Abstract
Rhesus macaque models have been instrumental for the development and testing of vaccines prior to human studies and have provided fundamental insights into the determinants of immune efficacy in a variety of infectious diseases. However, the characterization of antigen-specific T cell receptor (TCR) repertoires during adaptive immune responses in these models has previously relied on human TCR gene assignments. Here, we extracted and characterized TCR β-chain (TRB) genes from the recently sequenced rhesus macaque genome that are homologous to the human TRB genes. Comparison of the rhesus macaque TRB genes with the human TRB genes revealed an average best-match similarity of 92.9%. Furthermore, we confirmed the usage of most rhesus macaque TRB genes by expressed TCRβ sequences within epitope-specific TCR repertoires. This primary description of the rhesus macaque TRB genes will provide a standardized nomenclature and enable better characterization of TCR usage in studies that utilize this species.
Keywords: T cell, T cell receptor, T cell receptor repertoire
INTRODUCTION
The rhesus macaque is widely used as a non-human primate model to study infection and immunity due to the close genetic relationship with humans (∼93% average human-macaque sequence identity1) and the homology between human and rhesus pathogen genomes2, 3. Indeed, rhesus macaques have been used to study fundamental aspects of immunology, including the development and maintenance of T cell memory 4, immunodominance5 and the aging immune system6. There have also been many studies of immune responses in rhesus macaque models of human infections such as human immunodeficiency virus (HIV)7, influenza virus8, 9, tuberculosis10, Epstein-Barr virus (EBV)11, 12, cytomegalovirus (CMV)4, 13–15, smallpox16, measles17 and severe acute respiratory syndrome (SARS)18. Furthermore, rhesus macaques have been instrumental in the design and testing of vaccines against infections such as HIV19 and smallpox 16.
The various roles of T lymphocytes in adaptive immune responses to infection, which include the provision of helper functions to other immune cells and cytolytic control of infected cells, require that T cell populations recognize a large variety of foreign peptides bound to major histocompatibility complex (MHC) molecules. This recognition is facilitated by a diverse repertoire of T cell receptors (TCRs). The TCR repertoires that respond to different peptide-MHC epitopes can vary greatly. Indeed, diversity estimates range from ∼10 to >1000 different TCRs responding to a specific epitope20–23. Moreover, some epitope-specific TCR repertoires can feature biased usage of TCR Vβ (TRBV) or Jβ (TRBJ) genes, or distinct patterns of amino acid usage within the third complementarity-determining region (CDR3)24. Studies of the TCR repertoire can provide valuable information about the molecular evolution of an immune response and the factors that shape clonotype selection in vivo25. Furthermore, it is becoming increasingly apparent that the clonotypic structure of an epitope-specific T cell response can have important implications for the immune control of some viral infections. For example, one issue of current debate that has important consequences for the rational design of immunotherapeutic and vaccination strategies24, 26 is whether a restricted TCR repertoire responding to a highly variable pathogen could be associated with the emergence of viral mutants that escape T cell recognition at this epitope27–31.
Many studies of T cell immunity in rhesus macaque models of infection have utilized TCR repertoire data to gain additional insights5, 14, 30, 32–43. In particular, a large number of studies have characterized the TCR repertoires of target CD4+ T cell populations or CD8+ T cell populations involved in the control of simian immunodeficiency virus (SIV) in rhesus macaques5, 30, 32–39, 41–43. Most of these studies have relied on human TCR gene homology to identify V and J gene usage. Although the rhesus macaque TCR Dβ (TRBD) and TRBJ genes have previously been sequenced44, the TRBV genes were not previously available. Here, we present the TRBV, TRBD and TRBJ genes extracted from the rhesus macaque genome1 on the basis of their homology with the human TRB genes. In addition, we demonstrate extracted TRB gene usage in expressed TCRβ sequences by using an existing database of 7218 TCRβ sequences involved in CD8+ T cell responses specific for the immunodominant Mamu-A*01-restricted SL8/TL8 (S/TTPESANL; Tat, residues 28–35) and CM9 (CTPYDINQM; Gag, residues 181–189) epitopes derived from SIV30, 45. The TRB genes extracted from the rhesus macaque genome will enable more accurate characterization of rhesus macaque TCRβ repertoires.
RESULTS
Rhesus macaque TRBV genes
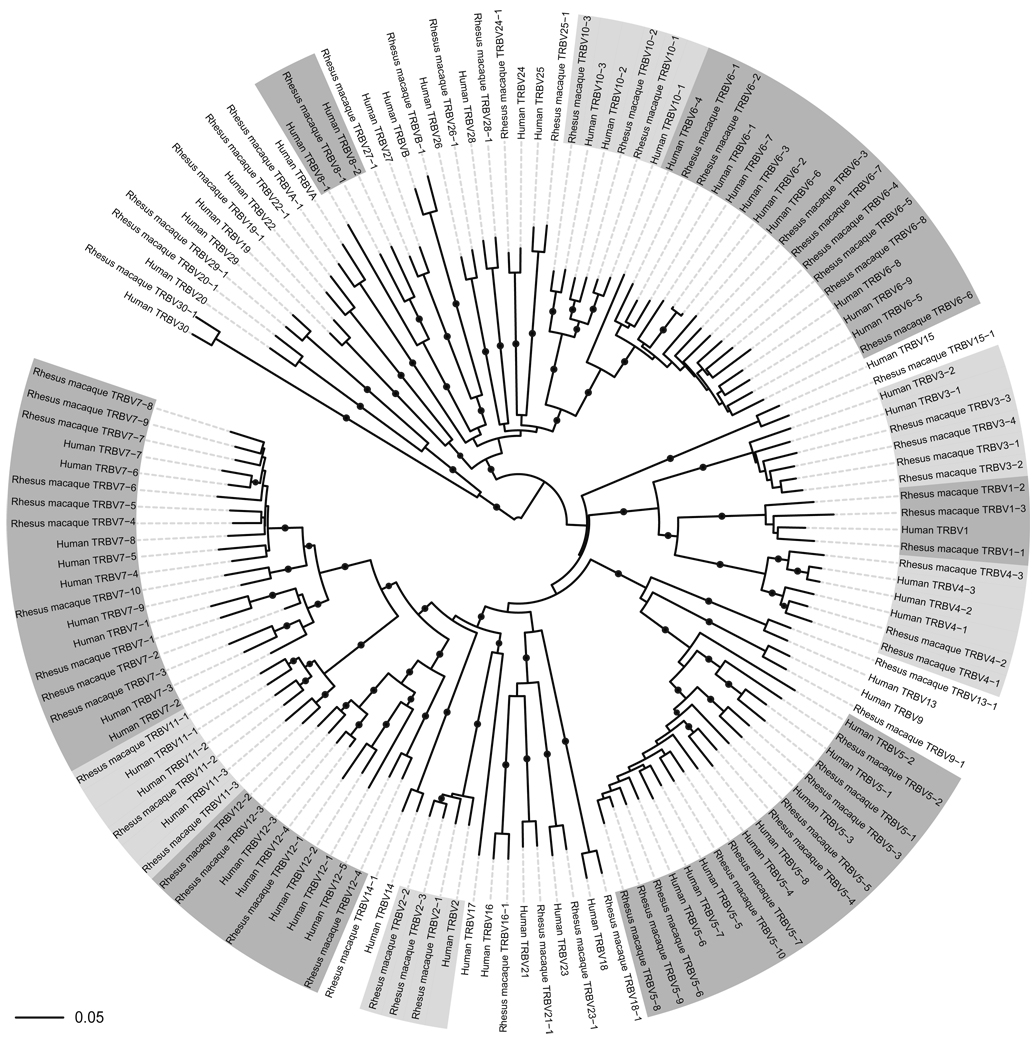
A total of 72 TRBV genes were extracted from chromosome 3 of the rhesus macaque genome (Table 1 and Rhesus_macaque_TRBV.fsa in Supporting Information). The human TRBV gene corresponding most precisely to each rhesus macaque TRBV gene was identified on the basis of the highest percentage match between the nucleotide sequences for the TRBV genes (i.e. V-GENE in the IMGT standarized labels). The percent similarity between the nucleotide sequences for the rhesus macaque and the best-match human TRBV genes ranged between 78.3% and 96.5%, with an average similarity of 92.2%. We could not identify a one-to-one correspondence between all rhesus macaque and human TRBV genes (Figure 1). In many cases, one human TRBV gene was found to be the best match to more than one of the TRBV genes extracted from the rhesus macaque genome. For example, the human TRBV6-5 gene had the highest percent similarity of all human TRBV genes to five of the rhesus macaque TRBV genes; in contrast, the human TRBV6-6 gene was not the best match to any of the rhesus macaque TRBV genes. For five of the 72 TRBV genes, only partial sequences were available from the rhesus macaque genome (Table S1 in Supporting Information) and only two of these partial TRBV genes were incomplete at the 3’ end, which would influence their use in analysis of the CDR3. The human TRBV17 subgroup, consisting of just one gene, was the only one for which no corresponding TRB gene was found in the rhesus macaque genome (using a cutoff of 75% similarity).
Table 1.
Comparison of the rhesus macaque TRBV genes and their best human homologues.
| Gene nucleotide sequence |
Exon nucleotide sequence |
Exon amino acid sequence3 |
Note | |||||
|---|---|---|---|---|---|---|---|---|
| Rhesus macaque gene |
Best human homologue1 |
Percent identity |
Alignment length2 |
Percent identity |
Alignment length |
Percent identity |
Alignment length |
|
| TRBV1-1 | TRBV1 | 93.3 | 489 | 93.1 | 333 | 86.5 | 111 | 8 |
| TRBV1-2 | TRBV1 | 91.0 | 490 | 90.1 | 334 | 64.0 | 111 | 8 |
| TRBV1-3 | TRBV1 | 85.5 | 496 | 82.8 | 337 | 8 | ||
| TRBV2-1 | TRBV2 | 93.5 | 475 | 94.8 | 347 | 89.6 | 115 | |
| TRBV2-2 | TRBV2 | 93.7 | 475 | 93.7 | 347 | 89.6 | 115 | |
| TRBV2-3 | TRBV2 | 94.3 | 475 | 93.9 | 347 | 87.8 | 115 | |
| TRBV3-1 | TRBV3-1 | 92.6 | 499 | 93.6 | 344 | 86.8 | 114 | |
| TRBV3-2 | TRBV3-1 | 92.8 | 499 | 92.4 | 344 | 84.2 | 114 | |
| TRBV3-3 | TRBV3-1 | 90.8 | 499 | 93.9 | 344 | 87.7 | 114 | |
| TRBV3-4 | TRBV3-1 | 92.6 | 499 | 93.6 | 344 | 86.0 | 114 | |
| TRBV4-1 | TRBV4-1 | 92.7 | 493 | 93.6 | 344 | 89.5 | 114 | |
| TRBV4-2 | TRBV4-1 | 94.7 | 493 | 94.8 | 344 | 93.0 | 114 | |
| TRBV4-3 | TRBV4-3 | 91.2 | 502 | 93.0 | 344 | 91.2 | 114 | |
| TRBV5-1 | TRBV5-1 | 93.5 | 509 | 93.9 | 343 | 7 | ||
| TRBV5-2 | TRBV5-1 | 84.8 | 512 | 83.7 | 343 | 7, 8 | ||
| TRBV5-3 | TRBV5-3 | 93.1 | 509 | 93.4 | 346 | 86.1 | 115 | |
| TRBV5-4 | TRBV5-8 | 93.1 | 506 | 93.6 | 343 | 88.6 | 114 | |
| TRBV5-5 | TRBV5-6 | 90.9 | 507 | 91.6 | 344 | 81.6 | 114 | |
| TRBV5-6 | TRBV5-6 | 94.7 | 505 | 95.0 | 343 | 91.2 | 114 | |
| TRBV5-7 | TRBV5-5 | 93.7 | 506 | 95.0 | 343 | 91.2 | 114 | |
| TRBV5-8 | TRBV5-6 | 93.1 | 506 | 93.9 | 343 | 90.4 | 114 | |
| TRBV5-9 | TRBV5-6 | 94.5 | 505 | 95.3 | 343 | 92.1 | 114 | |
| TRBV5-10 | TRBV5-5 | 92.1 | 506 | 92.1 | 343 | 87.7 | 114 | |
| TRBV6-1 | TRBV6-1 | 91.9 | 472 | 93.6 | 344 | 87.7 | 114 | |
| TRBV6-2 | TRBV6-1 | 94.3 | 351 | 93.9 | 295 | 87.6 | 97 | 4 |
| TRBV6-3 | TRBV6-5 | 93.9 | 475 | 94.2 | 344 | 88.6 | 114 | |
| TRBV6-4 | TRBV6-5 | 92.0 | 475 | 92.4 | 344 | 84.2 | 114 | |
| TRBV6-5 | TRBV6-9 | 91.5 | 424 | 90.8 | 355 | 84.7 | 111 | 5 |
| TRBV6-6 | TRBV6-5 | 92.2 | 475 | 94.5 | 344 | 87.7 | 114 | |
| TRBV6-7 | TRBV6-5 | 92.4 | 475 | 93.9 | 344 | 86.8 | 114 | |
| TRBV6-8 | TRBV6-5 | 91.0 | 390 | 91.3 | 298 | 83.8 | 99 | 5 |
| TRBV7-1 | TRBV7-1 | 83.6 | 329 | 93.1 | 290 | 83.3 | 96 | 4, 8 |
| TRBV7-2 | TRBV7-3 | 87.7 | 261 | 88.3 | 222 | 83.6 | 73 | 4 |
| TRBV7-3 | TRBV7-3 | 90.6 | 500 | 91.9 | 347 | 86.1 | 115 | |
| TRBV7-4 | TRBV7-7 | 89.8 | 549 | 90.8 | 347 | 84.3 | 115 | |
| TRBV7-5 | TRBV7-8 | 91.8 | 525 | 92.8 | 347 | 87.0 | 115 | |
| TRBV7-6 | TRBV7-6 | 89.7 | 532 | 92.8 | 347 | 87.8 | 115 | |
| TRBV7-7 | TRBV7-4 | 89.6 | 509 | 90.8 | 347 | 82.6 | 115 | |
| TRBV7-8 | TRBV7-6 | 87.4 | 546 | 89.2 | 361 | 37.2 | 121 | 8 |
| TRBV7-9 | TRBV7-6 | 92.8 | 538 | 94.5 | 347 | 89.6 | 115 | |
| TRBV7-10 | TRBV7-9 | 90.7 | 516 | 92.2 | 347 | 82.6 | 115 | |
| TRBV8-1 | TRBV8-2 | 86.7 | 511 | 85.1 | 336 | 30.4 | 115 | 8 |
| TRBV9-1 | TRBV9 | 94.7 | 514 | 96.2 | 343 | 93.9 | 114 | |
| TRBV10-1 | TRBV10-1 | 96.5 | 489 | 96.2 | 344 | 93.9 | 114 | |
| TRBV10-2 | TRBV10-2 | 95.1 | 489 | 95.3 | 344 | 93.9 | 114 | |
| TRBV10-3 | TRBV10-3 | 95.5 | 489 | 96.5 | 344 | 93.0 | 114 | |
| TRBV11-1 | TRBV11-1 | 93.3 | 490 | 93.9 | 347 | 87.0 | 115 | |
| TRBV11-2 | TRBV11-2 | 94.4 | 480 | 94.5 | 347 | 91.3 | 115 | |
| TRBV11-3 | TRBV11-3 | 93.9 | 477 | 94.2 | 347 | 88.7 | 115 | |
| TRBV12-1 | TRBV12-2 | 92.7 | 482 | 93.0 | 344 | 85.2 | 115 | |
| TRBV12-2 | TRBV12-3 | 91.8 | 486 | 93.1 | 347 | 87.8 | 115 | |
| TRBV12-3 | TRBV12-3 | 91.8 | 486 | 92.5 | 347 | 87.8 | 115 | |
| TRBV12-4 | TRBV12-5 | 94.9 | 486 | 94.5 | 347 | 92.2 | 115 | |
| TRBV13-1 | TRBV13 | 94.5 | 524 | 95.7 | 374 | 89.5 | 124 | |
| TRBV14-1 | TRBV14 | 93.6 | 482 | 95.1 | 347 | 89.6 | 115 | |
| TRBV15-1 | TRBV15 | 94.3 | 508 | 94.8 | 344 | 89.5 | 114 | |
| TRBV16-1 | TRBV16 | 94.7 | 493 | 94.8 | 347 | 88.7 | 115 | |
| TRBV18-1 | TRBV18 | 95.0 | 658 | 94.8 | 347 | 90.4 | 115 | |
| TRBV19-1 | TRBV19 | 95.5 | 516 | 95.6 | 344 | 94.7 | 114 | |
| TRBV20-1 | TRBV20-1 | 91.9 | 713 | 92.5 | 335 | 83.8 | 111 | |
| TRBV21-1 | TRBV21-1 | 94.0 | 500 | 94.0 | 348 | 19.5 | 118 | |
| TRBV22-1 | TRBV22-1 | 78.3 | 493 | 72.7 | 348 | 6, 8 | ||
| TRBV23-1 | TRBV23-1 | 92.0 | 536 | 93.9 | 347 | 91.3 | 115 | |
| TRBV24-1 | TRBV24-1 | 95.2 | 517 | 95.9 | 345 | 90.4 | 115 | |
| TRBV25-1 | TRBV25-1 | 94.7 | 507 | 95.1 | 344 | 90.4 | 114 | |
| TRBV26-1 | TRBV26 | 94.5 | 524 | 95.1 | 344 | 73.9 | 115 | 8 |
| TRBV27-1 | TRBV27 | 94.9 | 512 | 95.6 | 344 | 93.9 | 114 | |
| TRBV28-1 | TRBV28 | 93.3 | 523 | 96.2 | 344 | 93.9 | 114 | |
| TRBV29-1 | TRBV29-1 | 94.1 | 661 | 96.4 | 335 | 93.7 | 111 | |
| TRBV30-1 | TRBV30 | 94.3 | 743 | 96.1 | 335 | 93.7 | 111 | |
| TRBVA-1 | TRBVA | 92.7 | 493 | 91.1 | 316 | 6, 8 | ||
| TRBVB-1 | TRBVB | 88.3 | 563 | 87.4 | 422 | 8 | ||
| Average percent identity | 92.2 | 92.9 | 85.3 | |||||
The best human homologue had the highest percent identity with the rhesus macaque gene nucleotide sequence.
The alignment length is the total length across both the aligned rhesus macaque and human gene/exon sequences.
The exon amino acid sequence was translated in the frame that yielded a start codon at the 5’ end of the exon. Comparisons of the exon amino acid sequences were omitted for TRBV genes in which no start codon was found. For partial rhesus macaque exons missing a portion of sequence at the 5’ end, the sequences were translated in the frame in which the start codon was found in the human homologues.
The rhesus macaque gene is a partial sequence, with a missing portion of sequence at the 5’ end of the gene. The percent identities between rhesus macaque and human genes and exons are calculated with the missing portion of rhesus macaque gene excluded.
The rhesus macaque gene is a partial sequence, with a missing portion of sequence at the 3’ end of the gene. The percent identities between rhesus macaque and human genes and exons are calculated with the missing portion of rhesus macaque gene excluded.
The human NCBI reference gene was a partial sequence so the human IMGT reference gene sequence was used for these comparisons.
There was some ambiguity in the identification of the exons for the TRBV5-1 and TRBV5-2 genes (see Table 1 in Supporting Information).
The rhesus macaque exon amino sequence contained stop codons or, for those genes in which no start codon was found, the amino acid sequences in all three translation frames contained stop codons.
Figure 1. Unrooted circular phylogram showing the clustering relationships between all rhesus macaque and human TRBV gene sequences at the nucleotide level.
Exons, introns and recombination signal sequences have been included and gene families consisting of multiple genes are highlighted. All TRBV gene sequences were aligned using ClustalW and the tree was constructed in ClustalW using the neighbour-joining method49 and bootstrapped 1000 times. Branches with bootstrap values >80% are indicated with a black dot and branch lengths are those assigned by ClustalW. The tree was visualized using the Interactive Tree of Life50 (available at http://itol.embl.de/). Note that the tree has been rotated about the mid-point of the most distant nodes to assist visualization.
We also compared the TRBV exons (i.e. L-PART1+V-EXON in the IMGT standardized labels) between the rhesus macaque and best-match human TRBV genes (Table 1). The percent identities between the nucleotide sequences for the rhesus macaque and human TRBV exons ranged between 72.7% and 96.5%, with an average of 92.9%. The similarities between the rhesus macaque and human TRBV exons at the amino acid sequence level ranged between 19.5% and 94.7%, with an average of 85.3%.
Rhesus macaque TRBD genes
The two TRBD genes extracted from the rhesus macaque genome were found to have 95.0% and 92.8% agreements at the nucleotide level with the corresponding human TRBD genes (Table 2 and Rhesus_macaque_TRBD.fsa in Supporting Information). The percent similarities between the rhesus macaque and human TRBD exon (i.e. D-REGION in the IMGT standardized labels) nucleotide sequences were 84.6% and 75.0%.
Table 2.
Comparison of the rhesus macaque TRBD genes and their human homologues.
| Gene nucleotide sequence |
Exon nucleotide sequence |
||||
|---|---|---|---|---|---|
| Rhesus macaque gene |
Human gene homologue |
Percent identity |
Alignment length1 |
Percent identity |
Alignment length |
| TRBD1 | TRBD1 | 95.0 | 80 | 84.6 | 13 |
| TRBD2 | TRBD2 | 92.8 | 83 | 75.0 | 16 |
| Average percent identity | 93.9 | 79.8 | |||
The alignment length is the total length across both the aligned rhesus macaque and human gene/exon sequences.
The rhesus macaque TRBD genes have been sequenced in a previous study44. The TRBD1 gene extracted from the rhesus macaque genome does not differ from that reported in this previous study. A 1.2% difference was found between the TRBD2 gene reported here and that reported previously, with a single nucleotide difference occurring in the 5’ spacer. Thus, there are no differences in the TRBD2 D-REGION extracted from the rhesus macaque genome compared with that reported previously44.
Rhesus macaque TRBJ genes
For each of the 14 human TRBJ genes, there was one corresponding TRBJ gene found on chromosome 3 of the rhesus macaque genome (Table 3 and Rhesus_macaque_TRBJ.fsa in Supporting Information). The percent similarities between the rhesus macaque TRBJ genes and the corresponding human TRBJ genes are shown in Table 3 (range: 92.1% and 98.7%; average: 96.1%).
Table 3.
Comparison of the rhesus macaque TRBJ genes and their human homologues.
| Gene nucleotide sequence |
Exon nucleotide sequence |
Exon amino acid sequence2 |
|||||
|---|---|---|---|---|---|---|---|
| Rhesus macaque gene |
Human gene homologue |
Percent identity |
Alignment length1 |
Percent identity |
Alignment length |
Percent identity |
Alignment length |
| TRBJ1-1 | TRBJ1-1 | 98.7 | 76 | 97.9 | 48 | 93.3 | 15 |
| TRBJ1-2 | TRBJ1-2 | 92.1 | 76 | 91.7 | 48 | 86.7 | 15 |
| TRBJ1-3 | TRBJ1-3 | 94.9 | 78 | 92.0 | 50 | 87.5 | 16 |
| TRBJ1-4 | TRBJ1-4 | 98.7 | 79 | 100.0 | 51 | 100.0 | 16 |
| TRBJ1-5 | TRBJ1-5 | 94.9 | 78 | 92.0 | 50 | 87.5 | 16 |
| TRBJ1-6 | TRBJ1-6 | 97.5 | 81 | 96.2 | 53 | 100.0 | 17 |
| TRBJ2-1 | TRBJ2-1 | 97.4 | 78 | 96.0 | 50 | 100.0 | 16 |
| TRBJ2-2 | TRBJ2-2 | 93.7 | 79 | 90.2 | 51 | 87.5 | 16 |
| TRBJ2-2P | TRBJ2-2P | 96.2 | 53 | 95.7 | 46 | 86.7 | 15 |
| TRBJ2-3 | TRBJ2-3 | 98.7 | 77 | 98.0 | 49 | 93.8 | 16 |
| TRBJ2-4 | TRBJ2-4 | 94.9 | 78 | 96.0 | 50 | 81.3 | 16 |
| TRBJ2-5 | TRBJ2-5 | 97.4 | 76 | 97.9 | 48 | 100.0 | 15 |
| TRBJ2-6 | TRBJ2-6 | 95.1 | 81 | 96.2 | 53 | 94.1 | 17 |
| TRBJ2-7 | TRBJ2-7 | 94.7 | 75 | 95.7 | 47 | 93.3 | 15 |
| Average percent identity | 96.1 | 95.4 | 92.3 | ||||
The alignment length is the total length across both the aligned rhesus macaque and human gene/exon sequences.
The TRBJ exons were translated in the frame that yielded the characteristic FGXG or LGXG motif.
A comparison of the rhesus macaque and human TRBJ exons (i.e. J-REGION in the IMGT standardized labels) revealed percent similarities of nucleotide sequences ranging between 90.2% and 100%, with an average similarity of 95.4% (Table 3). The similarities between the translated TRBJ exons of the rhesus macaque and human genes ranged between 81.3% and 100%, with an average similarity of 92.3% (Table 3).
We compared the TRBJ genes extracted from the rhesus macaque genome with those reported in a previous study44. The only differences found were in the TRBJ1-6 and TRBJ2-1 genes, which differed by 1.9% and 2%, respectively. A single nucleotide difference in the 20th nucleotide position of the TRBJ1-6 exon resulted in a difference of a single amino acid (i.e. the TRBJ1-6 exon from the rhesus macaque genome contained H in the 7th amino acid position instead of Y). In the TRBJ2-1 gene, a single nucleotide difference in the 31st nucleotide position of the exon did not result in any amino acid differences between the TRBJ2-1 exon extracted from the rhesus macaque genome and that reported by Cheynier et al.44
Use of the rhesus macaque TRB genes by expressed TCRβ sequences
To demonstrate the use of the TRB genes extracted from the rhesus macaque genome by expressed TCRβ sequences, we used an existing database of 7218 TCRβ sequences involved in CD8+ T cell responses specific for the immunodominant Mamu-A*01-restricted SIV-SL8/TL8 and SIV-CM9 epitopes in 20 rhesus macaques30, 45. Each of these TCRβ sequences was aligned with the TRB gene exons to determine the most likely TRBV, TRBJ and TRBD gene usage. In Table 4 and Table 5 we show the rhesus macaque TRB genes that were found to be most likely used by at least one of the TCRβ sequences. The genes used by the TCRβ sequences included 54 of the 72 TRBV genes, both TRBD genes, and 13 of the 14 TRBJ genes. The highest percent homology and longest match between each TRB gene and a TCRβ sequence is also shown. Of the 18 rhesus macaque TRBV genes not used by the TCRβ sequences, 12 either didn’t begin with a start codon or contained stop codons when translated (Table 1). The rhesus macaque TRBJ2-2P gene, which is homologous to the human TRBJ2-2P gene (qualified by IMGT as having an “Open Reading Frame” functionality), was the only TRBJ gene not used by the TCRβ sequences. Deviations between the rhesus macaque TRB genes and TCRβ sequences were mostly attributed to the full-length genes not being used by the TCRβ sequences, owing to nucleotides being cleaved during TCR gene recombination. However, allelic differences could also exist between the single rhesus macaque sequenced in the genome project and the 20 SIV-infected macaques from which the TCRβ sequences were obtained.
Table 4.
Usage of the rhesus macaque TRBV genes by expressed TCRβ sequences.
| Rhesus macaque gene |
Highest % usage by a TCRβ sequence |
Alignment length1 |
|---|---|---|
| TRBV2-1 | 100 | 182 |
| TRBV2-2 | 99.1 | 116 |
| TRBV2-3 | 100 | 185 |
| TRBV3-1 | 100 | 274 |
| TRBV3-2 | 100 | 278 |
| TRBV3-3 | 100 | 179 |
| TRBV3-4 | 100 | 278 |
| TRBV4-1 | 100 | 167 |
| TRBV4-2 | 100 | 282 |
| TRBV4-3 | 100 | 273 |
| TRBV5-1 | 98.7 | 232 |
| TRBV5-3 | 92.1 | 76 |
| TRBV5-4 | 98.9 | 281 |
| TRBV5-6 | 98.9 | 272 |
| TRBV5-7 | 98.1 | 159 |
| TRBV5-8 | 97.9 | 280 |
| TRBV5-9 | 98.7 | 307 |
| TRBV5-10 | 100 | 189 |
| TRBV6-1 | 100 | 277 |
| TRBV6-2 | 100 | 219 |
| TRBV6-3 | 100 | 274 |
| TRBV7-2 | 98.9 | 182 |
| TRBV7-3 | 98.7 | 297 |
| TRBV7-4 | 99.3 | 281 |
| TRBV7-5 | 98.6 | 289 |
| TRBV7-6 | 99.7 | 287 |
| TRBV7-7 | 97.1 | 68 |
| TRBV7-9 | 97.9 | 290 |
| TRBV7-10 | 98.6 | 284 |
| TRBV9-1 | 99.4 | 180 |
| TRBV10-1 | 96.7 | 276 |
| TRBV10-2 | 100 | 279 |
| TRBV10-3 | 99.2 | 238 |
| TRBV11-1 | 99.6 | 276 |
| TRBV11-2 | 98.6 | 282 |
| TRBV11-3 | 98.9 | 282 |
| TRBV12-2 | 99.3 | 277 |
| TRBV12-3 | 98.8 | 168 |
| TRBV12-4 | 95.9 | 74 |
| TRBV13-1 | 98.9 | 182 |
| TRBV14-1 | 99.7 | 294 |
| TRBV15-1 | 99.2 | 262 |
| TRBV16-1 | 94.6 | 74 |
| TRBV18-1 | 97.6 | 167 |
| TRBV19-1 | 100 | 279 |
| TRBV20-1 | 99.5 | 197 |
| TRBV21-1 | 98.2 | 274 |
| TRBV23-1 | 100 | 298 |
| TRBV24-1 | 97.7 | 214 |
| TRBV25-1 | 98.5 | 268 |
| TRBV27-1 | 100 | 281 |
| TRBV28-1 | 99.2 | 119 |
| TRBV29-1 | 100 | 282 |
| TRBV30-1 | 98.2 | 170 |
The alignment length between the TRBV gene and the TCRβ sequence is mostly determined by the length of the TCRβ sequence to the 5’ end of the CDR3.
Table 5.
Usage of the rhesus macaque TRBD and TRBJ genes by expressed TCRβ sequences.
| Rhesus macaque gene |
Highest % usage by a TCRβ sequence |
Alignment length1 |
|---|---|---|
| TRBD1 | 100 | 13 |
| TRBD2 | 100 | 14 |
| TRBJ1-1 | 100 | 48 |
| TRBJ1-2 | 100 | 48 |
| TRBJ1-3 | 100 | 50 |
| TRBJ1-4 | 100 | 51 |
| TRBJ1-5 | 100 | 50 |
| TRBJ1-6 | 96.2 | 53 |
| TRBJ2-1 | 100 | 50 |
| TRBJ2-2 | 100 | 51 |
| TRBJ2-3 | 100 | 49 |
| TRBJ2-4 | 100 | 49 |
| TRBJ2-5 | 100 | 48 |
| TRBJ2-6 | 100 | 53 |
| TRBJ2-7 | 100 | 47 |
The alignments were performed over the total length of the TRBD or TRBJ exon.
Possible allelic variants of the TRB genes used by the TCRβ sequences were not identified due to the level of uncertainty associated with distinguishing allelic variants from sequencing errors, in either the rhesus macaque genome or TCRβ sequences, when there were often small numbers of TCRβ sequences per rhesus macaque using a particular TRB gene. However, we investigated whether the nucleotide sequence variants of the TRBJ1-6 and TRBJ2-1 genes reported by Cheynier et al.44 were used in our collection of epitope-specific TCRβ sequences. The previously reported variant of the TRBJ1-6 gene was found to be used by some TCRβ sequences, suggesting that this is an allelic variant of the TRBJ1-6 gene extracted from the rhesus macaque genome. The TRBJ2-1 gene variant was not used by any of the TCRβ sequences. This TRBJ2-1 gene variant may be an allelic variant that was not present in any of the 20 rhesus macaques in which the Mamu-A*01-restricted SIV-SL8/TL8- and SIV-CM9-specific TCRβ repertoires were studied but it is also possible that the single nucleotide difference in the TRBJ2-1 gene reported Cheynier et al.44 is due to sequencing error.
DISCUSSION
The assembly of reference TCR gene data sets for many species has often relied on the ad hoc sourcing of different TCR genes from various studies over time. Here, we report a reference set of TRB genes extracted from the rhesus macaque genome, most of which were expressed by TCRβ sequences in our extensive database of TCRβ repertoires involved in CD8+ T cell responses to the immunodominant Mamu-A*01-restricted SL8/TL8 and CM9 epitopes derived from SIV. Although there is a high degree of similarity (93.0%) between the exons of the rhesus macaque and human TRB genes, important interspecies differences exist. These interspecies differences are emphasized by the lack of a one-to-one correspondence between the rhesus macaque and human TRBV genes, and could potentially limit the accuracy of studies that rely on human TCR genes to characterize rhesus macaque TCR repertoires.
The rhesus macaque TRB genes described herein will not only aid in the identification of the TRBV and TRBJ genes used by TCRβ sequences, they will also improve the accuracy of studies that aim to characterize the V(D)J recombination mechanisms that produce TCRβ repertoires. Indeed, several of the extracted rhesus macaque TRB genes have already been used in a study of TCRβ sequence sharing between macaques in the SIV-SL8/TL8-specific and SIV-CM9-specific CD8+ T cell responses39. This study required predictions of the potential V(D)J recombination mechanisms involved in producing the observed epitope-specific TCRβ repertoires, which were more reliable using the rhesus macaque TRB genes instead of the human TRB genes.
Rhesus macaques are frequently used to study fundamental aspects of immunology and investigate vaccine efficacy in a variety of infectious diseases. Increasing evidence, much of which has come from studies conducted with this non-human primate model, indicates that the clonotypic architecture of antigen-specific T cell populations is a fundamental determinant of immune control and disease outcome26, 45. Thus, the rhesus macaque TRB genes presented here provide a valuable tool for dissecting the molecular features of TCRβ repertoires that underlie such associations in this model.
METHODS
Extraction of TRB gene sequences from the rhesus macaque genome
The published rhesus macaque (Macaca mulatta) genome1 is available from the National Center for Biotechnology Information (NCBI) Rhesus Macaque Genome Resources website (http://www.ncbi.nlm.nih.gov/projects/genome/guide/rhesus_macaque/). The TRB gene locus is located on chromosome 3 (Accession number: NC_007860.1). The rhesus macaque chromosome 3 sequence was queried against all human TRB reference genes (obtained from the NCBI Human Resources website http://www.ncbi.nlm.nih.gov/projects/genome/guide/human/) using BLAST (Basic Local Alignment Search Tool)46 to identify regions in the rhesus macaque sequence that resembled human TRB genes. Results were filtered to those with e-value ≤ 0.001, total alignment length ≥ 35% of the human reference gene, and total percent identity ≥ 75% with the human reference gene. These parameters were chosen to minimize false positive search results. Overlapping regions were merged and all regions were extended in both the 5’ and 3’ directions to account for regions missed in BLAST’s local alignment search. Sequence alignments using ClustalW47 were then performed to compare each region of the rhesus macaque genome with each human TRB gene from the NCBI human reference set. The best human match to each macaque region was identified and then used as a guide to determine the exact length and terminal ends of the rhesus macaque TRB gene sequences, as well as intron and exon positions.
Comparison of rhesus macaque and human TRB gene sequences
We assessed the similarity between the rhesus macaque and the NCBI human TRB reference gene sequences (or the IMGT human TRB reference gene if the NCBI reference gene sequence was partial) by identifying the human TRB gene that had the highest overall percentage identity with each rhesus macaque TRB gene using a ClustalW alignment. We encountered the following scenarios: (i) a clearly identifiable one-to-one correspondence between a rhesus macaque and a human TRB gene; (ii) a rhesus macaque TRB gene with reasonable similarity to a group of human TRB genes; and, (iii) a human TRB gene with no reasonable correspondence to a rhesus macaque TRB gene. We therefore adopted the following approach to labelling the rhesus macaque TRB genes. For each rhesus macaque TRB gene, we first identified the group of human TRB genes to which it was most similar (e.g. TRBV1). We then numbered all rhesus macaque TRB genes which were most similar to this same group of human TRB genes according to the order in which the TRB sequences were found in the rhesus macaque genome (e.g. TRBV1-1, TRBV1-2, etc.). The ImMunoGeneTics (IMGT)48 nomenclature for TCR genes was used throughout.
Analysis of expressed epitope-specific TCRβ sequences using the rhesus macaque TRB genes
For all Mamu-A*01-restricted SIV-SL8/TL8-specific and SIV-CM9-specific TCRβ sequences, we performed a complete alignment analysis using the identified rhesus macaque TRB genes. This analysis determined for each epitope-specific TCRβ sequence the best-percentage-match TRBV, TRBD and TRBJ genes over the longest alignment length by initially aligning the TRBV gene at the 5’ end of the TCRβ sequence and then aligning the TRBJ gene at the 3’ end of the TCRβ sequence. A minimum percentage match of 77% over an alignment length of at least 50 nucleotides was required for alignment of the TRBV genes. For alignment of the TRBJ genes, a minimum percentage match of 70% was required over the length of the TRBJ exon. The TRBD genes were then aligned to the sequence interval between the identified TRBV and TRBJ regions. A match to a string of two or more nucleotides was considered to originate from the TRBD gene.
Supplementary Material
Supplementary information is available at the Immunology and Cell Biology website.
ACKNOWLEDGEMENTS
We thank Dr Mark Tanaka for assistance with the phylogenetic analysis and Associate Professor Andrew Collins for helpful discussions. This work was supported by the James S. McDonnell Foundation 21st Century Research Award/Studying Complex Systems, the Australian Research Council (ARC), and the National Institutes of Health (NIH). MPD is a Sylvia and Charles Viertel Senior Medical Research Fellow and DAP is a Medical Research Council (UK) Senior Clinical Fellow.
REFERENCES
- 1.Gibbs RA, Rogers J, Katze MG, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 2.Rivailler P, Jiang H, Cho YG, Quink C, Wang F. Complete nucleotide sequence of the rhesus lymphocryptovirus: genetic validation for an Epstein-Barr virus animal model. J Virol. 2002;76:421–426. doi: 10.1128/JVI.76.1.421-426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansen SG, Strelow LI, Franchi DC, Anders DG, Wong SW. Complete sequence and genomic analysis of rhesus cytomegalovirus. J Virol. 2003;77:6620–6636. doi: 10.1128/JVI.77.12.6620-6636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pitcher CJ, Hagen SI, Walker JM, et al. Development and homeostasis of T cell memory in rhesus macaque. J Immunol. 2002;168:29–43. doi: 10.4049/jimmunol.168.1.29. [DOI] [PubMed] [Google Scholar]
- 5.Hasegawa A, Moriya C, Liu H, et al. Analysis of TCRalphabeta combinations used by simian immunodeficiency virus-specific CD8+ T cells in rhesus monkeys: implications for CTL immunodominance. J Immunol. 2007;178:3409–3417. doi: 10.4049/jimmunol.178.6.3409. [DOI] [PubMed] [Google Scholar]
- 6.Cicin-Sain L, Messaoudi I, Park B, et al. Dramatic increase in naive T cell turnover is linked to loss of naive T cells from old primates. Proc Natl Acad Sci U S A. 2007;104:19960–19965. doi: 10.1073/pnas.0705905104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lackner AA, Veazey RS. Current concepts in AIDS pathogenesis: insights from the SIV/macaque model. Annu Rev Med. 2007;58:461–476. doi: 10.1146/annurev.med.58.082405.094316. [DOI] [PubMed] [Google Scholar]
- 8.Fan J, Liang X, Horton MS, et al. Preclinical study of influenza virus A M2 peptide conjugate vaccines in mice, ferrets, and rhesus monkeys. Vaccine. 2004;22:2993–3003. doi: 10.1016/j.vaccine.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 9.Kobasa D, Jones SM, Shinya K, et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445:319–323. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- 10.Lewinsohn DM, Tydeman IS, Frieder M, et al. High resolution radiographic and fine immunologic definition of TB disease progression in the rhesus macaque. Microbes Infect. 2006;8:2587–2598. doi: 10.1016/j.micinf.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Rivailler P, Carville A, Kaur A, et al. Experimental rhesus lymphocryptovirus infection in immunosuppressed macaques: an animal model for Epstein-Barr virus pathogenesis in the immunosuppressed host. Blood. 2004;104:1482–1489. doi: 10.1182/blood-2004-01-0342. [DOI] [PubMed] [Google Scholar]
- 12.Fogg MH, Garry D, Awad A, Wang F, Kaur A. The BZLF1 homolog of an Epstein-Barr-related gamma-herpesvirus is a frequent target of the CTL response in persistently infected rhesus macaques. J Immunol. 2006;176:3391–3401. doi: 10.4049/jimmunol.176.6.3391. [DOI] [PubMed] [Google Scholar]
- 13.Sequar G, Britt WJ, Lakeman FD, et al. Experimental coinfection of rhesus macaques with rhesus cytomegalovirus and simian immunodeficiency virus: pathogenesis. J Virol. 2002;76:7661–7671. doi: 10.1128/JVI.76.15.7661-7671.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price DA, Bitmansour AD, Edgar JB, et al. Induction and Evolution of Cytomegalovirus-Specific CD4+ T Cell Clonotypes in Rhesus Macaques. J Immunol. 2008;180:269–280. doi: 10.4049/jimmunol.180.1.269. [DOI] [PubMed] [Google Scholar]
- 15.Powers C, Fruh K. Rhesus CMV: an emerging animal model for human CMV. Med Microbiol Immunol. 2008;197:109–115. doi: 10.1007/s00430-007-0073-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edghill-Smith Y, Venzon D, Karpova T, et al. Modeling a safer smallpox vaccination regimen, for human immunodeficiency virus type 1-infected patients, in immunocompromised macaques. J Infect Dis. 2003;188:1181–1191. doi: 10.1086/378518. [DOI] [PubMed] [Google Scholar]
- 17.El Mubarak HS, Yuksel S, van Amerongen G, et al. Infection of cynomolgus macaques (Macaca fascicularis) and rhesus macaques (Macaca mulatta) with different wild-type measles viruses. J Gen Virol. 2007;88:2028–2034. doi: 10.1099/vir.0.82804-0. [DOI] [PubMed] [Google Scholar]
- 18.Rowe T, Gao G, Hogan RJ, et al. Macaque model for severe acute respiratory syndrome. J Virol. 2004;78:11401–11404. doi: 10.1128/JVI.78.20.11401-11404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMichael AJ. HIV vaccines. Annu Rev Immunol. 2006;24:227–255. doi: 10.1146/annurev.immunol.24.021605.090605. [DOI] [PubMed] [Google Scholar]
- 20.Maryanski JL, Jongeneel CV, Bucher P, Casanova JL, Walker PR. Single-cell PCR analysis of TCR repertoires selected by antigen in vivo: a high magnitude CD8 response is comprised of very few clones. Immunity. 1996;4:47–55. doi: 10.1016/s1074-7613(00)80297-6. [DOI] [PubMed] [Google Scholar]
- 21.Pewe L, Heard SB, Bergmann C, Dailey MO, Perlman S. Selection of CTL escape mutants in mice infected with a neurotropic coronavirus: quantitative estimate of TCR diversity in the infected central nervous system. J Immunol. 1999;163:6106–6113. [PubMed] [Google Scholar]
- 22.Kedzierska K, Day EB, Pi J, et al. Quantification of repertoire diversity of influenza-specific epitopes with predominant public or private TCR usage. J Immunol. 2006;177:6705–6712. doi: 10.4049/jimmunol.177.10.6705. [DOI] [PubMed] [Google Scholar]
- 23.Pewe LL, Netland JM, Heard SB, Perlman S. Very diverse CD8 T cell clonotypic responses after virus infections. J Immunol. 2004;172:3151–3156. doi: 10.4049/jimmunol.172.5.3151. [DOI] [PubMed] [Google Scholar]
- 24.Turner SJ, Doherty PC, McCluskey J, Rossjohn J. Structural determinants of T-cell receptor bias in immunity. Nat Rev Immunol. 2006;6:883–894. doi: 10.1038/nri1977. [DOI] [PubMed] [Google Scholar]
- 25.Nikolich-Zugich J, Slifka MK, Messaoudi I. The many important facets of T-cell repertoire diversity. Nat Rev Immunol. 2004;4:123–132. doi: 10.1038/nri1292. [DOI] [PubMed] [Google Scholar]
- 26.Davenport MP, Price DA, McMichael AJ. The T cell repertoire in infection and vaccination: implications for control of persistent viruses. Curr Opin Immunol. 2007;19:294–300. doi: 10.1016/j.coi.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Cornberg M, Chen AT, Wilkinson LA, et al. Narrowed TCR repertoire and viral escape as a consequence of heterologous immunity. J Clin Invest. 2006;116:1443–1456. doi: 10.1172/JCI27804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gillespie GM, Stewart-Jones G, Rengasamy J, et al. Strong TCR conservation and altered T cell cross-reactivity characterize a B*57-restricted immune response in HIV-1 infection. J Immunol. 2006;177:3893–3902. doi: 10.4049/jimmunol.177.6.3893. [DOI] [PubMed] [Google Scholar]
- 29.Meyer-Olson D, Shoukry NH, Brady KW, et al. Limited T cell receptor diversity of HCV-specific T cell responses is associated with CTL escape. J Exp Med. 2004;200:307–319. doi: 10.1084/jem.20040638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price DA, West SM, Betts MR, et al. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 2004;21:793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 31.Yu XG, Lichterfeld M, Chetty S, et al. Mutually exclusive T-cell receptor induction and differential susceptibility to human immunodeficiency virus type 1 mutational escape associated with a two-amino-acid difference between HLA class I subtypes. J Virol. 2007;81:1619–1631. doi: 10.1128/JVI.01580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen ZW, Kou ZC, Shen L, Reimann KA, Letvin NL. Conserved T-cell receptor repertoire in simian immunodeficiency virus-infected rhesus monkeys. J Immunol. 1993;151:2177–2187. [PubMed] [Google Scholar]
- 33.Chen ZW, Li Y, Zeng X, et al. The TCR repertoire of an immunodominant CD8+ T lymphocyte population. J Immunol. 2001;166:4525–4533. doi: 10.4049/jimmunol.166.7.4525. [DOI] [PubMed] [Google Scholar]
- 34.Chen ZW, Kou ZC, Lekutis C, et al. T cell receptor V beta repertoire in an acute infection of rhesus monkeys with simian immunodeficiency viruses and a chimeric simian-human immunodeficiency virus. J Exp Med. 1995;182:21–31. doi: 10.1084/jem.182.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen ZW, Shen L, Regan JD, Kou Z, Ghim SH, Letvin NL. The T cell receptor gene usage by simian immunodeficiency virus gag-specific cytotoxic T lymphocytes in rhesus monkeys. J Immunol. 1996;156:1469–1475. [PubMed] [Google Scholar]
- 36.Chen ZW, Shen Y, Kou Z, et al. Prolonged dominance of clonally restricted CD4(+) T cells in macaques infected with simian immunodeficiency viruses. J Virol. 2000;74:7442–7450. doi: 10.1128/jvi.74.16.7442-7450.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen L, Chen ZW, Letvin NL. The repertoire of cytotoxic T lymphocytes in the recognition of mutant simian immunodeficiency virus variants. J Immunol. 1994;153:5849–5854. [PubMed] [Google Scholar]
- 38.Sen P, Charini WA, Subbramanian RA, et al. Clonal focusing of epitope-specific CD8+ T lymphocytes in rhesus monkeys following vaccination and simian-human immunodeficiency virus challenge. J Virol. 2008;82:805–816. doi: 10.1128/JVI.01038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venturi V, Chin HY, Price DA, Douek DC, Davenport MP. The role of production frequency in the sharing of simian immunodeficiency virus-specific CD8+ TCRs between macaques. J Immunol. 2008;181:2597–2609. doi: 10.4049/jimmunol.181.4.2597. [DOI] [PubMed] [Google Scholar]
- 40.Currier JR, Stevenson KS, Kehn PJ, Zheng K, Hirsch VM, Robinson MA. Contributions of CD4+, CD8+, and CD4+CD8+ T cells to skewing within the peripheral T cell receptor beta chain repertoire of healthy macaques. Hum Immunol. 1999;60:209–222. doi: 10.1016/s0198-8859(98)00109-8. [DOI] [PubMed] [Google Scholar]
- 41.Zhou D, Kou Z, Ibegbu C, et al. The disruption of macaque CD4+ T-cell repertoires during the early simian immunodeficiency virus infection. J Med Primatol. 1999;28:174–180. doi: 10.1111/j.1600-0684.1999.tb00267.x. [DOI] [PubMed] [Google Scholar]
- 42.Bostik P, Stephenson ST, Lynch RM, Cardona A, Ansari AA. Maintenance of CD4+ T cell TCR Vbeta repertoire heterogeneity is characteristic of apathogenic SIV infection in non-human primate model of AIDS. Virology. 2007;369:324–328. doi: 10.1016/j.virol.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manuel ER, Charini WA, Sen P, et al. Contribution of T-cell receptor repertoire breadth to the dominance of epitope-specific CD8+ T-lymphocyte responses. J Virol. 2006;80:12032–12040. doi: 10.1128/JVI.01479-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheynier R, Henrichwark S, Wain-Hobson S. Sequence of the rhesus monkey T-cell receptor beta chain diversity and joining loci. Immunogenetics. 1996;43:83–87. doi: 10.1007/BF00186610. [DOI] [PubMed] [Google Scholar]
- 45.Price DA, Asher TE, Wilson NA, et al. Public clonotype usage identifies protective Gag-specific CD8+ T cell responses in SIV infection. J Exp Med. 2009;206:923–936. doi: 10.1084/jem.20081127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 47.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lefranc MP, Giudicelli V, Ginestoux C, et al. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 1999;27:209–212. doi: 10.1093/nar/27.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 50.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information is available at the Immunology and Cell Biology website.