Abstract
Stomatin, prohibitin, flotillin, and HflK/C (SPFH) domain proteins are membrane proteins that are widely conserved from bacteria to mammals. The molecular functions of these proteins have not been established. In mammals, the domain is often found in raft-associated proteins such as flotillin and podocin. We determined the structure of the SPFH domain of PH0470 derived from Pyrococcus horikoshii using NMR. The structure closely resembles that of the SPFH domain of the paralog PH1511, except for two C-terminal helices. The results show that the SPFH domain forms stable dimers, trimers, tetramers, and multimers, although it lacks the coiled-coil region for oligomerization, which is a highly conserved region in this protein family. The oligomers exhibited unusual thermodynamic behavior, as determined by circular dichroism, NMR, gel filtration, chemical cross-linking, and analytical ultracentrifugation. The oligomers were converted into monomers when they were heated once and then cooled. This transition was one-way and irreversible. We propose a mechanism of domain swapping for forming dimers as well as successive oligomers. The results of this study provide what to our knowledge are new insights into the common molecular function of the SPFH domain, which may act as a membrane skeleton through oligomerization by domain swapping.
Abbreviations used: CD, circular dichroism; NfeD, nodulation formation efficiency D; SPFH domain, stomatin, prohibitin, flotillin, and HflK/C domain; Slp-3, stomatin-like protein 3; ASIC, acid-sensing, nonvoltage-gated Na+-channel; HSQC, heteronuclear single quantum coherence spectroscopy; NOESY, nuclear Overhauser effect spectroscopy; PDB, the Protein Data Bank; RMSD, root mean-square deviation; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; MW, molecular weight
Introduction
The lipid raft membrane microdomain contains high concentrations of cholesterol and glycosphingolipid. It is known to play important roles in a variety of cellular functions, such as cell signaling, intracellular transport, and maintenance of cell polarity (1). Many proteins related to signal transduction are concentrated at the lipid raft microdomain, including channels, receptors, kinases, and their substrates.
Stomatin, prohibitin, flotillin, and HflK/C (SPFH) domain proteins such as stomatin, podocin, and flotillin are among the various proteins localized at the lipid raft microdomain (2–6). These proteins are widely conserved in mammals and have disparate origins in bacteria, archaea, and yeast. The most common domain architecture of SPFH domain proteins is a single copy of the SPFH domain at the N-terminus, followed by coiled-coil regions (Fig. 1 B). In many cases, a single transmembrane helix precedes the SPFH domain (2). To date, a common biological function for the SPFH domain has not been established. In mammals, SPFH domain proteins are localized at many organelles and are related to various physiological functions (4). For example, stomatin is a major component of the membrane skeleton in red blood cells. Stomatin is believed to maintain the membrane structure and reduce leakage of monovalent cations from red blood cells. Inherited anemia, called stomatocytosis, results from a genetic defect of the stomatin gene (7–9). Stomatin has been shown to bind and regulate ASICs (10,11) and the GLUT-1 glucose transporter (12,13). These lines of experimental investigation suggest that the physiological roles of SPFH domain proteins are related to the regulation of channels and transporters. On the other hand, research on podocin suggests that cholesterol binding is one of the molecular functions of the SPFH domain (14,15). Nevertheless, an expanded hypothesis—that the common molecular function of the SPFH domain is cholesterol binding—is less convincing because SPFH domains are found not only in eukaryotes but also in bacteria and archaea, which do not possess the cholesterol biosynthetic pathway (8,16). To date, two SPFH domain structures have been reported in PDB: PDB ID 1win (mouse flotillin-2) and 3bk6 (PH1511) (17). However, these high-resolution SPFH domain structures offer little insight into the molecular function of the domain.
Figure 1.
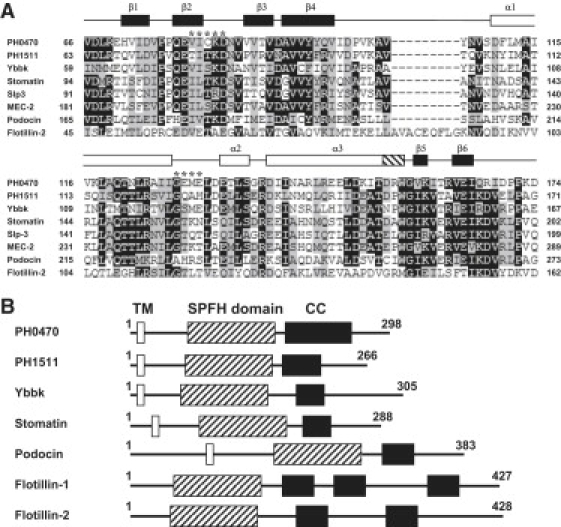
Sequence alignment and domain architecture of the SPFH domain. (A) Multiple sequence alignment domain-containing proteins. The secondary structure elements of PH0470SPFH are shown at the top of the diagram as open (α-helices), hatched (310-helix), and solid (β-strands) boxes. The residues corresponding to the subdomain latch are shown by an asterisk. Protein names and GenBank accession numbers are as follows: PH0470 (P. horikoshii; National Center for Biotechnology Information accession number NP_142449), PH1511 (P. horikoshii, NP_143371), YbbK (E. coli, YP_851665), Stomatin (human, NP_004090), Stomatin-like protein 3 (human, NP_660329), MEC-2 (C. elegans, AAA87552), Podocin (human, NP_055440), Flotillin 2 (human, NP_004466). (B) Domain arrangements and positions of the predicted coiled-coil region for SPFH domain-containing proteins. Open boxes are transmembrane helices, hatched boxes are the SPFH domains, and solid boxes show the coiled-coil region.
More than 350 prokaryotic and archaeal genomes have been shown to possess operons consisting of a gene pair of an NfeD homolog and a prokaryotic stomatin homolog, an SPFH domain protein (8). NfeD was originally identified as a genetic factor associated with the pSymB megaplasmid in the symbiotic bacterium Sinorhizobium meliloti, which enhances the rate of nodulation formation in host plants (18). Previously, we discovered the enzyme-substrate relationship between the NfeD/stomatin gene pair from the hyperthermophilic archaeon Pyrococcus horikoshii. The NfeD homolog PH1510 is a membrane-incorporated serine protease with a ClpP domain at the N-terminus. It cleaves the hydrophobic region of PH1511, a substrate and stomatin homolog (19). In addition, the genetic function of the NfeD/stomatin gene pair in Escherichia coli ybbJ/ybbK was shown to be involved in the heat stress response (20). Overexpression of ybbK partially reverses the growth retardation of Escherichia coli at 42°C, which is induced by the dual disruption of the nonessential membrane proteases ftsH and htpX. Growth retardation is suppressed more efficiently when both YbbK and YbbJ are overproduced.
On the other hand, several lines of physicochemical experimentation show that SPFH domain proteins form oligomers. For example, stomatin is known to form 9–12 mers using the region next to the C-terminal coiled-coil region (residues 264–272) (21). It has been demonstrated that prohibitins 1 and 2 form heterodimers through their SPFH domains; the dimers successively form a ring-shaped supercomplex through coiled-coil regions at the C-termini (22,23). In this oligomeric form, SPFH domain proteins have been proposed to perform a scaffolding function.
In this study, we examine the SPFH domain of PH0470 derived from P. horikoshii (residues 66–174, hereafter denoted as PH0470SPFH). We determined the solution structure of PH0470SPFH using standard NMR methods. In addition, PH0470SPFH was found to form oligomers even without the coiled-coil region C-terminal to the domain. We isolated several stable oligomeric forms (dimers, trimers, tetramers, and multimers). These oligomers displayed unusual behavior in that they irreversibly disassembled into monomers when heated to temperatures greater than 70°C and then cooled. This transition was one-way: the oligomers never formed from the monomers again. We further examined the oligomers using NMR, CD, chemical cross-linking, and analytical ultracentrifugation. Based on these results, we propose a hypothetical model in which PH0470SPFH forms oligomers through a domain swap rearrangement in which a region of secondary structure is exchanged between two or more monomeric protein domains.
Materials and methods
Protein techniques
Vectors for heterologous expression of GST fusion proteins of the SPFH domain of P. horikoshii (residues 66–174) were constructed using the PRESAT-vector methodology (24), as derived from the pGEX-4T3 vector (GE-Healthcare Biosciences, Piscataway, NJ). The 13C-15N-labeled recombinant proteins, for NMR spectroscopy, were expressed in E. coli BL21(DE3) codonplus RIL cells grown in M9 minimal medium at 30°C in the presence of 15NH4Cl and 13C-glucose as the sole nitrogen and carbon source. The cell lysate, after sonication, was cleared by centrifugation and then applied to a DEAE-Sepharose column (GE-Healthcare Biosciences). It was further purified using glutathione Sepharose affinity chromatography (GE-Healthcare Biosciences). The GST tag was removed by thrombin “on-beads”, the protease was removed using benzamidine Sepharose (GE-Healthcare Biosciences), and the thrombin-cleaved protein was purified by gel filtration. Each resultant construct contained a linker of four additional residues (corresponding to residues 62–65; see Fig. 3 A).
Figure 3.
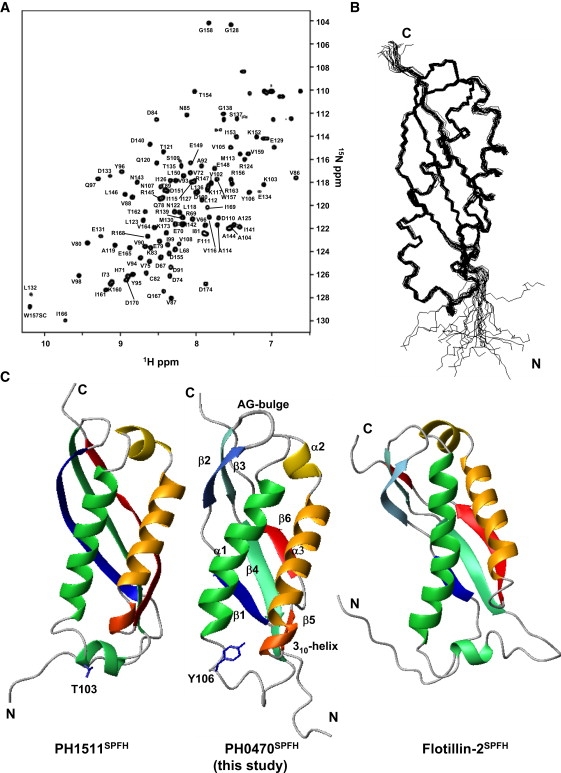
Solution structure of PH0470SPFH. (A) 1H-15N HSQC spectrum of PH0470SPFH. Numbers show residue assignments. (B) The best fit superposition of the 20 structures with the fewest structural violations. (C) Structural comparison of SPFH domain: left, PH1511SPFH; middle, PH0470SPFH; and right, FlotillinSPFH. The residues, which may destabilize the putative helix between β4 and α1 (Y106) in PH0470SPFH and the corresponding residue in PH1511SPFH (T103) is shown in blue.
Gel-filtration analysis
Gel-filtration analyses of purified protein samples were performed using a Superdex 75 Hiload (26/60) column that had been preequilibrated with 50 mM Tris-HCl and 150 mM NaCl (pH 7.5). The elution was carried out at a flow rate of 0.5 mL/min and was monitored continuously. Absorbance was measured at 280 nm.
NMR spectroscopy
Samples for NMR spectroscopy contained either 15N- or 13C/15N-labeled PH0470SPFH at concentrations of 0.7–1.0 mM in 10% D2O/90% H2O, 20 mM HEPES-K, pH 7.5. Backbone and side-chain assignments were obtained from 15N-HSQC, 13C-HSQC, HNCA, HNCO, HNCACB, CBCACONH, HCC(CO)NH, CC(CO)NH, and HCCH-TOCSY spectra recorded at 30°C using Bruker Avance spectrometers (500 MHz and 800 MHz, Avance; Bruker Biospin, Rheinstetten, Germany) equipped with cryomagnetic probes and an additional NMR spectrometer (900 MHz, Inova; Varian, Lexington, MA) (25,26). Data were processed using NMRPipe (27) and SPARKY (28) softwares. Interproton distances were obtained from three-dimensional (3D) 13C-edited and 15N-edited NOESY spectra recorded with 80 ms and 120 ms mixing times, respectively. Structures were calculated using the standard seven-iterative cycle protocol of the program CYANA, version 2.0.17 (29,30). All NOE cross peaks were picked manually using SPARKY. In total, 2229 meaningful NOE upper distance restraints were obtained, including 737 long-range distances. In addition, 18 hydrogen-bond restraints were obtained from the data of the H/D-exchange experiment, and confirmed by the presence of NOE signals between the corresponding residues. Dihedral angle restraints were calculated using the TALOS program on the basis of backbone atom chemical shifts (31). Model building of the domain-swapped dimer was performed with CYANA. The dihedral angle constraints and the distance constraints for the residues, excluding the hinge regions, which were obtained from the NMR experiments with the monomer, were used to generate the domain-swapped model.
Chemical cross-linking
The PH0470SPFH (0.8 mg/mL) was treated with 0% and 0.05% (wt/vol) glutaraldehyde for 60 min at room temperature in a buffer containing 50 mM HEPES-Na (pH 7.5) and 0.1 M NaCl. Each reaction was quenched using 0.2 M glycine-NaOH (pH 9.5) for 5 min at room temperature. Samples were resolved using SDS-PAGE (12.5% polyacrylamide gel) and stained with Coomassie brilliant blue.
Analytical ultracentrifugation
Sedimentation velocity experiments were carried out using an analytical ultracentrifuge (Optima XL-I; Beckman Coulter, Fullerton, CA) with a Ti rotor (An-50; Beckman). For sedimentation velocity experiments, cells with a standard Epon two-channel centerpiece and sapphire windows were used. The sample (400 μL) and reference buffer (420 μL) were loaded into the cells. The rotor temperature was equilibrated at 20°C in the vacuum chamber for 1–2 h before startup. Absorbance (OD280) scans were collected at 10 min intervals during sedimentation at 50 × 103 rpm. The sedimentation velocity experiments for PH0470SPFH were conducted at concentrations of 0.6 mg/mL. Partial specific volume of the protein, solvent density, and solvent viscosity were calculated from standard tables using the SEDNTERP program (32) (version 1.08). The resulting scans were analyzed using the continuous distribution (c(s)) analysis module in the SEDFIT program (33) (version 9.3). Sedimentation coefficient increments of 50 or 200 were used in the appropriate range for each sample. The frictional coefficient was allowed to float during fitting. The c(s) distribution was converted to a molar mass distribution c(M). In our experiments, the c(M) distribution gave a very good molar mass estimate because only one main species was observed in the c(s) distribution.
Sedimentation equilibrium experiments were also carried out in cells with a six-channel centerpiece and quartz windows. The sample concentrations were 0.15, 0.30, and 0.60 mg/mL. The absorbance wavelength was set at 280 nm, and data were acquired at 20°C. Data were obtained at 10, 15, 20, 25, and 30 × 103 rpm. A total equilibration time of 14 h was used for each speed, with a scan taken at 12 h to ensure that equilibrium had been reached. The optical baseline was determined by accelerating at 50 × 103 rpm at the end of data collection. Data analysis was performed by global analysis of data sets obtained at different loading concentrations and rotor speeds using UltraSpin software (MRC Center for Protein Engineering, Cambridge, UK, http://www.mrc-cpe.cam.ac.uk/ultraspin).
Electron microscopy
For negatively stained electron microscopy, the sample was gently resuspended by pipetting on a glow-discharged, carbon-coated copper grid and stained with chilled 2% uranyl acetate. The JEM-1010 microscope (JEOL, Tokyo, Japan) was operated at an acceleration voltage of 100 kV and images were recorded with a 1k × 1k slow scan charge-coupled device camera (TVIPS). During the entire procedure, samples were kept at 4°C.
PDB and Biological Magnetic Resonance Bank accession numbers
The atomic coordinates of the 29 best PH0470SPFH NMR structures have been deposited in the PDB under the accession number 2rpb (http://www.wwpdb.org/). Chemical shift data are available from the Biological Magnetic Resonance Bank under accession number 11060 (http://www.bmrb.wisc.edu/).
Results
Oligomer formation of PH0470SPFH
We obtained two mutually distinct 1H-15N HSQC spectra of PH0470SPFH according to the method of protein preparation either with heat treatment after thrombin digestion or without heat treatment (see Fig. S1 in the Supporting Material). PH0470SPFH without heat treatment gave few HSQC signals after eight measurement scans on a 500 MHz NMR spectrometer with 0.1 mM samples, whereas the heat-treated protein gave an ideal “NMR-ready” HSQC spectrum with finely dispersed amide proton signals. Though both samples were of near-identical purity, we further aimed to determine any difference by gel-filtration analysis. In the chromatograph of unheated PH0470SPFH several peaks, corresponding to MWs higher than that of monomer protein, were repeatedly observed. In contrast, only two peaks were detected for the heat-treated protein corresponding to monomers and multimers of a larger MW (Fig. 2 A).
Figure 2.
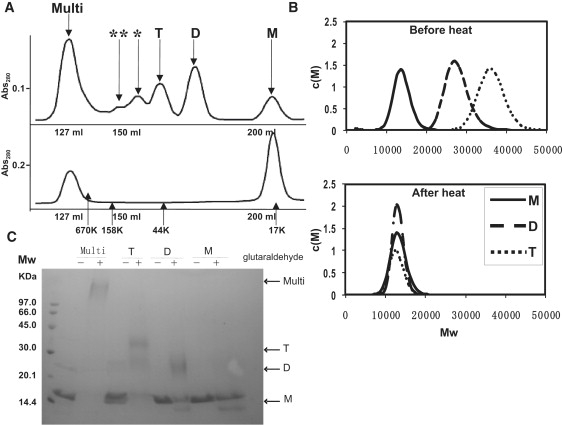
Oligomer formation of PH0470SPFH. (A) Gel-filtration profiles of PH0470SPFH. The upper panel shows the elution pattern without heat treatment; the lower panel shows the pattern with heat treatment. (B) Distribution of molar mass (c(M)) for PH0470SPFH. The c(s) distribution was converted to a molar mass distribution c(M). The upper panel shows the c(M) distribution without heat treatment; the lower panel shows the distribution with heat treatment. (C) Chemical cross-linking, PH0470SPFH (0.8 mg/mL) with glutaradehyde (0.05%) for 60 min at room temperature. Each sample was resolved using SDS-PAGE and stained with Coomassie brilliant blue. Numbers on the left indicate MW markers. The size of each oligomer state is shown to the right of the gel.
Unheated protein samples were examined using analytical ultracentrifugation. Three protein samples were isolated from the unheated protein samples by gel filtration chromatography and then analyzed by sedimentation equilibrium analysis. Their MWs were calculated as 13.6 (±0.1) × 103, 27.0 (±0.1) × 103, and 37.5 (±0.1) × 103 using a simple monomeric analysis model (data not shown), given that the theoretical MW of PH0470SPFH is 12,853. This corresponds to monomers, dimers, and trimers, respectively. Moreover, the MW determined by sedimentation velocity measurements agreed very well with those of the sedimentation equilibrium analysis (Fig. 2 B). These analytical ultracentrifugation experiments suggested that isolated monomers, dimers, and trimers were very stable and that equilibration among them was very slow. Subsequently, the monomers, dimers, and trimers were heat-treated and resubjected to ultracentrifugation. Both the dimers and trimers had disassembled into monomers (Fig. 2 B). Notably, peaks likely attributable to tetramers and pentamers were also observed in the unheated preparation of PH0470SPFH (Fig. 2 A). These oligomers also appeared to be disassembled into monomers upon heating (Fig. 2 A). The dimer, trimer, and multimer formation was further confirmed using a chemical cross-linking experiment with 0.05% glutaraldehyde. Complexes corresponding to dimers, trimers, and multimers larger than MW 600 k were isolated by gel-filtration and resolved on SDS-PAGE (Fig. 2 C).
We performed CD analysis to investigate the secondary structure of these oligomeric forms. The monomer, dimer, and trimer spectra were identical, which confirms that the secondary structure of PH0470SPFH was preserved among all samples (Fig. S6 A). These results suggest that the PH0470SPFH dimers and trimers are not denatured proteins; rather, they comprise folded protomers with identical secondary structure.
Next, we examined the critical temperatures of the oligomer-to-monomer transition using NMR. Since the oligomer-to-monomer transition was one-way, we cannot refer to this as the melting temperature. Heating for 10 min at 40°C, 50°C, and 60°C did not affect the HSQC spectra. The disassembly of the oligomer to monomer occurred partially at 70°C and was almost completed at 80°C (data not shown). Long-term storage of the isolated dimers at 4°C promoted partial formation of monomers, although conversely, the long storage of monomers at the same temperature did not result in dimer or oligomer formation (data not shown).
Taken together, the data suggest that 1), the monomer is the most stable species; 2), the transition from oligomer to monomer is irreversible; 3), the equilibrium between the monomer and dimer/trimer is very slow; and 4), the transition rate from the dimer/trimer to monomer is temperature-dependent. As a continuation of our studies, we decided to solve the tertiary structure of the monomer. The monomeric form of PH0470SPFH was separated by gel filtration from the heat-treated mixture of oligomers and subjected to NMR measurements.
Structural determination of PH0470SPFH
The NMR signals of PH0470SPFH were assigned using a combination of standard 3D NMR techniques (25,26). The assignment of the backbone 1H-15N HSQC peaks is portrayed in Fig. 3 A. The side-chain signals were also assigned according to a standard protocol, and 99% of the nonexchangeable protons were assigned. An ensemble of 20 structures of low CYANA (29,30) target function was generated from 2229 experimental NMR constraints. The resultant 20 structures satisfy the experimental constraints very well (Table 1). For unknown reasons, the stereochemical quality of the members of the ensemble is slightly worse than the standard, with all backbone ϕ/ψ angles occupying the most favored (80%) or slightly less favored (19%) regions of the Ramachandran plot (Fig. S2). Excluding the disordered regions—the N-terminal region (for residues from the expression vector + residues 66–69) and the C-terminal residues (residues 171–174)—the RMSD values are 0.33 Å for backbone heavy atoms and 0.78 Å for all heavy atoms (Fig. 3 B).
Table 1.
Experimental restraints and statistics for 20 structures of PH0470SPFH
| Distance restraints | |
|---|---|
| Total number of restraints | 2229 |
| Intraresidue | unused |
| Sequential restraints [ | i-j | = 1 ] | 1053 |
| Medium-range restraints [ 1 < | i-j | ≤ 4 ] | 439 |
| Long-range restraints [ | i-j | >4 ] | 737 |
| Dihedral angle restraints | 118 |
| ϕ/ψ/χ | 59/59/0 |
| Hydrogen-bond restraints | 18 |
| Statistics used for and obtained from the structure calculations | |
| Final statistics (20/200) | |
| Cutoffs: distance (0.3 Å) and angle (3.0 deg) | |
| Maximum target function | 0.14 |
| Maximum violations | |
| Distance violation (Å) | 0.17 |
| Angle violation (deg) | 7.83 |
| Coordinate precision (residues 70–170) | |
| Backbone RMSD (Å) | 0.33 |
| Heavy atoms RMSD (Å) | 0.78 |
| Ramachandran plot statistics (%) (residues 62–174) | |
| Residues in most favored regions | 80.0 |
| Residues in additionally allowed regions | 19.3 |
| Residues in generously allowed regions | 0.7 |
| Residues in disallowed regions | 0.0 |
Comparison with other SPFH domain structures
PH0470SPFH is an α/β-fold domain composed of three α-helices (α1(residues 110–127), α2(134–137), and α3(139–157)) and six antiparallel β-strands (β1(71–74), β2(78–81), β3(87–90), β4(92–98), β5 (159–160), and β6 (164–166); Fig. 3 C). In addition, a short 310-helix from 155 to 157 (D155, R156, and W157) and the residues C82, N85, and V86 participating in the AG-type bulge are involved. We compared the structures of PH0470SPFH with other known SPFH structures. To date, two structures of the SPFH domain are known: mouse flotillin-2 (43–172) (PDB 1win) and PH1511 (56–234) (PDB 3bk6), the paralog of PH0470 (17). PH0470SPFH shares 19% sequence identity with mouse flotillinSPFH, and these structures are quite similar (RMSD 2.4 Å). A short loop (residues I99-S109 in PH0470SPFH) makes PH0470SPFH fold more compactly than flotillinSPFH (Fig. 3 C). In addition, the structure of PH0470SPFH superimposes well onto that of PH1511SPFH (51.9% sequence identity and RMSD 2.2 Å). The residues in PH1511SPFH corresponding to the 310-helix (D152, P153, and W154) and the AG bulge (T79, N82, and V83) are conserved in PH0470SPFH, although one major difference is the absence of the first short α-helix in PH0470SPFH. This difference is probably attributable to the bulky aromatic side chain of Y106 (T103 in PH1511), a nonconserved residue in all other SPFH domain proteins, causing collapse of the α-helix (Fig. 3 C).
Several factors might determine the molecular interaction of PH0470SPFH and its putative target. We compared arrangements of residues on the protein surface of three SPFH domain structures with respect to charge distribution and amino acid conservation by producing surface electric potential maps (Fig. 4 A and Fig. S3). The genetically conserved residues on the surface of PH0470SPFH assigned by the program Consurf (34) were mapped onto the molecular surface and their charges and hydrophobicity were examined (Fig. 4 B). From this analysis, two evolutionarily conserved regions among prokaryotic stomatin homologs were found. Region I, surrounding α1, is mainly hydrophobic with several positively charged residues. Among them, R124 is particularly well conserved and exposed to the solvent. Region II, located between the sheet composed by β4 and β6, contains a cluster of negative charges. In this region, R139 is also well conserved and may form an ionic bridge between the relatively conserved residues, such as D140 and/or E165. Because PH0470 is a membrane-bound protein with a single transmembrane helix preceding the SPFH domain, PH0470SPFH exists near the plasma membrane. We assume that region I with its nonpolar residues is directed toward the membrane, whereas the polar residues of region II are exposed to the cytoplasm.
Figure 4.
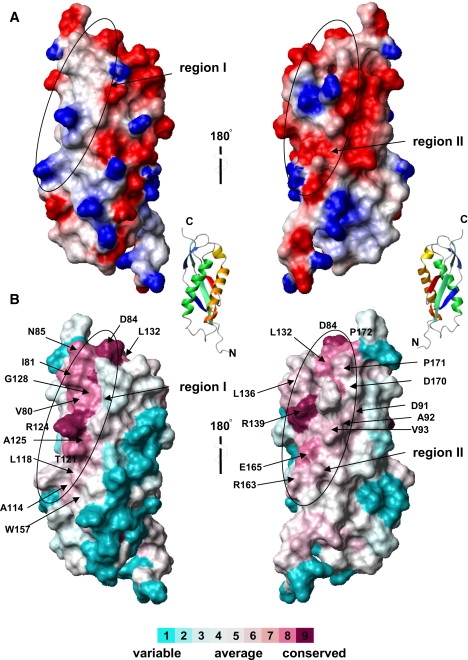
Surface characteristics of PH0470SPFH. (A) Electrostatic surface potential diagrams with positive (blue) and negative (red) electrostatic potentials mapped onto a van der Waals surface diagram of the conserved surface patch. The color scale ranges between −20 kBT (red) and +20 kBT (blue), where kB is Boltzmann's constant and T is temperature. (B) Sequence conservation with variable (cyan) and conservative (purple) residues is mapped onto the surface. This figure was produced by ConSurf (34) and MOLMOL (38). A front view (left) and rear view (right) are shown. Ribbon diagram of PH0470SPFH in the same molecular orientations as in A and B.
Comparison of HSQC spectra of the monomer and dimer of PH0470SPFH
We measured 1H-15N HSQC spectra of the heat-treated monomers, and isolated dimers and trimers of PH0470SPFH. Only a faint 1H-15N HSQC spectrum with a poor signal/noise ratio was obtained for the trimers, probably due to low sample concentration and increased MW. For the dimers, we were able to observe a substantial number of sharp amide proton signals (Fig. 5 A). In comparison, in the HSQC spectra of monomers, we found that many signals originating from the residues within the loop (I99-S109) and turn (R156-G158) regions changed their resonance positions dramatically (Fig. 5, B and C). The loop and turn regions were therefore inferred to be important for dimer formation.
Figure 5.

NMR chemical shift differences between the backbone amide and 15N resonances of the PH0470SPFH monomer and those of the PH0470SPFH dimer. (A) 1H-15N HSQC spectrum of PH0470SPFH monomer and dimer. The monomer spectrum is shown in black; the dimer spectrum is shown in red. (B) NMR chemical shift differences between the backbone amide and 15N resonances of PH0470SPFH monomer and dimer. The chemical shift changes were calculated using the equation {(δN × 0.2)2 + δHN2}1/2, where δN and δHN are the chemical shift differences between the backbone nitrogen resonances and the amide proton resonances, respectively. (C) The surface residues indicated in light and dark green showed medium and large signal changes (δ > 0.05 and δ > 0.10, respectively) mapped on the surface of PH0470SPFH.
Discussion
SPFH domains are widely found in the genomes of various organisms, including bacteria, archaea, and eukaryotes. Although the common molecular function of the SPFH domain has not been established, many lines of experimentation indicate that SPFH domain proteins function as scaffolding proteins on lipid membranes. Several binding partners of SPFH domains have been reported, including cholesterol (podocin and MEC-2 (14)) and actin (flotillin and stomatin) (35,36). In this study, we discovered two conserved regions on the surface of the SPFH domain, hydrophobic (region I; Fig. 4) and hydrophilic with conserved charged residues (region II). The hydrophobic nature of region I is evolutionarily conserved over prokaryotic stomatins, mammalian stomatin, podocin, and MEC-2, but not in flotillins (data not shown). The charged surface of region II is more widely conserved, and also occurs in flotillins. This observation is consistent with convergent evolution of flotillins among other SPFH domain proteins (37). Assuming that lipid binding is a key function of SPFH domains, the hydrophobic interface (region I) may provide a binding site for lipids such as cholesterol. This putative lipid-binding site may involve the conserved 310-helix and the bulge that surrounds the hydrophobic region. In contrast, the charged surface (region II), opposite the putative lipid-binding site, may interact with the target proteins. We were unable to find a narrow pocket or cleft, such as the cleft found in the PDZ or WW domains, in any of the three SPFH domain structures. The protein-protein interaction of SPFH domains is assumed to be a rigid-body rather than an induced-fit contact. The latter is commonly seen in protein-peptide interactions, in which an induced fit of a flexible peptide ligand occurs upon binding to the protein. Actin is the major candidate as the target of eukaryotic SPFH domain proteins (35,36). The actin-like cytoskeleton protein Mre is widely conserved in bacteria and archaea, and may be a candidate binding partner of the prokaryotic stomatins.
Many reports have described oligomer formation by SPFH domain proteins under physiological conditions. Among the interfaces involved in oligomerization are two distinct classes: the coiled-coil region and the SPFH domain itself. A typical example of the former case is flotillin. It is widely believed that the coiled-coil region is involved in homo- and heterophilic protein oligomerization, and indeed almost all SPFH domain proteins have a coiled-coil region in their C-terminal halves (Fig. 1). Nevertheless, there are numerous examples of the SPFH domain itself forming oligomers, including podocin, MEC-2, and prohibitins (14,22,23). Moreover, the SPFH domain from PH1511, a paralog of PH0470, forms trimers utilizing contacts within the SPFH domain without the coiled-coil formation. In the crystal structure of PH1511SPFH (56–234), PH1511SPFH forms head-to-tail trimers provided by the interaction between one β-sheet (residues 57–59) and the β-sheet (154–168) of a neighboring molecule, and the interaction between one helix (96–103) and the helix (178–186) of a neighboring molecule. However, PH0470SPFH comprises residues 66–174, whose corresponding residues are 63–171 in PH1511SPFH, and PH0470SPFH lacks the important region for forming head-to-tail type trimers, such as the N-terminal short β-sheet and the C-terminal helical regions (Fig. S4). The nature of oligomerization of PH0470SPFH is therefore expected to be different from that observed in the PH1511 crystal.
The assumption described above was partly supported by the comparison of 1H-15N HSQC spectra from monomers and dimers of PH0470, in which residues belonging to the loop region (I99-S109) and the peripheral residues showed signal changes (Fig. 5). There are usually two possible explanations for signal changes of amide groups upon protein interactions: 1), the surface contact between the two molecules; and 2), structural changes within the monomer subunit. Considering that the dimeric and monomeric forms are not in equilibrium and that the dimers irreversibly form monomers after heat treatment, it is unlikely that PH0470SPFH forms dimers through surface contacts involving residues that produce signal changes in the 1H-15N HSQC spectra. Therefore, we propose what to our knowledge is a new alternative mechanism for the oligomerization of this domain: the domain swap model. This model is based on the two-layer sandwich structure, the common structure of SPFH domains. SPFH domains can be divided into an α-helical subdomain and a β-sheet subdomain, which are connected by a hinge region and folded together. We hypothesize that PH0470SPFH opens once between the α-helical and β-sheet subdomains, and then form domain swap dimers with nearby molecules by swapping subdomains with each other (Fig. 6 and Fig. S5). In this model, the two hinge regions corresponding to loop 2 (I99-S109) and the turn (residues R156-G158) are the regions with the largest structural changes. The atomic environments of most residues, except for those of the hinge region, would be preserved between the monomers and the oligomers, which is consistent with NMR data. Indeed, many residues in the hinge region showed substantial signal changes in the HSQC spectra (Fig. 5, A–C), whereas those in the core region were mostly unchanged.
Figure 6.
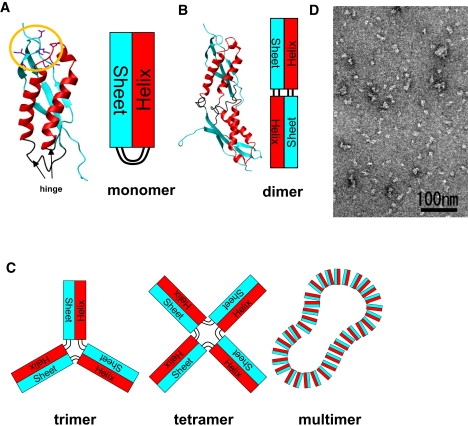
Domain swap oligomer model of PH0470SPFH. (A) PH0470SPFH monomer structure. Left: The α-helical region is shown in red, and β-sheet is shown in cyan. The region shown by an arrow corresponds to the hinge region. The region enclosed in the orange circle corresponds to the latch region. Right: Simplified model of PH0470SPFH monomer structure. Red and cyan boxes represent the α-helical and β-sheet subdomains, respectively. (B) PH0470SPFH dimer model structure by 3D-domain swapping (left) with simplified block diagram (right). (C) Simplified model of trimer, tetramer, and multimer formation. (D) Negatively stained electron micrograph of the PH0470SPFH multimer.
Although oligomers are possibly artifacts of overexpression of the archaeal protein in E. coli, oligomer formation of PH0470SPFH was reproducible in two distinct expression systems: 1), the GST-fusion system; and 2), His6-tag fusion system. His6-tagged PH0470SPFH was demonstrated to display identical oligomerization characteristics (data not shown). When an SPFH domain forms oligomers from monomers, the first step involves opening of the two subdomains, which probably occurs cotranslationally in E. coli. It is noteworthy that residues V80–D84 and G128-E131, corresponding to the “latch” that fastens the α-helical and β-sheet subdomains, are well conserved throughout SPFH domains (Fig. 1 A). Furthermore, this domain swap model can explain the uncommon behavior of the oligomers, such as the temperature-dependent, one-way transition from dimer to monomer. A high-energy barrier between monomers and dimers is assumed to hinder the disassembly process, in addition to the opening of subdomains in the monomers (Fig. S5). During transitions, interactions between many residues must be rearranged. Consequently, the transition from oligomer to monomer requires that this high-energy barrier be overcome by heating to temperatures greater than 70°C. No reformation of any oligomers occurs upon cooling. Although we attempted to form dimers from monomers at various speeds of annealing, we remained unsuccessful after many trials. Consequently, the structure determination of dimer by NMR became unachievable because we were unable to isolate the isotopically labeled and nonlabeled combination of the protein dimer. Work is therefore in progress to obtain structural information using crystallographic and small-angle x-ray scattering techniques.
Our domain swap model is also consistent with many unique phenomena observed during this study. For example, it can explain the formation not only of dimers but also trimers, tetramers, and various oligomers of indefinite number. This differs greatly from the other surface contact models, which cannot explain the formation of oligomers larger than dimers. The domain swap model is consistent with observations of successive multimers of MW > 600 k. We examined these ∼600 k oligomers using negatively stained electron microscopy. We observed particles of similar sizes but varying shapes (Fig. 6 D), although we did not find ring-shaped oligomers, such as prohibitins (22,23). Finally, the domain swap model can explain why the dimer-to-monomer transition is not reversible. This unusual behavior is inconsistent with the model of surface contact. The domain swap model predicts the presence of a putative “open” high-energy conformation as a transition state for oligomer disassembly. The presence of this “open” conformation was partially supported by a combination of CD spectra (Fig. S6 B) and “pulse-chased” H/D exchange NMR experiments (Fig. S7). The CD spectra of PH0470SPFH at 60°C, 70°C, and 80°C are mostly identical to that at 30°C. In the control H/D exchange experiment, amide protons at the interface between the α- and β- subdomains were resistant to H/D exchange after 15 min at 30°C and pH 7.5. Increasing the temperature to 80°C for a short period also increased the H/D exchange rate of those amide protons. These results are consistent with the hypothesis that a very small part of PH0470SPFH may adopt a putative “open” form at 80°C. Monomer formation is kinetically more advantageous than that of oligomers when the transition state closes.
As described previously, many SPFH domains, such as stomatin and podocin, are considered to function as scaffold proteins, with the oligomers possibly promoting recruitment of other proteins at the membrane surface. One idea is that the SPFH protein oligomer may form a patch on the lipid raft microdomain. In general, it is thought that the interface of oligomerization is located at the coiled-coil region. However, in this study, we demonstrate that an SPFH domain can form a metastable oligomer, probably through a domain swap structure rearrangement. This suggests that through multivalent protein interactions, SPFH domain proteins can form more complex supramolecular structures, such as membrane covering 2D sheets. Furthermore, the results of this study suggest a possible switch mechanism between different oligomeric states of the SPFH domain at high temperature; that is, the metastable SPFH domain oligomer acts as a sensor of heat or other physicochemical stresses. Of interest, such a function is seen for Slp-3, a protein that is known to regulate the mechanosensitive channel in mouse (11).
In conclusion, the results of this study reveal unusual thermosensitive oligomeric forms of the SPFH domain, which we propose are formed by a domain swap mechanism. To our knowledge, this is the first report of oligomers formed via the core of the SPFH domain without involving the coiled-coil or helical regions. In the case of P. horikoshii, PH0470SPFH may form oligomers only when the archaea are subjected to environmental stresses, such as decreased temperatures in hydrothermal vents, whereas the coiled-coil region of PH0470 is supposed to continue forming oligomer, thereby changing PH0470 to the different oligomeric state. The roles of the two possible oligomeric states may be related to the heat stress response that induces thermal switching, which is due to the disassembly of PH0470SPFH oligomers. Finally, a putative molecular function of SPFH proteins in which a large oligomeric complex is formed on the membrane surface, known as the membrane skeleton, is suggested.
Supporting Material
Seven figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(09)01297-1.
Supporting Material
Acknowledgments
We are grateful to Drs. M. Shirakawa, H. Tochio, and S.-Y. Park for their helpful suggestions. We thank Ms. C. Addy (YCU) for her critical reading of the manuscript.
This work was supported by grants to H.H. from the Japanese Ministry of Education, Culture, Sports, Science and Technology, and the Japan Science and Technology Agency.
References
- 1.Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 2.Morrow I.C., Parton R.G. Flotillins and the PHB domain protein family: rafts, worms and anaesthetics. Traffic. 2005;6:725–740. doi: 10.1111/j.1600-0854.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- 3.Tavernarakis N., Driscoll M., Kyrpides N.C. The SPFH domain: implicated in regulating targeted protein turnover in stomatins and other membrane-associated proteins. Trends Biochem. Sci. 1999;24:425–427. doi: 10.1016/s0968-0004(99)01467-x. [DOI] [PubMed] [Google Scholar]
- 4.Browman D.T., Hoegg M.B., Robbins S.M. The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol. 2007;17:394–402. doi: 10.1016/j.tcb.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Salzer U., Prohaska R. Stomatin, flotillin-1, and flotillin-2 are major integral proteins of erythrocyte lipid rafts. Blood. 2001;97:1141–1143. doi: 10.1182/blood.v97.4.1141. [DOI] [PubMed] [Google Scholar]
- 6.Dermine J.F., Duclos S., Garin J., St-Louis F., Rea S. Flotillin-1-enriched lipid raft domains accumulate on maturing phagosomes. J. Biol. Chem. 2001;276:18507–18512. doi: 10.1074/jbc.M101113200. [DOI] [PubMed] [Google Scholar]
- 7.Stewart G.W., Argent A.C., Dash B.C. Stomatin: a putative cation transport regulator in the red cell membrane. Biochim. Biophys. Acta. 1993;1225:15–25. doi: 10.1016/0925-4439(93)90116-i. [DOI] [PubMed] [Google Scholar]
- 8.Green J.B., Fricke B., Chetty M.C., von During M., Preston G.F. Eukaryotic and prokaryotic stomatins: the proteolytic link. Blood Cells Mol. Dis. 2004;32:411–422. doi: 10.1016/j.bcmd.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 9.Delaunay J. The hereditary stomatocytoses: genetic disorders of the red cell membrane permeability to monovalent cations. Semin. Hematol. 2004;41:165–172. doi: 10.1053/j.seminhematol.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Price M.P., Thompson R.J., Eshcol J.O., Wemmie J.A., Benson C.J. Stomatin modulates gating of acid-sensing ion channels. J. Biol. Chem. 2004;279:53886–53891. doi: 10.1074/jbc.M407708200. [DOI] [PubMed] [Google Scholar]
- 11.Wetzel C., Hu J., Riethmacher D., Benckendorff A., Harder L. A stomatin-domain protein essential for touch sensation in the mouse. Nature. 2007;445:206–209. doi: 10.1038/nature05394. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J.Z., Hayashi H., Ebina Y., Prohaska R., Ismail-Beigi F. Association of stomatin (band 7.2b) with Glut1 glucose transporter. Arch. Biochem. Biophys. 1999;372:173–178. doi: 10.1006/abbi.1999.1489. [DOI] [PubMed] [Google Scholar]
- 13.Montel-Hagen A., Kinet S., Manel N., Mongellaz C., Prohaska R. Erythrocyte Glut1 triggers dehydroascorbic acid uptake in mammals unable to synthesize vitamin C. Cell. 2008;132:1039–1048. doi: 10.1016/j.cell.2008.01.042. [DOI] [PubMed] [Google Scholar]
- 14.Huber T.B., Schermer B., Muller R.U. Podocin and MEC-2 bind cholesterol to regulate the activity of associated ion channels. Proc. Natl. Acad. Sci. USA. 2006;103:17079–17086. doi: 10.1073/pnas.0607465103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schermer B., Benzing T. Lipid-protein interactions along the slit diaphragm of podocytes. J. Am. Soc. Nephrol. 2009;20:473–478. doi: 10.1681/ASN.2008070694. [DOI] [PubMed] [Google Scholar]
- 16.Pearson A., Budin M., Brocks J.J. Phylogenetic and biochemical evidence for sterol synthesis in the bacterium Gemmata obscuriglobus. Proc. Natl. Acad. Sci. USA. 2003;100:15352–15357. doi: 10.1073/pnas.2536559100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yokoyama H., Fujii S., Matsui I. Crystal structure of a core domain of stomatin from Pyrococcus horikoshii illustrates a novel trimeric and coiled-coil fold. J. Mol. Biol. 2008;376:868–878. doi: 10.1016/j.jmb.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Rodriguez F.M., Toro N. Sinorhizobium meliloti nfe (nodulation formation efficiency) genes exhibit temporal and spatial expression patterns similar to those of genes involved in symbiotic nitrogen fixation. Mol. Plant Microbe Interact. 2000;13:583–591. doi: 10.1094/MPMI.2000.13.6.583. [DOI] [PubMed] [Google Scholar]
- 19.Yokoyama H., Matsui I. A novel thermostable membrane protease forming an operon with a stomatin homolog from the hyperthermophilic archaebacterium Pyrococcus horikoshii. J. Biol. Chem. 2005;280:6588–6594. doi: 10.1074/jbc.M411748200. [DOI] [PubMed] [Google Scholar]
- 20.Chiba S., Ito K., Akiyama Y. The Escherichia coli plasma membrane contains two PHB (prohibitin homology) domain protein complexes of opposite orientations. Mol. Microbiol. 2006;60:448–457. doi: 10.1111/j.1365-2958.2006.05104.x. [DOI] [PubMed] [Google Scholar]
- 21.Snyers L., Umlauf E., Prohaska R. Oligomeric nature of the integral membrane protein stomatin. J. Biol. Chem. 1998;273:17221–17226. doi: 10.1074/jbc.273.27.17221. [DOI] [PubMed] [Google Scholar]
- 22.Tatsuta T., Model K., Langer T. Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol. Biol. Cell. 2007;16:248–259. doi: 10.1091/mbc.E04-09-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merkwirth C., Langer T. Prohibitin function within mitochondria: essential roles for cell proliferation and cristae morphogenesis. Biochim. Biophys. Acta. 2009;1793:27–32. doi: 10.1016/j.bbamcr.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 24.Goda N., Tenno T., Takasu H., Hiroaki H., Shirakawa M. The PRESAT-vector: asymmetric T-vector for high-throughput screening of soluble protein domains for structural proteomics. Protein Sci. 2004;13:652–658. doi: 10.1110/ps.03439004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamazaki T., Lee W., Arrowsmith C.H., Muhandiram D.R., Kay L.E. A suite of triple resonance NMR experiments for the backbone assignment of 15N, 13C, 2H labeled proteins with high sensitivity. J. Am. Chem. Soc. 1994;116:11655–11666. [Google Scholar]
- 26.Cavanagh J., Fairbrother W.J., Palmer A.G., Skelton N.J. Academic Press; San Diego, CA: 1996. Protein NMR Spectroscopy. [Google Scholar]
- 27.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 28.Goddard T.D., Kneller D.G. University of California; San Francisco, CA: 2004. Sparky 3. [Google Scholar]
- 29.Herrmann T., Guntert P., Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 30.Guntert P. Automated NMR protein structure calculation. Prog. Nucl. Magn. Reson. Spectrosc. 2003;43:105–125. [Google Scholar]
- 31.Cornilescu G., Delaglio F., Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 32.Laue T.M., Shah B.D., Ridgeway T.M., Pelletier S.L. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S.E., Rowe A.J., Horton L.C., editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; Cambridge, UK: 1992. pp. 90–125. [Google Scholar]
- 33.Schuck P., Perugini M.A., Gonzales N.R., Howlett G.J., Schubert D. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys. J. 2002;82:1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langhorst M.F., Solis G.P., Hannbeck S., Plattner H., Stuermer C.A. Linking membrane microdomains to the cytoskeleton: regulation of the lateral mobility of reggie-1/flotillin-2 by interaction with actin. FEBS Lett. 2007;581:4697–4703. doi: 10.1016/j.febslet.2007.08.074. [DOI] [PubMed] [Google Scholar]
- 36.Wilkinson D.K., Turner E.J., Parkin E.T., Garner A.E., Harrison P.J. Membrane raft actin deficiency and altered Ca2+-induced vesiculation in stomatin-deficient overhydrated hereditary stomatocytosis. Biochim. Biophys. Acta. 2008;1778:125–132. doi: 10.1016/j.bbamem.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 37.Rivera-Milla E., Stuermer C.A., Malaga-Trillo E. Ancient origin of reggie (flotillin), reggie-like, and other lipid-raft proteins: convergent evolution of the SPFH domain. Cell. Mol. Life Sci. 2006;63:343–357. doi: 10.1007/s00018-005-5434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koradi R., Billeter M., Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


