 23 primary breast tumors was used to show a general increase in miR-10b expression in patients with metastasis.
Interestingly, the BRMS1 metastasis suppressor that regulates miR-146a/b also regulates TWIST, miR-10b and RHOC expression (7). Whether the regulation of these genes by BRMS1 is direct or indirect is still not known. Regardless, the data all point to common pathways impacted by these metastasis regulatory molecules. Perhaps hopefully, with additional experimentation, we will be able to find a point of convergence for several signaling cascades important in metastasis.
miR-373 and -520c
Huang, Agami and colleagues transduced a non-metastatic MCF7 human breast cancer cells with a miRNA expression library and screened the transductants using a transwell migration assay (15). Both miR-373 and -520c promoted migration and were subsequently found to increase in vivo metastasis at least in part by targeting the adhesion molecule, CD44. Clinically, miR-373 expression was higher in lymph-node metastasis compared with the primary tumors from 11 pairs of matched samples.
miR-21, -143 and -182
Invasion and migration are increased while apoptosis is decreased by miR-21 expression in multiple model systems (breast cancer, colon cancer, and glioma) (16-18). miR-21 was found to target TPM1 (tropomyosin 1), PDCD4 (programmed cell death 4) and regulators of matrix metalloproteinases. miR-143 and miR-182 promoted hepatocellular carcinoma and melanoma, respectively (19, 20). miR-143 is up-regulated by NFkB and decreases adhesion. miR-182’s effects can be reversed by re-expression of MITF (microphthalamia-associated transcription factor M) or FOXO3. miRNA-182 is part of a cluster (miR-183-96-182). Many miRNA are encoded as genetically linked clusters and are expressed as a single pri-miRNA. As a result it is not always possible to distinguish biological effects that are the result of a single miRNA or the collective actions of multiple miRNA.
Metastamir pathways, concepts and future directions
While metastamir have only been recognized for slightly more than two years, the explosive rate of discovery of this important family of molecules is impressive. Metastamirs are components of complex pathways and are often expressed downstream of pro-or anti-metastatic signals, including pathways regulated by NFkB, EGFR, TWIST1, BRMS1, ZEB1/2 and HIF1α (8). Unfortunately, the mechanisms by which miRNA are regulated still remain relatively ill-defined (21), and will be the subject of intense future investigation.
Interestingly, positive and negative feedback loops have been found whereby the upstream effectors are themselves targets of the miRNA that they regulate. This implies an important role for metastamir in modulating key signaling pathways involved in metastasis. Because of their position as nodes within these pathways and their promiscuity with regard to downstream targets, each metastamir can (and probably does) multiply pro- and anti-metastatic signaling events. It is likely that metastamir regulation of these signaling events are context dependent, relying on microenvironmental cues in both directions. We predict that yet-to-be-discovered co-factors will lead to specificity of miRNA effects on selected pathways; however, their existence is speculation at this time.
We find ourselves in the midst of a revolution with regard to the biochemical and molecular regulation of cancer metastasis. Old notions of identity equating tumorigenicity with metastasis have to be discarded. There are clear distinctions between the phenotypes; biologically, biochemically and genetically. Understanding the interrelationships between regulatory genes and gene products (proteins and non-coding RNA) and how these are modulated by the microenvironmental context is beginning to unravel the complex tapestry that is cancer metastasis.
Acknowledgments
Grant support: USPHS grants CA87728 (D.R. Welch) and F32CA113037 (D.R. Hurst); predoctoral fellowship from the U.S. Army Medical Research and Materiel Command W81-XWH-08-1-0786 (M.D. Edmonds); and National Foundation for Cancer Research, Center for Metastasis Research (D.R. Welch).
References
- (1).Negrini M, Nicoloso MS, Calin GA. MicroRNAs and cancer--new paradigms in molecular oncology. Curr Opin Cell Biol. 2009;21:470–9. doi: 10.1016/j.ceb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- (2).Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nature Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- (3).Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- (4).Eccles SA, Welch DR. Metastasis: recent discoveries and novel treatment strategies. Lancet. 2007;369:1742–57. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nature Rev Cancer. 2009;9:274–U65. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- (6).Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 BRMS1 up-regulates miR-146 that suppresses breast cancer metastasis. Cancer Res. 2009;69:1279–83. doi: 10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Edmonds MD, Hurst DR, Vaidya KS, Stafford LJ, Chen D, Welch DR. Int J Cancer. 2009. Breast Cancer Metastasis Suppressor 1 (BRMS1) coordinately regulates metastasis-associated microRNA expression. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs - the micro steering wheel of tumour metastases. Nature Rev Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- (9).Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang QQ, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147. doi: 10.1038/nature06487. NIL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nature Immunol. 2008;9:839–45. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- (11).Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008;42:5643–7. doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Lin SL, Chiang A, Chang D, Ying SY. Loss of mir-146a function in hormone-refractory prostate cancer. RNA. 2008;14:417–24. doi: 10.1261/rna.874808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–46. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- (14).Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- (15).Huang QH, Gumireddy K, Schrier M, LeSage C, Nagel R, Nair S, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nature Cell Biol. 2008;10:202. doi: 10.1038/ncb1681. NIL. [DOI] [PubMed] [Google Scholar]
- (16).Asangani IA, Rasheed SAK, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- (17).Zhu SM, Wu HL, Wu FT, Nie DT, Sheng SJ, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–9. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- (18).Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–80. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zhang X, Liu S, Hu T, Liu S, He Y, Sun S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 2009 doi: 10.1002/hep.23008. [DOI] [PubMed] [Google Scholar]
- (20).Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci. 2009;106:1814–9. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nature Cell Biol. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]