Abstract
The rapid proliferation of cancer cells mandates a high protein turnover. The endoplasmic reticulum (ER) is intimately involved in protein processing. An accumulation of unfolded or misfolded proteins in the ER leads to a cascade of transcriptional and translational events collectively referred to as the unfolded protein response (UPR). Protein disulfide isomerase (PDI) is one of the most abundant ER proteins and maintains a sentinel function in organizing accurate protein folding. Treatment of cells with PABA/NO (O2- [2,4-dinitro-5- (N-methyl-N-4-carboxyphenylamino) phenyl] 1-N, N-dimethylamino) diazen-1-ium-1, 2-diolate) resulted in a dose dependent increase in intracellular NO that caused S-glutathionylation of various proteins. Within 4h, PABA/NO activated the UPR and led to translational attenuation as measured by the phosphorylation and activation of the ER transmembrane kinase, PERK, and its downstream effector eIF2 in human leukemia (HL60) and ovarian cancer cells (SKOV3). Cleavage of the transcription factor, XBP-1 and transcriptional activation of the ER resident proteins, BiP, PDI, GRP94 and ERO1 (5-10 fold induction) also occurred. Immunoprecipitation of PDI showed that while nitrosylation was undetectable, PABA/NO treatment caused S-glutathionylation of PDI. Mass spectroscopy analysis showed that single cysteine residues within each of the catalytic sites of PDI had a mass increase [+305.3 Da] consistent with S-glutathionylation. Circular dichroism confirmed that S-glutathionylation of PDI results in alterations in the alpha-helix content of PDI and is concurrent with inhibition of its isomerase activity. Thus, it appears that S-glutathionylation of PDI is an upstream signaling event in the UPR and may be linked with the cytotoxic potential of PABA/NO.
Introduction
Protein S-glutathionylation is increasingly recognized as an important mechanism underlying redox regulation of signaling pathways with downstream impact on numerous cell functions (1, 2). Cysteine residues localized in a basic environment (vicinal to lys, arg or his) can have low pKa values, making them targets for the addition of glutathione (GSH), i.e. can be S-glutathionylated. This modification adds ∼305Da and introduces a net negative charge to the protein (as a consequence of the addition of glu). Critical in ascribing any regulatory function to this process is the reversibility of S-glutathionylation by small molecule proteins such as glutaredoxin and sulfiredoxin (3-5). Our recent studies suggest that glutathione S-transferase pi (GSTP) is involved in mediating the forward reaction of this cycle (6). Glutaredoxin (Grx) is reportedly involved in both the forward and reverse steps (7). Protein disulfide isomerase (PDI) is a CXXC-motif containing protein implicated (albeit in a minor role) in deglutathionylation, (8). PDI is a 57 kDa member of the thioredoxin superfamily organized into 5 domains (a, b, b', a' and c) with a C-terminal KDEL sequence that targets it to the ER (9) where it is involved in protein folding through disulfide bond formation and isomerization. PDI contains two active sites in the a and a' domains each having two conserved cysteine residues within the motif CGHC that cycle between oxidized (disulfide) and reduced (dithiol) states (10). PDI has enzymatic functions that facilitate correct folding of proteins in the ER, e.g. isomerase activity that catalyzes the rearrangement of incorrectly formed disulfide bonds and oxidase activity that introduces disulfides into proteins. PDI can also participate in multimeric protein clusters as a subunit of prolyl-4-hydroxylase and microsomal triglyceride transfer protein. In addition, cell surface PDI can be involved in cellular entry antigen processing (11), complexes with integrin receptor in regulating cell adhesion (12), glioma cell invasion (13), attachment and infectivity of Chlamydia (14), promotion of disulfide bond rearrangements in HIV-1 envelope protein that accompanies viral entry (15) and may even be a determinant of drug resistance in malignant B lymphocytes (16).
PABA/NO is among a series of prodrugs that incorporate diazeniumdiolate anions that release nitric oxide (NO) in a kinetically favorable manner resulting in nitrosative stress in cells (17, 18). While NO has significantly broad biological consequences in mammalian cells, we have been interested in its capacity to induce S-glutathionylation of proteins. Moreover, PABA/NO has antitumor potential both in vitro and in mice (19), suggesting that there is value in developing this agent as a chemotherapeutic. As such, there is a need to understand how its pharmacological effects may confer cytotoxicity.
Many anticancer drugs have electrophilic centers that can cause direct and indirect oxidative or nitrosative stress. These agents tend to have relatively non-specific intracellular targets that when effected result in apoptotic cell death. Recently, a new consensus is emerging that the endoplasmic reticulum (ER) may be a common target for unrelated electrophilic species [1]. Dysregulation of ER homeostasis may cause a disruption of protein folding with the consequence that mis-folded proteins may accumulate in the ER lumen and trigger activation of the unfolded protein response (UPR). Drug induced disruption of the components of the various steps that constitute this pathway may play a pivotal role in inducing cell death, particularly in tumor cells. As a consequence, there is a need to clarify those molecular events that may link drug effects with aberrations in the UPR and produce cytotoxicity.
Stress emanating from the ER results in the accumulation of unfolded or misfolded proteins such that hydrophobic amino acid side chains are exposed on the protein's surface. This “misfolding” can result in a protein that does not maintain native interactions or alternatively, forms insoluble aggregates. Protein folding in the ER is a crucial process and multiple signaling pathways have evolved to ensure quality control. The UPR leads to a cascade of transcriptional and translational events that include three mechanisms, transcriptional activation of UPR-induced proteins (including PDI and other chaperones); translational attenuation to decrease the flux of nascent polypeptides imported; ER associated degradation (ERAD) of terminally misfolded proteins via translocation to the cytosol for proteosomal degradation. The ER membrane contains three signal-transducing proteins that modulate the UPR: pancreatic ER kinase (PERK); activating transcription factor (ATF); Inositol requirement 1 (Ire1). Each is negatively regulated during homeostasis by a glucose regulated protein 78 (GRP78 / BiP) through protein:protein interactions. An accumulation of unfolded or misfolded proteins results in the dissociation of BiP and activation of the membrane proteins. PERK activates an immediate response to ER stress that results in a transient attenuation of mRNA translation through phosphorylation of eukaryotic initiation factor, eIF2. PERK is associated with activation of two transcription factors, ATF4 and NF-E2 related factor (Nrf2), which regulate downstream UPR genes and antioxidant cellular defense pathways. IRE1 is a site-specific endonuclease that cleaves a small intron in the mRNA of X-box protein 1 (XBP-1), a bZIP transcription factor, leading to mRNA stabilization. Both IRE1 and ATF6 mediate the UPR through regulation of gene products with ER and antioxidant stress response elements.
When the UPR is unable to counter-balance the protein folding capacity, apoptotic pathways are activated (20). UPR-induced apoptosis involves activation of JNK and dissociation of the TRAF2-apoptosis signal-regulating kinase1 (ASK1) complex and transcriptional activation of the gene for C/EBP homologous protein (CHOP). IRE1 and ATF6 bind to the promoter of the apoptotic protein, CHOP. CHOP is a 29 kDa protein that was also described as growth arrest- and DNA-damage-inducible gene 153 (GADD153). CHOP can be phosphorylated and activated on Ser 79 and 82 by the p38 MAP kinase family and lead to cell cycle arrest (21). Overexpression of CHOP leads to Bax translocation from the cytosol to the mitochondria. IRE1 has also been shown to be activated itself by proapototic Bcl-2 members, Bax and Bak through protein-protein interactions in the cytoplasmic domain (22). Bax and Bak have been localized in the mitochondria and ER membrane where they are proposed to contribute to membrane permeability (23).
Using a broad-spectrum proteomic approach, we have shown previously that PDI is S-glutathionylated when HL60 cells are exposed to PABA/NO (24). In the present study, we examine the specific susceptible molecular targets within PDI and how that may alter structure and function with cellular consequences that may directly invoke the UPR.
Materials and Methods
Chemicals
Reduced glutathione (GSH), oxidized glutathione (GSSG), and the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reagent were purchased from Sigma (St. Louis, MO). DAF-FM-DA (3-Amino, 4-aminomethyl-2′,7′-difluorofluorescein Diacetate), was purchased from Invitrogen (Carlsbad, CA). Thapsigargin was purchased from Calbiochem (San Diego, CA). PABA/NO was a gift from Dr. Larry Keefer from the Chemistry Section, Laboratory of Comparative Carcinogenesis, NCI at Frederick (Maryland). Antibodies were purchased from the following sources: anti-PDI (Affinity BioReagents, Golden, CO); anti-glutathionylation (Virogen, Watertown, MA); anti-phospho-c-Jun N-terminal kinase (JNK) (Promega, Madison, WI); anti-PKR-like ER kinase (PERK), anti-eukaryotic initiation factor2 (eIF2) and phospho-specific PERK and eIF2 (Santa Cruz Biotechnology, Santa Cruz, CA); anti-JNK1/2, BiP, Chop and LC3 (BD Biosciences PharMingen, San Diego, CA).
Cell lines and cell culture
The human ovarian (SKOV3) and leukemia (HL60) cancer cell lines were purchased from American Type Tissue Company (Manassas, VA) and grown as recommended by the supplier. Cells were maintained in RPMI (HL60) or McCoys (SKOV3) containing 10% fetal calf serum, 100 μg/ml streptomycin, 100 U/ml penicillin, and 2 mM L-glutamine at 5% CO2 and 37°C. 5×105 cells per treatment group were plated 24h prior to drug treatment.
Protein preparation
Cells (5×105 per treatment group) were harvested and washed with phosphate-buffered saline (PBS). Cell pellets were suspended in lysis buffer (20 mM Tris-HCl, pH 7.5, 15mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, and 1 mM β-glycerophosphate with freshly added protease and phosphatase inhibitors, 5 mM NaF and 1 mM Na3VO4) and incubated for 30 min on ice. Lysates were sonicated for 10 sec and centrifuged for 30 min at 10,000g at 4°C. Protein concentrations in the supernatant were assayed with the Bradford reagent (Bio-Rad Laboratories, Hercules, CA) using IgG as a standard.
Immunoblot Analysis
Equivalent amounts of protein were electrophoretically resolved under non-reducing conditions on 10% SDS-polyacrylamide gels (SDS-PAGE); unmodified proteins were separated under reducing conditions. Proteins were transferred onto nitrocellulose membranes (Bio-Rad, Hercules, CA). Non-specific binding was reduced by incubating the membrane in blocking buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Tween 20, 1μM protease inhibitors, 5 mM NaF, and 1 mM Na3VO4) containing 10% non-fat dried milk for 1 h. Membranes were incubated with the indicated antibody (blocking buffer containing 5% non-fat dried milk) at stated dilutions overnight at 4°C, washed 3x with PBS for 15 min, and incubated with the appropriate secondary antibody conjugated to horseradish peroxidase for 1 h. The membranes were washed 3 times and developed with enhanced chemiluminescence detection reagents (Bio-Rad). The blots were scanned with a BioRad ChemiDoc system and visualized with a transilluminator. The images were stored in a TIFF format. The relative intensity of bands was evaluated using Quantity One software (ver. 4.5.2; Bio-Rad) and plotted as arbitrary units (a.u.) in relation to actin.
Immunoprecipitation of PDI
SKOV3 cells were treated with 0 or 25 μM PABA/NO for 4h. 500 μg cell lysates from control and treated cells were precleared with protein A/G Plus-Agarose (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) in the presence or absence of non-specific IgG. PDI was precipitated with the anti-PDI antibody (1:100) at 4°C overnight. Prior to SDS-PAGE, the samples were heated to ∼95°C for 10 min in non-reducing sample loading buffer.
In Vitro S-glutathionylation of PDI
Recombinant human PDI in the expression vector pET-28a was provided by Dr. Lana Lee (Univ. of Windsor, Ontario) and expressed using the Escherichia coli strain BL21 (DE3) as previously described (25). 2 mg/mL PDI (> 95% homogeneous) in 20 mM phosphate buffer (pH 7.4) was incubated for 1h at room temperature as follows: a) control; b) 25 μM PABA/NO and 1 mM GSH; c) 25 μM PABA/NO; d) 1 mM GSH. Excess PABA/NO and GSH were eliminated through Biospin-6 Size exclusion micro-spin columns (Bio-Rad, Hercules, CA) with 20 mM PBS. Nitrosylation and S-glutathionylation were detected by immunoblot. Control and drug treated samples were evaluated by mass spectrometry, circular dichroism and fluorescence scanning.
Identification of the target cysteine modifications of PDI
PDI from control and drug treated cells were separated by SDS-PAGE under non-reducing conditions. Protein bands of interest were excised and subject to LysC digestion. The peptides were analyzed by mass spectrometry at the Proteomics Core Facility of the Medical University of South Carolina. The digested fragments were separated by HPLC and 20uL of the digested sample was injected with a Famous autosampler (LC Packings) onto a 75um Microtech Scientific C18 RP column. The peptides were eluted during a 30min gradient from 2-70% AcN with 0.2% formic acid using a 180 nl/min flow rate with an Ultimate pump (LC Packings). The eluted peptides were detected on a Finnigan LTQ linear ion trap mass spectrometer operated in data dependent acquisition mode with dynamic exclusion enabled. Protein identification was performed using software from the National Center for Biotechnology Information protein database.
Spectroscopic Analysis of PDI in vitro
The effect of PDI S-glutathionylation on enzyme secondary structure was examined by Circular Dichroism (CD) where measurements were carried out on a 202 AVIV Associates (Lakewood, NJ) using a semi-micro quartz rectangular 1 × 10 × 40 mm cuvette. Native and S-glutathionylated PDI samples were maintained at 22°C using a Pelletier element. Spectra were recorded while scanning in the far-ultraviolet region (190–260 nm), with bandwidth of 1.0 nm, step size of 1.0 nm, integration time of 30 s, and with three repeats. The output of the CD spectrometer was recalculated according to the protein concentration, amino acid content, and cuvette thickness into molecular ellipticity units (degrees/cm2/dmol). Protein tryptophan fluorescence was recorded on an F 2500 spectrofluorometer (Hitachi) using 10×10×40 mm quartz cuvette, excitation and emission slits were 2.5 and 5.0 nm respectively. The excitation wavelength was 295 nm to minimize an effect of protein tyrosines and phenylalanines. Background spectra were subtracted from final emission of the protein.
Nitric Oxide Measurement
Cells (5.0*105cells/ml) were incubated with 5 μM of DAF-FM-DA for 1 hour in complete media at 37°C. The labeled cells were washed 3 times with PBS. The dynamics of intracellular NO changes were measured using a Modulus™ Microplate Multimode Reader (Turner BioSystems, Sunnyvale, CA) with a “Blue” optical kit (Ex = 490 nm, Em = 510-570 nm) and standard kinetics mode. The measurements were performed at room temperature in 96-well Fluotrac 200 black microplates (VWR, Greiner Bio-One North America, Inc. Monroe, NC). The starting point for the kinetic experiments began ∼ 2 min after the addition of PABA/NO (e.g. the time necessary for additions of drug to all wells). All fluorescent measurements were corrected using the following controls: fluorescence of unlabeled cells, fluorescence changes in labeled cells without any addition of PABA/NO, the effect of addition of solvents on the cellular fluorescence without probes.
PDI activity assay
PDI was incubated with various concentrations of PABA/NO (0-100μM) and 1mM GSH for 30 minutes at 37°C. The activity of PDI was monitored using the turbidimetric assay of insulin disulfide reduction. Briefly, 0.25 μM purified human PDI was added to a solution of insulin (0–250 μM) and GSH (500 μM) in a 0.1 M K2HPO4 buffer containing 2 mM EDTA (pH 7.0). The increase in turbidity was monitored (λ=630) at 30 s intervals for 30 min. Three independent experiments were conducted. Mean values and S.E. were computed for each group.
Cytotoxicity Assays
10,000 cells were seeded in 96 well plates in 50 μl medium. Increasing drug concentrations were added to a final volume of 100 μl and maintained in drug for 72 h. Following drug exposure, cell viability was assayed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) conversion assay. Each drug concentration was represented in quadruplicate and three independent experiments were conducted. Mean values and S.E. were computed for each group.
RNA isolation and quantitative real-time RT-PCR
Total RNA was isolated and DNase I treated using the RNeasy mini kit according to the protocol of the manufacturer (Qiagen, Valencia, CA). cDNA was prepared using either the iScript™ cDNA synthesis kit from BioRad (Hercules, CA) or the RT2 First Strand Kit from SABiosciences (Frederick, MD) according to the protocols of the manufacturers. All quantitative real-time RT-PCR (qPCR) reactions were performed using the MyiQ™ system from BioRad, and iQ™ SYBR green supermix from BioRad or SYBR Green/Fluorescein PCR Master Mix from SABiosciences according to the protocols of the manufacturers. Actin mRNA levels were used to normalize differences in cDNA levels and the data are expressed using the ΔΔCt method (26). Primers were designed using Primer3 software (27)and primer sequences are listed in Table 1. All reactions were performed in triplicate and data were analyzed using the 2 −ΔΔCT method (26). GAPDH and actin was used as a gene control to normalize differences in cDNA between groups.
Table 1.
Primer sequences used in the real-time PCR analyses
| Name | Sequence | |
|---|---|---|
| Actin | Forward | 5′ CCT GGC ACC CAG CAC AAT 3′ |
| Reverse | 5′ GCC GAT CCA CAC GGA GTA CT 3′ | |
| PDI | Reverse | 5′ TCC TTG CCC TGT ATC AAA TCT T 3′ |
| Forward | 5′ TGA CCA GTG GCA AAA TTA AAA A 3′ | |
| ERO1 | Forward | 5′ TAA ACC TGA AGA GGC CGT GT 3′ |
| Reverse | 5′ TGA CAT GGT TTG ACA GCA CA 3′ | |
| HSP40 | Forward | 5′ TGA CCA TCG AAG TGA AGA AGG 3′ |
| Reverse | 5′ TGT TGG AGG TCT GGT CTC CT 3′ | |
| CHOP | Forward | 5′ CTG AAT CTG CAC CAA GCA TGA 3′ |
| Reverse | 5′ AAG GTG GGT AGT GTG GCC C 3′ | |
| Bip | Forward | 5′ AAG AAG CTA TTC AGT TGG ATG GA 3′ |
| Reverse | 5′ TTC TGT TAA CTT CGG CTT GGA 3′ | |
| HSP70 | Forward | 5′ TGC AGC AGG ACA TCA AGT TC 3′ |
| Reverse | 5′ ATG TCT TTG TTT GCC CAC CT 3′ | |
| GADD34 | Forward | 5′ CTG GGG ACT TTT GGA TGA TG 3′ |
| Reverse | 5′ ATT GAC TTC CCT GCC CTC TA 3′ | |
| XBP-1 | Forward | 5′ CCA AAA ACT TTT GCT AGA AAA TCA GC 3′ |
| Reverse | 5′ CAT CCC CAA GCG CTG TCT TA 3′ | |
| CDKN1A | Forward | 5′ ATG AAA TTC ACC CCC TTT CC 3′ |
| Reverse | 5′ CCC TAG GCT GTG CTC ACT TC 3′ | |
| IL8 | Forward | 5′ AAG AAA CCA CCG GAA GGA AC 3′ |
| Reverse | 5′ AAA TTT GGG GTG GAA AGG TT 3′ | |
| JUN | Forward | 5′ CCC CAA GAT CCT GAA ACA GA 3′ |
| Reverse | 5′ CCG TTG CTG GAC TGG ATT AT 3′ | |
| VEGFA | Forward | 5′ CCA ACT TCT GGG CTG TTC TC 3′ |
| Reverse | 5′ CCC CTC TCC TCT TCC TTC TC 3′ |
Statistical analysis
Mean values, S.D. and S.E. for at least three experiments were calculated for each treatment group. These data were analyzed for statistically significant differences between groups with Student's t test using SigmaStat 3.5 (Systat Software Inc., MA). Differences were considered statistically significant if the p value was <0.05.
Results
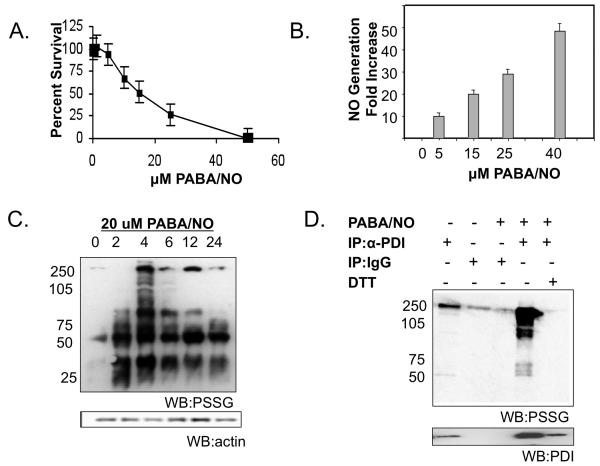
PABA/NO treatment leads to NO generation, cytotoxicity and S-glutathionylation of PDI in SKOV3 cells
PABA/NO elicits antitumor activity in cisplatin sensitive and resistant ovarian cancer xenografts (19). The following studies were designed to validate that PDI is a pharmacodynamic marker of PABA/NO in ovarian cancer cells. Figure 1 shows that PABA/NO treatment leads to dose-dependent cytotoxicity and NO generation in human ovarian cancer cells, SKOV3 (Figure 1A-B). We have previously reported that the toxic effects of PABA/NO treatment are attributed to S-glutathionylation of cellular proteins in HL60 cells, among which is PDI (24). Consistent with these findings, PABA/NO treatment caused S-glutathionylation of a number of proteins in SKOV3 cells, Figure 1C. One of the high molecular weight proteins had a mobility characteristic of PDI. Immunoprecipitation of PDI, followed by immunoblot for S-glutathionylated moiety confirmed that PDI is S-glutathionylated, Figure 1D. Lane 4 shows that PABA/NO treatment enhanced the extent of PDI S-glutathionylation in a manner that was reversible by the reducing agent DTT (lane 5). The lower panel shows that PDI itself was present only in lanes 1, 4 and 5.
Figure 1. The cytotoxic effects of PABA/NO treatment are concurrent with NO generation and S-glutathionylation of PDI in SKOV3 cells.
SKOV3 cells were treated with varying concentrations of PABA/NO. Cell viability was measured at 72h with the MTT assay (A). Cells were labeled with 5 μM DAF-FM to detect NO. NO generation was assessed following 0-50 μM PABA/NO treatment (B). At various time points cells were harvested and analyzed by immunoblot for S-glutathionylation (C). SKOV3 cells were treated with 25μM PABA/NO for 4h. PDI was immunoprecipitated from 500μg lysate and separated under non-reducing conditions by SDS-PAGE (D). Specific antibodies were used to detect S-glutathionylation of PDI and subsequently the membrane was stripped and probed for PDI. Data are the mean for 3 independent experiments +/− S.D.
The active site cysteines in hPDI are targets for S-glutathionylation
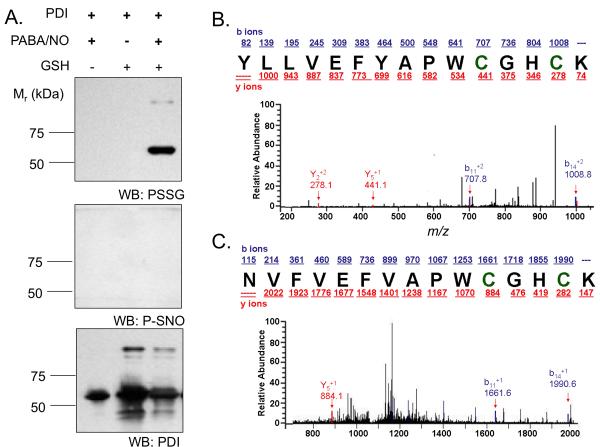
Using human full-length PDI, we confirmed via immunoblot and mass spectroscopy (MS) that drug treatment leads to S-glutathionylation of cysteine residues. PDI (1 μM) was incubated in 50 mM NaPO4 buffer (pH7.4) at 37°C for 30 min with 100 μM PABA/NO or 10 mM GSH or both. The samples were separated by SDS-PAGE under non-reducing conditions. The gels were transferred overnight to PVDF membrane and immunoblotted with anti-S-glutathionylation, anti-S-nitrosylation, and anti-PDI antibodies. Nitrosylation was not detected by western blot. Native and S-glutathionylated PDI was subjected to LysC digestion, the resultant fragments analyzed under non-reducing conditions by tandem MS. Within two distinct peptides (residues 43-57 and 387-401) a mass increase of 305.6 was consistent with the S-glutathionylation of one cysteine residue in each fragment (see asterisks).
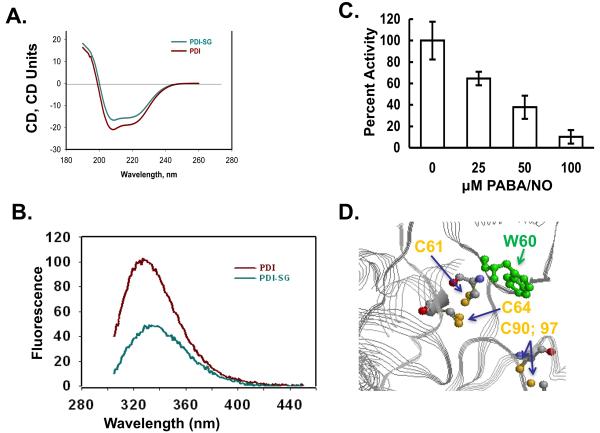
S-glutathionylation of the active site cysteines on PDI alters structure
We evaluated the effects of S-glutathionylation of PDI on its secondary structure. The CD spectrum (200-240 nm) of S-glutathionylated PDI was quite similar to native protein (Figure 3A), but consistent with a small decrease in the α-helical content (206-220nm) of the protein. Based on the crystal structure of PDI, the sulfhydryls of Cys residues 61 and 64 are proximal to a tryptophan at position 60. Consistent with the known quenching effect of a disulfide bond on adjacent tryptophanyl fluorescence, the intrinsic trytophanyl fluorescence of PDI was measured (6). Our data showed a substantial PDI tryptophanyl fluorescence decrease corresponding with S-glutathionylation (Figure 3B).These results indicate the dynamic nature of S-glutathionylation resulting in spatial disulfide/sulfhydryl equilibria. Moreover, a hypsochromic shift of emission maximum ∼4 nm after protein S-glutathionylation indicates some shielding of Trp38 from a polar environment through Cys61 and/or 64 S-glutathionylation (Figure 3D). The possible disulfide that may form at positions Cys90 and 97 is too distal from the Trp residue to have an impact. These fluorescent analyses confirm that S-glutathionylation impacts the tertiary structure of PDI.
Figure 3. S-glutathionylation of the active site cysteines on PDI alters protein structure.
Spectroscopic analysis of native (red) and PABA/NO + GSH-treated (green) PDI in vitro was performed using CD (A) and tryptophanyl fluorescence (B) of purified protein. According to the published crystal structure (39), the relative positions of PDI's C61 and 64 and W60 are depicted using RasMol 2.7.4.2 (http://rasmol.org) (D). The enzymatic activity of PDI was assessed using the insulin turbidity assay(C). Data are the mean for 3 independent experiments +/− S.D.
PDI reductase activity was assessed by insulin turbidity assay, Figure 3C. Native and S-glutathionylated PDI were applied to Micro Bio-Spin columns to remove excess PABA/NO and GSH. PDI proteins (250 nM) were then incubated at 25°C with 1 mg/ml insulin in NaPO4 buffer in the presence of 1 mM DTT. The precipitation of reduced insulin was recorded every 2 min for 30 min at 650 nm. After subtracting the basal rate of DTT-mediated insulin reduction, the specific activity of native PDI in this assay was 1518 ± 258 (units/min/mgP).Moreover, the catalytic activity of PDI is diminished in a dose-dependent manner following PABA/NO treatment (Figure 3C). We propose that S-glutathionylation of PDI may be an upstream event that is sufficient to contribute to the accumulation of unfolded proteins and thereby trigger activation of the UPR.
S-glutathionylation of PDI is concurrent with activation of the UPR
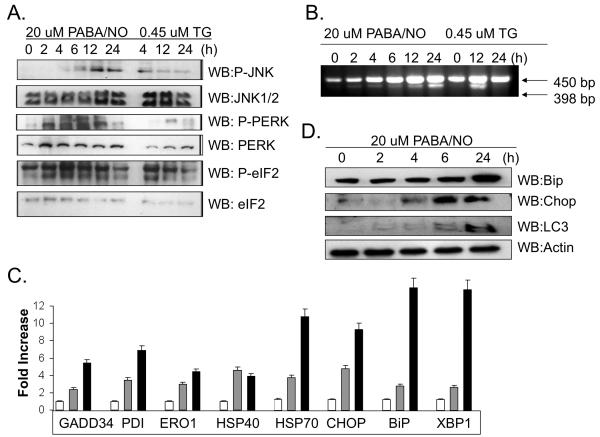
Translational attenuation is a cellular consequence of ER stress and is mediated by phosphorylation of PERK and eIF2. By treating SKOV3 cells with either PABA/NO or the UPR-inducing agent, Thapsigargin (TG) at approximate IC50 concentrations (Figure 4A) it was possible to show that cytotoxic effects were temporally accompanied by increases in phosphorylation of JNK, PERK and eIF2. Total protein levels of these products were unaffected (Figure 4B).
Figure 4. S-glutathionylation of PDI is concurrent with activation of the UPR in SKOV3 cells.
Thapsigargin (TG) was used as a positive control to induce ER stress and activation of the UPR. SKOV3 cells were treated with 20μM PABA/NO or 0.45μM TG for 0-24h. Translational attenuation through activation of PERK and eIF2 was evaluated in SKOV3 cells following drug treatment (A). 50μg protein lysate was separated by SDS-PAGE and analyzed by immunoblot for levels of JNK, PERK and eIF2. The membranes were stripped and probed for total levels of phosphorylated proteins. Activation of XBP-1 was detected by PCR (B). RNA was extracted at various time points and analyzed with primers specific to XPB-1. A 398 bp fragment corresponds to the cleaved and active form of XBP-1. Transcriptional activation of ER stress proteins was evaluated with SYBR-green RT-PCR methods (C). SKOV3 cells treated with 20 μM PABA/NO for 0h (no shading); 6h (gray shading)or 24h (black shading). The relative quantity was measured as the ratio of the mRNA expression of the target gene to that of GAPDH. Immunoblot analysis confirmed that the protein levels of UPR-inducible genes are also elevated following treatment with PABA/NO for 0-24h. Data are the mean for 3 independent experiments +/− S.D.
Paradoxically, transcriptional activation and translation of UPR genes also occurs under ER stress. We monitored drug-induced changes in transcript levels for a number of UPR genes at 0, 4 and 24h following PABA/NO treatment. Cleavage of the transcription factor, XBP-1 was detected as early as 2h following treatment (Figure 4B). Using real-time PCR, it was possible to quantitate a time-dependent transcriptional activation of the ER resident proteins BiP, GRP94, ERO1 and PDI, all of which showed between a 5 and 10 fold induction, Figure 4D. PABA/NO is cytotoxic to cancer cells and we show a 9-fold induction of the pro-apoptotic transcription factor, CHOP, which is also a UPR effector. Using immunoblot analysis we confirm that the protein levels for BiP and CHOP are upregulated, Figure 4D.
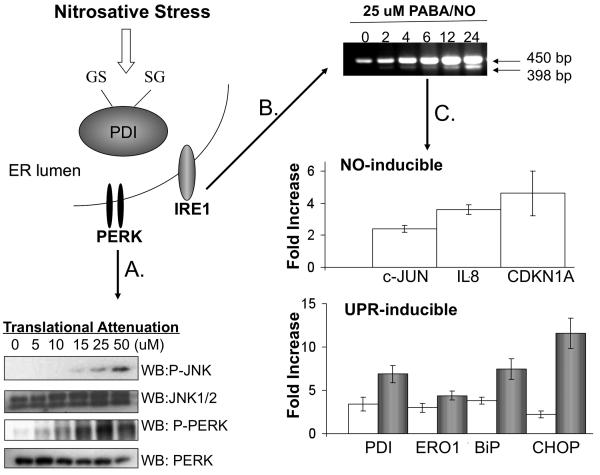
In the preclinical characterization of PABA/NO, we discovered that PDI is S-glutathionylated in HL60 cells following treatment (24). These studies showed that PDI is a molecular target of PABA/NO in SKOV3 cells and suggest that the mechanism of cell death is attributed to activation of the UPR. These studies were extended to HL60 cells and it is illustrated in Figure 5 how PABA/NO treatment can lead to nitrosative stress and cell death through induction of the UPR.
Figure 5. The mechanism of PABA/NO-induced antitumor activity is attributed to activation of the UPR.
The mechanistic studies were extended to HL60 cells in which S-glutathionylation of PDI was originally identified following PABA/NO treatment. HL60 cells were treated with 0-50 μM PABA/NO for 1h. Translational attenuation through activation of PERK was evaluated in HL60 cells following drug treatment (A). 50μg protein lysate was separated by SDS-PAGE and analyzed by immunoblot for levels of JNK and PERK. The membranes were stripped and probed for total levels of phosphorylated proteins. Activation of XBP-1 was detected by PCR (B). RNA was extracted at various time points and analyzed with primers specific to XPB-1. A 398 bp fragment corresponds to the cleaved and active form of XBP-1. Transcriptional activation of NO- and UPR-inducible proteins was evaluated with SYBR-green RT-PCR methods (C). HL60 cells treated with 25 μM PABA/NO for 4-6h (no shading) or 24h (gray shading). The relative quantity was measured as the ratio of the mRNA expression of the target gene to that of GAPDH. Data are the mean for 3 independent experiments +/− S.D.
Discussion
In designing these investigations, we have sought to link the mechanism of cytotoxic action of an NO releasing drug with the induction of an ER stress response in target cells. At the same time, drug-induced S-glutathionylation of PDI regulates its activity and facilitates upstream regulation of the UPR. It seems plausible that this post-translational modification of PDI may contribute to how the ER detects stress and may determine whether a cell survives a treatment that can produce a UPR event. Activation of the UPR is mediated by at least three ER proximal sensors (IRE1, PERK and ATF6) and our data demonstrate that PABA/NO impacts the expression of each of these. The coordination of the PDI and downstream signaling events provides an integration point for the functional determination of cell survival pathways. A number of recent reports have suggested that cancer pathology and the response of cancer cells to drugs may be dependent upon an integration of ER stress response pathways (28). It is also worth noting that PDI can have important chaperone functions that depend upon formal protein:protein interactions (29, 30).
When S-glutathionylation and PERK activation by PABA/NO were compared to a known ER stress-inducing agent TG, some significant similarities were observed. The mechanism of action of TG is attributed to inhibition of the Ca2+ pump protein located in ER membranes (SERCA; (31)). As such, it is intriguing that TG induces S-glutathionylation with kinetics similar to PABA/NO, notably with proteins in the 50 to 60 kDa range. This is consistent with the hypothesis that S-glutathionylation is an important signaling event associated with stress emanating from the ER. A significant pool of PDI in SKOV3 cells undergoes S-glutathionylation in a time-dependent manner following PABA/NO. Immunoblots detected 3 bands in the PABA/NO treated sample that correspond to monomeric PDI, suggesting that multiple sites on the protein may be modified. Interestingly, larger molecular weight species were also present suggesting multimers might undergo redox modification.
The cytotoxic effect of PABA/NO induced nitrosative stress has been linked to JNK activation that leads to apoptosis (24). Our present studies provide further insight into the mechanism and suggest that nitrosative stress elicits cell damage through ER stress. To demonstrate mechanism, two of the three signaling events that initiate the UPR, PERK and IRE1 activation were up-regulated by PABA/NO. An accumulation of unfolded ormis-folded proteins leads to the dissociation of BiP from PERK and IRE, allowing phosphorylation and activation of both proteins. The kinase activity is dependent upon conformational changes that occur when PERK is phosphorylated and this occurred in a dose- and time- dependent manner in SKOV3 cells following PABA/NO. Validation that PABA/NO activates UPR responsive genes was provided by the 2-10 fold increased expression of PDI, ERO1, BiP and CHOP. Collectively these data suggest that S-glutathionylation of PDI diminishes the folding capacity within the ER and mediates the UPR.
The link between S-glutathionylation of PDI and the UPR presents an important mechanistic connection with respect to cellular cytotoxic effects of PABA/NO and TG. We envision the following model for how PABA/NO may induce cytotoxic effects. Activation of PABA/NO occurs intracellularly in the presence of GST and GSH (17, 32). This releases NO that through nitrosative stress causes direct or indirect S-glutathionylation of a number of protein clusters, amongst which are those involved in protein folding (1). By comparing PABA/NO with TG, it is possible to draw parallels between the mechanisms of action of the two drugs. Recently, we have shown that TG and PABA/NO share the capacity to alter calcium flux from the ER to cytosol (Manevich et al, submitted). TG has been categorized as an inhibitor of SERCA but there is no obvious metabolite of the drug that is likely to produce either oxidative or nitrosative stress (33). Thus, it is unexpected that TG causes S-glutathionylation and has implied a cause:effect relationship of this post-translational modification and calcium homeostasis, a subject of another study (Manevich et al, submitted).
Implications for PDI function should also consider that our present study has not drawn distinction between the cellular localization of PDI. While the majority of PDI is located in the ER, there are reports that PDI may also regulate the cell surface redox environment (16, 34). This can be important in cell adhesion, antigen processing or viral entry (11, 12, 15). S-glutathionylation of cell surface proteins by oxidized glutathione alters both extracellular and intracellular redox homeostasis in HL60 cells (35).
PDI is involved in both rapid metabolism of GSNO and intracellular NO delivery and during this process PDI is itself altered by thiol modification (36). Following treatment of cells with diethylamine-NO, PDI was shown to be subject to nitrosylation (37). Although, we did not detect nitrosylation of PDI following PABA/NO (data not shown), such a modification may readily be subject to rapid conversion to S-glutathionylation (2, 38). The functional implication of this post-translational modification is that PDI has a reduced activity for the period of time that the cysteine residue is S-glutathionylated. Our previous results estimate that the half-life of the S-glutathionylation modification approximates 3 hours (6). In cataloguing existing literature, at least six clusters of proteins seem to be subject to S-glutathionylation, cytoskeletal/structural, energy metabolism/glycolysis, calcium homeostasis, signaling kinases and phosphatases, redox homeostasis and protein folding (1). Similar to phosphorylation, cysteine modification is critical to normal cellular signaling and its dysregulation is linked with multiple diseases. From this perspective, there may well be a generallink between diseases associated with ER stress and the accumulation of S-glutathionylated PDI. Finally, it is worth noting that drugs such as Velcade essentially have an indirect impact on the UPR through interference with the proteosomal machinery. It appears that PABA/NO has a mechanism of action that induces a UPR cell death more directly through PDI as an intermediary.
Figure 2. The active site cysteines in hPDI are targets for S-glutathionylation.
hPDI was treated with 1 mM GSH and / or 25 μM PABA/NO for 1h. S-glutathionylation or nitrosylation of PDI was detected by immunoblot (A). To identify specific cysteine residues that are targets for S-glutathionylation, control and PABA/NO-treated hPDI were digested with LysC under non-reducing conditions and analyzed by tandem MS. Two fragments corresponding to the active sites (43-57 and 387-401) were detected in the treated samples (B-C, respectively). Each result was consistent with a single S-glutathionylated modification [+305.6] with a probability of 8.7 e-002 and 7.4 e-002, respectively.
Acknowledgements
Grant Support: National Cancer Institute grants CA08660 and CA117259.
We thank the Drug Metabolism and Pharmacokinetics, X-ray Crystallography and Proteomics Core Facilities of the Hollings Cancer Center.
Abbreviations
- GSH
glutathione reduced
- GSSG
oxidized
- GSTp
glutathione S-transferase pi
- JNK
c-Jun N-terminal kinase
- ATF
activating transcription factor
- PERK
PKR-like ER kinase
- IRE1
inositol requiring enzyme 1
- XBP-1
X-box binding protein-1
- BiP
binding protein
- CHOP
C/EBP homologous protein
- IRE1
eukaryotic initiation factor 2
- PDI
protein disulfide isomerase
- ROS-RNS
reactive oxygen and nitrogen species
- UPR
unfolded protein response
References
- 1.Townsend DM. S-Glutathionylation: What are the Consequences to Cell Injury and the Unfolded Protein Response? Molecular Interventions. 2007;7:313–24. doi: 10.1124/mi.7.6.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tew KD. Redox in redux: Emergent roles for glutathione S-transferase P (GSTP) in regulation of cell signaling and S-glutathionylation. Biochem Pharmacol. 2007;73:1257–69. doi: 10.1016/j.bcp.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 3.Beer SM, Taylor ER, Brown SE, et al. Glutaredoxin 2 catalyzes the reversible oxidation and glutathionylation of mitochondrial membrane thiol proteins: implications for mitochondrial redox regulation and antioxidant defense. J Biol Chem. 2004;279:47939–51. doi: 10.1074/jbc.M408011200. [DOI] [PubMed] [Google Scholar]
- 4.Shelton MD, Chock PB, Mieyal JJ. Glutaredoxin: role in reversible protein s-glutathionylation and regulation of redox signal transduction and protein translocation. Antioxid Redox Signal. 2005;7:348–66. doi: 10.1089/ars.2005.7.348. [DOI] [PubMed] [Google Scholar]
- 5.Findlay VJ, Townsend DM, Morris TE, Fraser JP, He L, Tew KD. A novel role for human sulfiredoxin in the reversal of glutathionylation. Cancer research. 2006;66:6800–6. doi: 10.1158/0008-5472.CAN-06-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Townsend DM, Manevich Y, He L, Hutchens S, Pazoles CJ, Tew KD. Novel role for glutathione S-transferase {pi}: regulator of protein S-glutathionylation following oxidative and nitrosative stress. J Biol Chem. 2009;284:436–45. doi: 10.1074/jbc.M805586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gravina SA, Mieyal JJ. Thioltransferase is a specific glutathionyl mixed disulfide oxidoreductase. Biochemistry. 1993;32:3368–76. doi: 10.1021/bi00064a021. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura S, Matsushima M, Song H, Kikuchi M. A role of PDI in the reductive cleavage of mixed disulfides. J Biochem. 1996;120:525–30. doi: 10.1093/oxfordjournals.jbchem.a021445. [DOI] [PubMed] [Google Scholar]
- 9.Pihlajaniemi T, Helaakoski T, Tasanen K, et al. Molecular cloning of the beta-subunit of human prolyl 4-hydroxylase. This subunit and protein disulphide isomerase are products of the same gene. EMBO J. 1987;6:643–9. doi: 10.1002/j.1460-2075.1987.tb04803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrari DM, Soling HD. The protein disulphide-isomerase family: unravelling a string of folds. Biochem J. 1999;339(Pt 1):1–10. [PMC free article] [PubMed] [Google Scholar]
- 11.Park B, Lee S, Kim E, et al. Redox regulation facilitates optimal peptide selection by MHC class I during antigen processing. Cell. 2006;127:369–82. doi: 10.1016/j.cell.2006.08.041. [DOI] [PubMed] [Google Scholar]
- 12.Swiatkowska M, Szymanski J, Padula G, Cierniewski CS. Interaction and functional association of protein disulfide isomerase with alphaVbeta3 integrin on endothelial cells. FEBS J. 2008;275:1813–23. doi: 10.1111/j.1742-4658.2008.06339.x. [DOI] [PubMed] [Google Scholar]
- 13.Goplen D, Wang J, Enger PO, et al. Protein disulfide isomerase expression is related to the invasive properties of malignant glioma. Cancer Res. 2006;66:9895–902. doi: 10.1158/0008-5472.CAN-05-4589. [DOI] [PubMed] [Google Scholar]
- 14.Conant CG, Stephens RS. Chlamydia attachment to mammalian cells requires protein disulfide isomerase. Cell Microbiol. 2007;9:222–32. doi: 10.1111/j.1462-5822.2006.00783.x. [DOI] [PubMed] [Google Scholar]
- 15.Ou W, Silver J. Role of protein disulfide isomerase and other thiol-reactive proteins in HIV-1 envelope protein-mediated fusion. Virology. 2006;350:406–17. doi: 10.1016/j.virol.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 16.Tager M, Kroning H, Thiel U, Ansorge S. Membrane-bound proteindisulfide isomerase (PDI) is involved in regulation of surface expression of thiols and drug sensitivity of B-CLL cells. Exp Hematol. 1997;25:601–7. [PubMed] [Google Scholar]
- 17.Saavedra JE, Srinivasan A, Buzard GS, et al. PABA/NO as an anticancer lead: analogue synthesis, structure revision, solution chemistry, reactivity toward glutathione, and in vitro activity. J Med Chem. 2006;49:1157–64. doi: 10.1021/jm050700k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saavedra JE, Billiar TR, Williams DL, Kim YM, Watkins SC, Keefer LK. Targeting nitric oxide (NO) delivery in vivo. Design of a liver-selective NO donor prodrug that blocks tumor necrosis factor-alpha-induced apoptosis and toxicity in the liver. J Med Chem. 1997;40:1947–54. doi: 10.1021/jm9701031. [DOI] [PubMed] [Google Scholar]
- 19.Findlay VJ, Townsend DM, Saavedra JE, et al. Tumor cell responses to a novel glutathione S-transferase-activated nitric oxide-releasing prodrug. Mol Pharmacol. 2004;65:1070–9. doi: 10.1124/mol.65.5.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao RV, Bredesen DE. Misfolded proteins, endoplasmic reticulum stress and neurodegeneration. Curr Opin Cell Biol. 2004;16:653–62. doi: 10.1016/j.ceb.2004.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang XZ, Lawson B, Brewer JW, et al. Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153) Mol Cell Biol. 1996;16:4273–80. doi: 10.1128/mcb.16.8.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hetz C, Bernasconi P, Fisher J, et al. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science. 2006;312:572–6. doi: 10.1126/science.1123480. [DOI] [PubMed] [Google Scholar]
- 23.Scorrano L, Oakes SA, Opferman JT, et al. BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science. 2003;300:135–9. doi: 10.1126/science.1081208. [DOI] [PubMed] [Google Scholar]
- 24.Townsend DM, Findlay VJ, Fazilev F, et al. A glutathione S-transferase pi-activated prodrug causes kinase activation concurrent with S-glutathionylation of proteins. Mol Pharmacol. 2006;69:501–8. doi: 10.1124/mol.105.018523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raturi A, Vacratsis PO, Seslija D, Lee L, Mutus B. A direct, continuous, sensitive assay for protein disulphide-isomerase based on fluorescence self-quenching. Biochem J. 2005;391:351–7. doi: 10.1042/BJ20050770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 28.Moenner M, Pluquet O, Bouchecareilh M, Chevet E. Integrated endoplasmic reticulum stress responses in cancer. Cancer Res. 2007;67:10631–4. doi: 10.1158/0008-5472.CAN-07-1705. [DOI] [PubMed] [Google Scholar]
- 29.Kimura T, Hosoda Y, Sato Y, et al. Interactions among yeast protein-disulfide isomerase proteins and endoplasmic reticulum chaperone proteins influence their activities. J Biol Chem. 2005;280:31438–41. doi: 10.1074/jbc.M503377200. [DOI] [PubMed] [Google Scholar]
- 30.Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochim Biophys Acta. 2004;1699:35–44. doi: 10.1016/j.bbapap.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 31.Vangheluwe P, Raeymaekers L, Dode L, Wuytack F. Modulating sarco(endo)plasmic reticulum Ca2+ ATPase 2 (SERCA2) activity: cell biological implications. Cell Calcium. 2005;38:291–302. doi: 10.1016/j.ceca.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 32.Saavedra JE, Shami PJ, Wang LY, et al. Esterase-sensitive nitric oxide donors of the diazeniumdiolate family: in vitro antileukemic activity. J Med Chem. 2000;43:261–9. doi: 10.1021/jm9903850. [DOI] [PubMed] [Google Scholar]
- 33.Xu C, Ma H, Inesi G, Al-Shawi MK, Toyoshima C. Specific structural requirements for the inhibitory effect of thapsigargin on the Ca2+ ATPase SERCA. J Biol Chem. 2004;279:17973–9. doi: 10.1074/jbc.M313263200. [DOI] [PubMed] [Google Scholar]
- 34.Zai A, Rudd MA, Scribner AW, Loscalzo J. Cell-surface protein disulfide isomerase catalyzes transnitrosation and regulates intracellular transfer of nitric oxide. J Clin Invest. 1999;103:393–9. doi: 10.1172/JCI4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Townsend DM, He L, Hutchens S, VandenBerg TE, Pazoles CJ, Tew KD. NOV-002, a glutathione disulfide mimetic, as a modulator of cellular redox balance. Cancer Research. 2008;68:2870–7. doi: 10.1158/0008-5472.CAN-07-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shah CM, Bell SE, Locke IC, Chowdrey HS, Gordge MP. Interactions between cell surface protein disulphide isomerase and S-nitrosoglutathione during nitric oxide delivery. Nitric Oxide. 2007;16:135–42. doi: 10.1016/j.niox.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Uehara T, Nakamura T, Yao D, et al. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–7. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- 38.Jourd'heuil D, Jourd'heuil FL, Feelisch M. Oxidation and nitrosation of thiols at low micromolar exposure to nitric oxide. Evidence for a free radical mechanism. J Biol Chem. 2003;278:15720–6. doi: 10.1074/jbc.M300203200. [DOI] [PubMed] [Google Scholar]
- 39.Tian G, Xiang S, Noiva R, Lennarz WJ, Schindelin H. The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell. 2006;124:61–73. doi: 10.1016/j.cell.2005.10.044. [DOI] [PubMed] [Google Scholar]