Abstract
Cryptochromes are blue light absorbing photoreceptors found in many organisms and involved in numerous developmental processes. At least two highly similar cryptochromes are known to affect branching during gametophytic development in the moss Physcomitrella patens. We uncovered a relationship between these cryptochromes and the expression of particular members of the SBP-box genes, a plant specific transcription factor family. Transcript levels of the respective moss SBP-box genes, all belonging to the LG1-subfamily, were found to be dependent, albeit not exclusively, on blue light. Moreover, disruptant lines generated for two moss representatives of this SBP-box gene subfamily, both showed enhanced caulonema side branch formation, a phenotype opposite to that of the ppcry1a/1b double disruptant line. In this report we show that PpCRY1a and PpCRY1b act negatively on the transcript levels of several related moss SBP-box genes and that at least PpSBP1 and PpSBP4 act as negative regulators of side branch formation.
Electronic supplementary material
The online version of this article (doi:10.1007/s00425-007-0661-5) contains supplementary material, which is available to authorized users.
Keywords: Blue light, Cryptochromes, Photomorphogenesis, Physcomitrella, SBP-box gene, Side branch formation
Introduction
Changes in light quantity and quality provide important environmental stimuli to plants to adapt their growth and morphology (for recent review see Jiao et al. 2007). Different classes of photoreceptors have evolved that enable plants to sense light of different wavelengths. Among these are the cryptochromes, flavin-type photoreceptors that are active at the blue end of the spectrum. A cryptochrome-encoding gene could first be cloned from Arabidopsis thaliana (Ahmad and Cashmore 1993) and the developmental functions of this photoreceptor are also best studied in this model species. A. thaliana actually has two highly similar and well-characterised cryptochromes, AtCRY1 and AtCRY2, and a third less characterised one, AtCRY3 (Chen et al. 2004). AtCRY1 and AtCRY2 are known to control inhibition of hypocotyl elongation, cotyledon and leaf expansion (Ahmad and Cashmore 1993; Lin et al. 1998), induction of anthocyanin synthesis (Ahmad et al. 1995), flowering time (Guo et al. 1998; Mockler et al. 1999) and the circadian clock (Somers et al. 1998; Devlin and Kay 2000).
Intrinsic to light signalling are transcription factors, necessary to effectuate changes in development through activation and repression of specific downstream genes. Well-studied examples in A. thaliana are HY5 and its homologs HYH and HFR1, transcription factors known to be involved in both cryptochrome-mediated blue light and phytochrome-mediated far-red light signalling (Mathews 2006; Vandenbussche et al. 2007).
At least two highly similar cryptochrome genes with redundant function also exist in the genome of Physcomitrella patens, PpCRY1a and PpCRY1b (Imaizumi et al. 2002). Their transcripts can be detected in protonema and gametophores at low levels and their proteins are found in the nucleus. Protonema represents the filamentous stage in the life cycle of P. patens and is characterised by two different cell types. The chloronema cells, on average 100 μm in length and dividing every 10–12 h, harbour many chloroplasts, which suggests a function in energy supply. The caulonema cells, however, are believed to function mainly in habitat capture as they are pale green, on average 250 μm in length and divide once every 5–6 h (Schumaker and Dietrich 1997). Both cell strands grow by tip growth and produce side branches in the dependence of light. Whereas protonemata grown in darkness or far-red light do not branch, blue light and red light synergistically strongly induce branch formation (Uneaka et al. 2005). Accordingly, the ppcry1a/1b double disruptant was found to be repressed in side branch formation, thereby underlining the role of the cryptochrome blue-light receptors in the control of moss development (Imaizumi et al. 2002; Ichikawa et al. 2004).
Transcription factors involved in light signalling in P. patens have not been reported to date. In a first effort to uncover the developmental role of the plant specific SBP-box transcription factors in the moss P. patens, we obtained disruptant lines with phenotypes opposite to those of the ppcry1a/1b disruptants. SBP-box genes encode proteins recognizing and binding specific DNA-sequences through the evolutionary conserved SBP-domain (Klein et al. 1996; Yamasaki et al. 2004; Birkenbihl et al. 2005). They are found in moderately sized families from the unicellular alga Chlamydomonas reinhardtii to mono- and eudicots as rice and A. thaliana. Based on mutant phenotypes, SBP-box genes have been found to be involved in lateral organ development (Moreno et al. 1997; Wang et al. 2005), developmental phase transitions (Cardon et al. 1997; Wu and Poethig 2006; Gandikota et al. 2007), fertility (Unte et al. 2003; Zhang et al. 2007), fruit ripening (Manning et al. 2006), fungal toxin sensitivity (Stone et al. 2005) and copper-signalling (Kropat et al. 2005).
Here we report that loss-of-function of the P. patens SBP-box genes PpSBP1 and PpSBP4 (Riese et al. 2007) results in an enhanced branching phenotype and that these, and closely related moss paralogs, become strongly upregulated in a cryptochrome disruptant line. Based on our results we propose that in moss SBP-box transcription factors of the LG1-subfamily are negatively regulated by the blue-light receptor PpCRY and as such involved in phototransduction.
Materials and methods
Plant material and growth conditions
Physcomitrella patens ssp. patens B.S.G. was routinely grown under standard conditions as described by D. G. Schaefer (http://www2.unil.ch/lpc/docs/pdf/PPprotocols2001.pdf). Osram (Munich, Germany) L30W/25 fluorescence light tubes provided white light with an intensity of approximately 250 Lux. Our media did not contain glucose.
Microscopy
Either a Leica binocular microscope (Wetzlar, Germany) or a Zeiss Axiophot microscope (Göttingen, Germany), both equipped with a KY-F5U 28CCD camera (JVC, Yokohama, Japan), were used to examine and photograph entire gametophores and colonies on petri dishes at different stages of development or branching of protonema after dissection in water, respectively.
Sequence alignments and phylogenetic reconstruction
Multiple alignments of amino acid sequences were generated by the program ClustalW of the MacVector 7.2.2 software package (Accelrys Ltd, Cambridge, UK) using the BLOSUM 30 matrix with an open gap penalty of 10 and an extend gap penalty of 0.05. Only the SBP-domain was used for the phylogenetic reconstruction. The tree was constructed using the neighbor-joining algorithm of the MacVector 7.2.2 software package.
Isolation of genomic DNA and RNA
Physcomitrella patens genomic DNA was isolated using the CTAB-method (Murray and Thompson 1980). Total RNA was routinely isolated with the RNeasy Kit according to the protocol of its supplier (Qiagen, Hilden, Germany). For RNA isolation in the experiments described below we followed the protocol as described by Markmann-Mulisch et al. (1999).
For the analysis of circadian and light quality dependent expression patterns, protonema was pre-cultured for 10 days in a long-day regime (16 h of light, 8 h of darkness), in a Rumed 5001 growth cabinet (Rubarth Apparate, Laatzen, Germany) equipped with Osram L36W/11-860 Lumilux Plus Daylight lamps, with a light intensity of approximately 250 Lux.
For the analysis of circadian dependent expression, RNA was prepared at particular time points throughout the day. At the end of 1 day of sampling (24 h after sunset), the cultures were shifted to continuous light conditions, and sampling was continued for another two time points. For the analysis of light quality dependent expression, RNA was prepared 90 min before sunrise and 90 min after sunrise. At the same time points, RNA was prepared from cultures that had been deprived of light at the subjective morning by wrapping in alu-foil, as well as from cultures that were not irradiated with white light at sunrise, but with blue light, red light, or far-red light in a Percival E-30 LED growth cabinet (CLF Laborgeraete, Emersacker, Germany), with light intensities of 240, 206, and 30 Lux for red light (600–700 nm), far-red light (700–750 nm) and blue light (400–500 nm), respectively.
Construction of PpSBP1 and PpSBP4 gene disruption vector constructs
As target sequences for homologous recombination, two adjacent DNA fragments encompassing, respectively, the 5′ and 3′ regions of the PpSBP1 and PpSBP4 loci were cloned into the pBTSK vector (Stratagene, Amsterdam, The Netherlands) after their amplification from genomic DNA by PCR using the following, for cloning adapted primer pairs (introduced restriction sites are underlined): MR71/MR72 (AATTGGTACCATGAACCCTTCTGTAAGTTCTAA and TTAGATCTCCACCTGAAGTAGAGGCATTTAG) and MR69/MR70 (ATTGAGATCTCGAACATCTGGTCCTGGCTATTG and ATAGGCTCGAGTGGTTACTTCCCATCGTTACCG) for the 5′ and 3′ region, respectively, of PpSBP1, MR74/MR73 (TTAAGGTACCATGCACTAATCATGTTAAAAGAGAAGAGG and TATAAGATCTTGCATTGTTGACAAAACCTCTGAG) and MR76/MR75 TAGTAGATCTTTCATGGTCCAGGGATCACG and TATAGCTCGAGGTCTTAACGCTTCAATATCTTGCGAG) for the 5′ and 3′ region, respectively, of PpSBP4. With the primers used, a suitable BglII restriction site had been created in between the 5′ and 3′ genomic sequences for the subsequent introduction of the following selection cassettes. The PpSBP1 disruptant vector containing a CaMV 35S promoter, the bleomycin (ble)resistance gene and a CaMV 35S polyadenylation signal, which was amplified from the vector p35S-Zeo (kindly provided by Dr. Yuji Hiwatashi, NIBB, Okazaki, Japan) using the primers MR110/MR111 (CCCCTGATCAGGTCGACGGTATCGATAAGC and AGAATGATCAGATCCCCGTCACCGGTGTGAG). The PpSBP4 disruptant vector containing a CaMV 35S promoter, the hygromycin B phosphotransferase (hpt) gene and a nopaline synthase (nos) polyadenylation signal, which was amplified from the vector pUC-Hyg (kindly provided by Dr. Bernd Reiss, MPIZ, Cologne, Germany) with help of the primers MR138/MR139 (GCTGTGATCACCTCTGGTAAGGTTGGGAAG and TCGATGATCATACCCGAAATATAAACAACTTGT). The PCR-fragments were digested with BclI and inserted into the compatible BglII restriction sites created between the respective PpSBP1 and PpSBP4 5′ and 3′ fragments cloned as described above in the pBTSK vector. The resulting plasmid vectors with the disruption constructs were named pSBP1-Zeo and pSBP4-Hyg.
After transformation, genomic integration on the 5′ and the 3′ region was confirmed by PCR on genomic DNA of the transformants and with the help of oligonucleotides priming in the selection cassette and outside the region of integration.
Protoplast isolation, transformation and regeneration
Protoplasts were isolated and transformed as described in Schaefer et al. (1991). According to the protocol, we transferred the protoplasts to solid medium with an appropriate selection marker but without ammonium-tartrate. Linearised plasmid DNA used for transformation was isolated and purified with the Plasmid-Midi Kit (Qiagen).
Real-time PCR
Relative quantification of transcript levels was performed by real-time PCR on an iQ5 thermal cycler (BioRad, Munich, Germany). Thereto, 2.5 μg of total RNA, isolated from selected tissues or developmental stages, were converted into a cDNA-pool using the SuperScriptII Reverse Transcription System (Invitrogen, Carlsbad, CA, USA) according to the supplier’s protocol. For each quantification, 1 μl out of the cDNA-pool was used as template for amplification with the appropriate primers in the iQ-SYBR-Green Supermix (BioRad). After normalization of all Ct values against 18S rRNA amplified with the primer pair MR311/MR312 (AGGAATTGACGGAAGGGCAC and GGACATCTAAGGGCATCACA) the Pfaffl method was followed for calculations of relative expression (Tichopad et al. 2003). The required amplification efficiencies for the primer pairs used were determined from standard curves generated from cDNA dilution series.
To determine the number of integrations in the genome of the disruptant lines, we performed a qPCR with oligonucleotides priming in the selection cassette. Ten nanograms of genomic DNA were used as a template for amplification. After normalization of all Ct values against the single copy genes PpSBP1 (oligonucleotides priming outside of the region of integration) and PpSBP3 we calculated the copy numbers of the selection cassettes with the help of the Pfaffl-method (Tichopad et al. 2003).
For all primers used that are not listed here, see Table 1 of the Electronic Supplementary Material.
DNA sequencing
Sequencing was done by the MPIZ DNA core facility on Applied Biosystems (Weiterstadt, Germany) Abi Prism 377 and 3700 sequencers using BigDye-terminator chemistry.
Remaining techniques and methods
Standard molecular biology techniques were performed as described by Sambrook et al. (1989). Digital photographic images were cropped and assembled using Adobe Photoshop (Adobe Systems, San Jose, CA, USA).
Gene accession numbers
The following accession numbers correspond to either GenBank entries or to the first release of the annotated P. patens genome sequence (http://genome.jgi-psf.org/Phypa1_1/Phypa1_1.home.html): PpSBP1, AJ968320; PpSBP4, AJ968319; PpSBP7, EF016493 and FGENESH1_PG.SCAFFOLD_252000020; PpSBP8, FGENESH1_PG.SCAFFOLD_194000034; PpSBP9, EF016494 and FGENESH1_PG.SCAFFOLD_337000032; PpSBP12, EF016496 and FGENESH1_PG.SCAFFOLD_50000085; PpCOL1, AJ890106; PpCOL2, AJ890107; PpCOL3, AJ890108; P. patensRAN, CJ972918; P. patens18S rRNA, X80986.
Results
Generation of PpSBP1 and PpSBP4 single disruptants
In a first effort to uncover the roles of SBP-box genes in P. patens development, we generated targeted knockouts for PpSBP1 and PpSBP4 through an efficient homologous recombination system unique to this model plant (see “Materials and methods”).
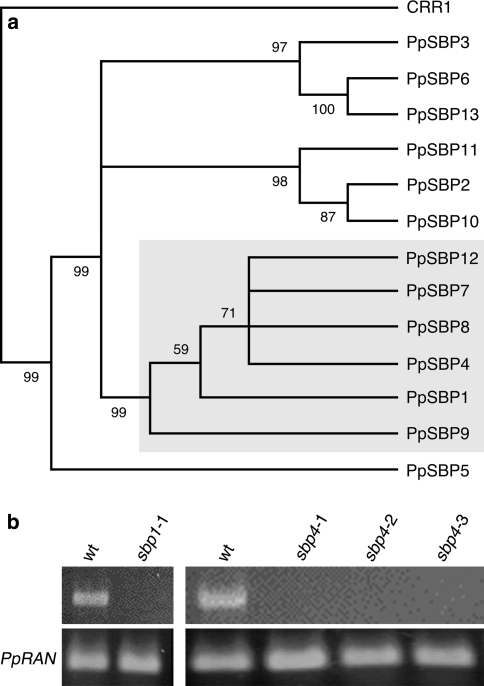
Our focus was on the LG1-subfamily of SBP-box genes and herein PpSBP1 and PpSBP4 were selected for functional analysis (Fig. 1a; Cardon et al. 1999; Riese et al. 2007). We found this subfamily to be constituted by a total of six paralogs, compared to only one in A. thaliana, AtSPL8, known to be involved in sporogenesis (Unte et al. 2003).
Fig. 1.
Physcomitrella patens SBP-box genes PpSBP1 and PpSBP4 and their disruption. a Phylogenetic relationship of PpSBP1 and PpSBP4 and other P. patens SBP-box genes as based on the conserved SBP-domain. C. reinhardtii CRR1 functioned as outgroup. The LG1-subfamily is boxed in grey. Only bootstrap values over 50% are shown. b Absence of transcript validation by RT-PCR of the ppsbp1-1 and ppsbp4-1, -2 and -3 disruptant lines. Presence of the respective transcripts in wild type (wt) is shown for comparison and amplification of PpRAN transcript as reference for quantification
One PpSBP1 gene disruptant strain (designated ppsbp1-1) and three independent PpSBP4 gene disruptant strains (designated ppsbp4-1, -2 and -3) could be obtained. That the endogenous loci of PpSBP1 and PpSBP4 of these strains were indeed replaced with the respective targeting constructs could be confirmed by PCR (data not shown). Integrated copy number determination based on qRT-PCR, proved that all disruptant lines reported in this paper are single integration lines (data not shown). In addition, RT-PCR analysis proved absence of PpSBP1 or PpSBP4 transcripts in the corresponding strains (Fig. 1b).
Abnormal protoplast regeneration in consequence of PpSBP1 and PpSBP4 disruption
As a first observation, the colonies formed from regenerating protoplasts of the disruptant lines ppsbp1 and ppsbp4 were found to be denser and smaller when grown on minimal medium supplemented with ammonium tartrate (PPNH4-medium) than the non-transformed wild-type colonies of the same age.
In order to analyse the regeneration behaviour of ppsbp1 and ppsbp4 in comparison with wild type in a more detailed manner, protoplasts were plated on minimal (PPNO3) and PPNH4 media, both with mannitol added as osmoregulator, following the protocol of Schaefer (http://www2.unil.ch/lpc/docs/pdf/PPprotocols2001.pdf). Their regeneration was followed for over 2 weeks.
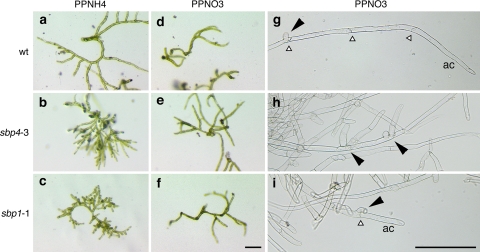
We observed a faster regeneration of wild-type protoplasts on PPNH4 in comparison to PPNO3 medium (Fig. 2a, d), whereas the disruptant lines showed the opposite tendency (Fig. 2b, e, c, f). On PPNO3 medium, no obvious difference in regeneration and growth could be observed between wild type and disruptant line protoplasts at this early stage of development. In contrast, ppsbp1 and ppsbp4 disruptant protoplasts regenerating on PPNH4 medium showed more densely branched, compact colonies in comparison to wild type (Fig. 2a–c). Altered side branch formation could also be revealed in ppsbp1 and ppsbp4 disruptant colonies at later stages of development on PPNO3 medium (Fig. 2g–i). Ammonium tartrate thus seems to act as an enhancer of the mutant phenotype.
Fig. 2.
Microscopy of protoplast regeneration and side branch formation of ppsbp1 and ppsbp4 disruptants. Left and middle panels, regenerating protoplasts after 14 days on PPNH4 medium (a–c) and PPNO3 medium (d–f). Right panels, protonema grown on PPNO3 10 days after propagation (g–i). Regenerating protoplasts and protonema of wild type (wt, upper row) are shown for comparison. White triangles indicate anticlinal cell walls, black arrowheads side branches; ac apical cell. Bar represents 200 μm
Side branch formation is affected in both ppsbp1 and ppsbp4 disruptants
The abnormal branching was studied in more detail using light microscopy on protonemata 10 days after propagation of ppsbp1 and ppsbp4 strains grown on PPNO3 medium and in comparison to identically treated wild type.
In wild type, initiation of protonema branching can be observed as a swelling occurring generally at the distal part of the second subapical cell close to its anticlinal cell wall. Occasionally this cell may initiate a second branch, again in its distal half (Schumaker and Dietrich 1997). In contrast to wild type, the ppsbp1 disruptant lines initiated side branch formation already in the first subapical cell and both the ppsbp1 and the ppsbp4 disruptant lines showed a more variable positioning, i.e. not always from the most distal part of the cell (Fig. 2h, i). For quantification, we defined branching in wild type and the disruptants as abnormal when (1) it was observed in already the first subapical cell or (2) it did not arise in the most distal part of the second subapical cell or (3) the second subapical cell showed double branching. Accordingly, of the protonemata tips analysed in wild type we found on average only 7% (n = 94) to branch abnormally and in all cases it concerned double branching. Of the ppsbp1 protonemata tips, however, 42% (n = 65) branched abnormally with 34% involved in double branching. For the three independent ppsbp4 lines we found on average 45% (n = 87 + 85 + 68) to branch abnormally with on average 32% double branching.
Loss of PpSBP1 function affects caulonema development
Both the ppsbp1 and ppsbp4 disruptant lines differed similarly from wild type with respect to protonema branching. Phenotypic differences between both SBP-box gene disruptants, however, became obvious during subsequent stages of the moss life cycle. We observed that the ppsbp1 disruptant line deviated from normal development when routinely grown in long day (16 h light/8 h dark; LD). Up till 4 months after propagation the ppsbp1 disruptant line did not develop any macroscopically visible leafy shoots. Normally, a gametophore develops from an initial cell specified in the caulonema after about 2–3 weeks after propagation and growth in LD (Schumaker and Dietrich 1997; own observation).
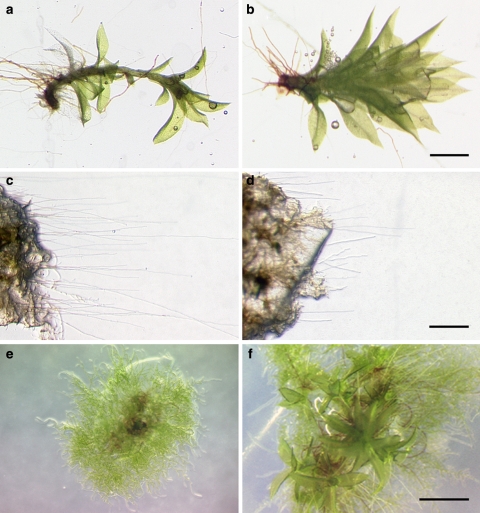
Stereomicroscopic inspection of the ppsbp1 disruptant revealed that the actual number of buds that were initiated, does not obviously differ from the wild type (data not shown). Most of these, however, remained small and only a few developed, more than 4 months after propagation, into leafy shoots that, in comparison to wild-type gametophores, looked bushier (Fig. 3a, b). However, the development of the reproductive organs and the sporophyte on these gametophores did not differ from wild type, but the germination rate of their spores did. Whereas under our standard conditions 80% of wild-type spores had germinated after 5 days in continuous light on PPNO3 medium, only 57% of ppsbp1 spores had germinated after this period (n > 200, representing four different spore capsules).
Fig. 3.
Caulonema and gametophore development in ppsbp1 and ppsbp4 disruptant lines. Mature gametophores of wild type (a) and ppsbp1-1 (b). Caulonema strands of wild type (c) and ppsbp1-1 (d) after 2 weeks in darkness. Four-week-old colonies of wild type (e) and ppsbp4-3 (f). Bar represents 1 mm
The denser colonies and the few mature gametophores suggest an abnormal development of caulonema cells. To analyse this developmental stage in more detail, we placed 4-week-old colonies derived from a single protoplast (isogenic lines) on solid PPNO3 medium in petri dishes and put them vertically in a dark growth cabinet. Because only caulonema cells are able to grown in complete darkness, where it lacks branching and displays negative gravitropism (Cove et al. 2006), this would be an ideal test system to analyse the development of caulonema. After 2 weeks, number and length of the newly formed caulonemata, i.e. filaments extending from the original colony, were determined. Interestingly, the number of caulonema strands did not differ between wild type and ppsbp1, indicating that, despite its enhanced branching phenotype in the light, branching is still suppressed in the ppsbp1 disruptant when grown in the dark. The length of the caulonema strands, were found to be significantly shorter in the ppsbp1 disruptant but the length of the single cells remained unaffected compared to those of the wild type (Fig. 3c, d).
Loss of PpSBP4 function affects development of gametophores and spore germination
Wild-type colonies grown on PPNO3 medium and in LD did not develop any gametophores within the first 4 weeks after regenerating from protoplasts. In contrast, regenerating isogenic lines of the three ppsbp4 disruptants under the same conditions had already developed several gametophores after this period (Fig. 3e, f). The frequency of gametophore formation and the phenotype of them, however, did not differ from wild type. Also in complete darkness the ppsbp4 disruptants behaved similar to wild type in that gametophore initiation became completely repressed (data not shown). After their appearance gametophores and sporophytes of the ppsbp4 disruptants developed normally. As described for the ppsbp1 mutant, the spore germination rate of ppsbp4 mutants is also affected. Only around 37% of the spores had germinated after 5 days in continuous light (n > 200, representing four different spore capsules).
PpSBP1 and PpSBP4 are under the control of cryptochromes
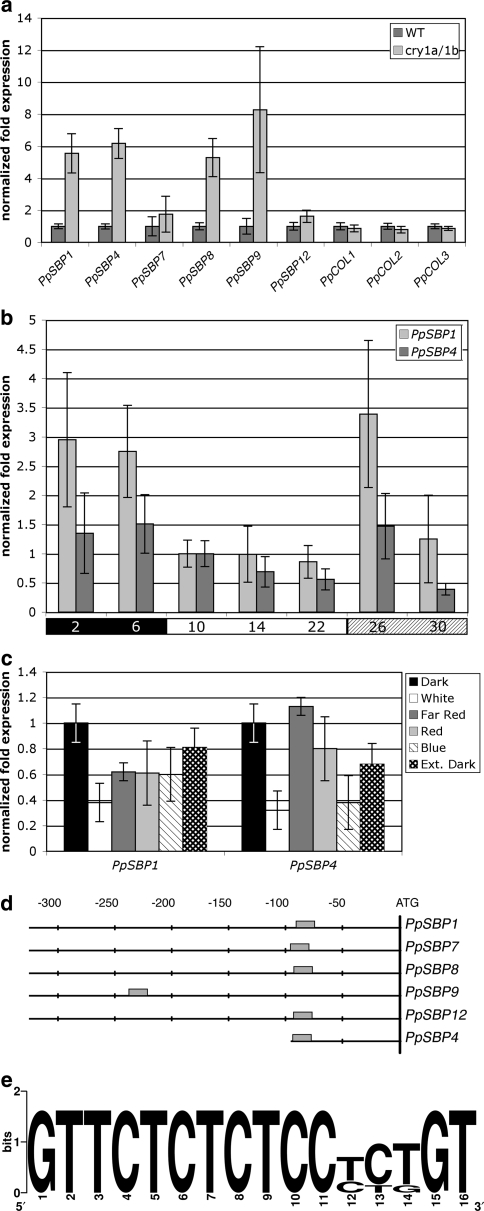
Interestingly, the disruptant lines ppsbp1 and ppsbp4 described here show an altered side branch phenotype opposite to that of the ppcry1a/1b double disruptant. To test the hypothesis that PpSBP1 and PpSBP4 are actually negatively regulated by cryptochromes, we compared their relative transcript levels during the day in wild type and the ppcry1a/1b double disruptant by qRT-PCR. In agreement with the hypothesis, transcript levels of both SBP-box genes were significantly raised in the ppcry1a/1b double disruptant line (Fig. 4a).
Fig. 4.
Responses of PpSBP1 and PpSBP4 to light and cryptochrome. a Relative quantification of transcript levels of PpSBP1, PpSBP4 and other members of the LG1-subfamily in 4-week-old LD cultures of wild type (WT) and of the ppcry1a/1b double disruptant line (cry1a/1b) at the end of the light period. Relative transcript levels of P. patensCONSTANS-like genes (PpCOL1 to -3) are shown for comparison. Expression in wild type is arbitrarily set at one. b Relative quantification of transcript levels of PpSBP1 and PpSBP4 in wild type during one full 8 h dark/16 h light cycle. The black and the white bars below the histogram indicate, respectively, the dark and light periods of the normal 24 h day. The shaded bar below the histogram marks the samples taken during extension of the white-light period. Numbers in the bars indicate isolation time points in hours after start of the night. Transcript levels are arbitrarily set at one at 10 h after start of the cycle, i.e. 2 h after start of the light period. c Relative quantification of transcript levels of PpSBP1 and PpSBP4 in wild type grown under different light qualities. Transcript levels in total RNA isolated 90 min before the subjective dawn (Dark) are arbitrarily set at one. Sampling in different light quality conditions was performed 90 min after the subjective dawn. d Schematic representation and alignment of the sequences upstream of the translational start codons in the Physcomitrella members of the LG1-subfamily of SBP-box genes. The grey box indicates the conserved motif shown in e. The positions, in nucleotides relative to the start codon (ATG), are shown above the alignment. e Nucleic acid sequence logo of the conserved motif found in the promoter region of the Physcomitrella LG1-subfamily members as identified by the program MotifSampler (Thijs et al. 2002). In the logo each stack corresponds to one position in the sequence. The overall height of a stack indicates the sequence conservation at that position, while the height of symbols within the stack indicates the relative frequency of each nucleotide at that position (Crooks et al. 2004)
To obtain a more general impression of the light dependence of PpSBP1 and PpSBP4 expression, we determined their relative expression levels over a complete day–night cycle. Transcript levels of both PpSBP1 and PpSBP4, albeit the latter to a lesser extent, were found to decrease after dawn and to rise again after dusk (Fig. 4b). When an anticipated night was postponed by an extension of the photoperiod of the preceding days, transcript levels of both genes temporally increased (Fig. 4b). This observation strongly suggests a circadian clock driven regulation of PpSBP1 and PpSBP4 transcriptional activity.
Our finding that PpSBP1 and PpSBP4 are negatively regulated by cryptochromes indicates an important role for blue light in the regulation of these genes. Therefore, we tested the expression levels of both genes in moss colonies that, after being cultivated in LD, were exposed to only blue light after the last dark period. In agreement with the hypothesis, transcript levels of both genes were found to be decreased in comparison to levels in colonies harvested before the light treatment, i.e. in the dark (Fig. 4c). Furthermore, in blue light PpSBP4 transcript abundance dropped to levels comparable to those in colonies exposed to white light. In the case of PpSBP1, however, transcript levels in blue light remained somewhat higher in comparison to white light. The reason for this behaviour could be determined when instead of to blue light, the colonies were shifted to red or far-red light after the last dark period. This experiment showed that, in addition to cryptochrome-mediated blue light, phytochrome mediated red and far-red light also negatively regulate PpSBP1. Interestingly, the transcript levels of both genes drop under extended night conditions, which again raises the hypothesis of an involvement of the circadian clock (Fig. 4c).
Cryptochrome controls the expression of other members of the moss LG1-subfamily of SBP-box genes
According to phylogenetic reconstructions, PpSBP1 and PpSBP4 are grouped in the LG1-subfamily of SBP-box genes. In P. patens, four more members, i.e. PpSBP7, PpSBP8, PpSBP9 and PpSBP12, represent this subfamily (Riese et al. 2007). To learn, if these closely related genes may also be under the control of cryptochrome, we determined their transcript levels in the ppcry1a/1b disruptant. Like for PpSBP1 and PpSBP4, we found these genes to be derepressed in the disruptant, although to different degrees (Fig. 4a).
For comparison, we analysed the ppcry1a/1b disruptant for transcript levels of a set of unrelated transcription factors that were also shown to be regulated by light, namely the CONSTANS-like genes PpCOL1, PpCOL2 and PpCOL3 (Zobell et al. 2005). Unlike the SBP-box mRNAs, we found mRNA levels of all three CONSTANS-like genes to be unaltered in comparison to wild type (Fig. 4a). As such, the latter represent a distinct case of light-regulated mRNA abundance (Zobell et al. 2005) that is not cryptochrome-specific.
As more or less all members of the LG1-subfamily in P. patens showed a similar transcriptional response to cryptochrome loss-of-function, we compared their promoter regions with the help of the program Motifsampler (Thijs et al. 2002), in order to uncover conserved and putative cis-acting elements possibly related to this response. As most striking conserved motif shared by all family members, “GTTCTCTCTCCYYKGT” was found approximately 80 nt upstream of the translational start codon (Fig. 4e). The exception is PpSBP9 where this motif is found much further upstream.
Discussion
The SBP-box genes PpSBP1 and PpSBP4 control side branch formation
In search for developmental functions of SBP-box genes in plants, we uncovered an important role for these transcription factors in P. patens side branch formation. Disruption of either PpSBP1 or PpSBP4 in each case led to an increased branching of caulonema filaments.
Cultured in darkness, P. patens develops fast growing caulonema filaments that do not form side branches. Growth rate of caulonemata is reduced upon exposure to light as side branches are initiated at subapical cells (Cove et al. 1978). In darkness, ppsbp1 and ppsbp4 disruptant lines produced unbranched caulonemata, just like the wild type. However, branches remained shorter in comparison to wild type, likely due to a reduction in cell division rate, since caulonema cell length was found to be unaffected in the ppsbp1 disruptant.
These observations suggest that both PpSBP1 and PpSBP4 act primarily in P. patens development through repression of branching on one hand and promotion of unidirectional cell division on the other. Together, this explains the relatively small and dense colonies that are formed by ppsbp1 and ppsbp4 disruptant lines. It remains to be determined whether there exists a causal relation between repressed branching and increased unidirectional growth.
PpSBP1 and PpSBP4 are components of a photomorphogenic response
Previous studies have shown that side-branch initials form as the first visible and irreversible response of dark-adapted caulonemata to a 1-min pulse of blue light (Russell et al. 1998). In agreement with this, the P. patensppcry1a/1b double disruptant displays a reduced branching phenotype, implicating that cryptochromes are major photoreceptor pigments for blue light in this process (Imaizumi et al. 2002; Uneaka et al. 2005). Interestingly, we found transcript levels of both PpSBP1 and PpSBP4 to be reduced in response to blue light as well as strongly elevated in the ppcry1a/1b double disruptant line. Together with the enhanced branching phenotype of the respective disruptants, this strongly suggests that the side branch promoting function of blue light is at least partially mediated through a cryptochrome dependent repression of the SBP-box transcription factors PpSBP1 and PpSBP4.
It may be noted from the data presented in Fig. 4, that the moderate reduction of PpSBP1 and PpSBP4 transcript levels in response to blue light does not quantitatively reflect their strongly elevated levels in the ppcry1a/1b double disruptant. An explanation of this seeming discrepancy between the outcome of both experiments is probably to be found in (1) different timing of RNA sampling during the day, (2) quantitative and qualitative differences in light and/or (3) different tissue composition of the samples. The material in Fig. 4c represents protonema only, whereas the material represented in Fig. 4a consisted of 4-week-old colonies already including small gametophores.
In addition to caulonema side branch formation, the functions of PpSBP1 and PpSBP4 seem to be required for proper spore germination and gametophore development, respectively. A role for blue light in P. patens gametophore development has been described before (Imaizumi et al. 2002). An effect of blue light on spore germination in P. patens is unknown, however, in the fern Adiantum capilus-veneris, it is known to severely affect the process (Imaizumi et al. 2000).
Generally in plants, multiple photoreceptors with sensitivities to different wavelengths of light contribute in concert to a particular photomorphogenic response, albeit in different ways (Gyula et al. 2003; Jiao et al. 2007). Branching in moss for instance, is initiated under both continuous blue and red light. In contrast to blue light, however, a 1-min pulse of red light is not able to induce branching in dark-adapted caulonemata (Russell et al. 1998). Similarly and in addition to blue light, we found PpSBP1 also to be repressed by red and far-red light. Transcript levels of PpSBP4 seem to be less dependent or not dependent on red light and far-red light, respectively.
As a consequence of their light dependence, we found PpSBP1 and PpSBP4 transcript levels to follow a diurnal variation, being highest during the night and lowest during the day. Interestingly, upon extension of the light or dark period both genes responded with an initial elevation of their transcript levels at respective dusk, as if they actually had anticipated night or day. This strongly suggests the involvement of an endogenous circadian pacemaker in their regulation. In A. thaliana both blue- and red-light receptors, i.e. cryptochromes and phytochromes, are involved in photic entrainment of the circadian clock. However, whereas AtPHYA requires AtCRY1 for signalling to the circadian clock in both red and blue light, cryptochromes seem not to be part of the central circadian oscillator (Devlin and Kay 2000). If this holds true in P. patens, it may explain why PpSBP1 transcript levels responded much stronger to the extended photoperiod than PpSBP4, since mRNA levels of the latter are also less influenced by red and far-red light.
Photomorphogenic responses are often controlled by more than one factor. In moss, for instance, the transition from chloronema to caulonema cells is controlled by both blue light and auxin (Imaizumi et al. 2002). Imaizumi and colleagues proposed that cryptochrome-mediated light signals control developmental changes by suppressing auxin signals at least at the point of transcription of auxin-regulated genes. Therefore, it might be interesting to analyse the auxin signalling pathway in the ppsbp1 and ppsbp4 disruptant lines as well.
Other LG1-SBP-box subfamily members may also play roles in blue light signalling
Together with four other SBP-box genes, PpSBP1 and PpSBP4 are members of the LG1-subfamily, which represents almost half of all SBP-box genes in P. patens. In the ppcry1a/1b double mutant, transcript levels of all subfamily members were found raised, albeit only weakly for two of them. Furthermore, all six LG1-subfamily members share a defined sequence motif in their promoter region. Querying the PLACE database (Higo et al. 1999) with this sequence did not reveal obvious similarities to any known plant cis-acting regulatory DNA element. Uncovering the biological relevance of the conserved motif may thus have to await future promoter deletion studies. Taken together, however, our observations suggest that the blue light dependent repression of the entire LG1-subfamily of SBP-box genes in P. patens is mediated through a common transcriptional repressor.
Based on their sequence similarity and common control, it is reasonable to assume that at least some P. patens LG1-subfamily members are functionally redundant with each other. To what extent they are functionally redundant has to await the generation of double as well as higher-order disruptants.
Whether any functional conservation within the LG1-subfamily exists between moss and higher plant members is highly speculative. It is, however, interesting to note that AtSPL8, the only LG1-subfamily homologue in A. thaliana, is one of the most differentially expressed transcription factors between blue light irradiated wild type seedlings and Atcry1 mutant seedlings, as revealed by micro-array analysis (Folta et al. 2003). Remarkably, AtSPL8 relative transcript levels were found to be reduced in the A. thaliana cryptochrome mutant, which is exactly the opposite of what we report here for the LG1-subfamily homologues in moss. During reproductive growth, AtSPL8 promotes sporogenesis in anthers and ovules (Unte et al. 2003) and, seen from this perspective, thus the sporophytic to gametophytic phase change. It would thus be interesting to learn if the seemingly contradictory response of LG1-subfamily members in moss and A. thaliana to blue light relates to the fact that in moss the gametophytic phase is dominant, whereas in higher plants this is the sporophytic phase (Cove et al. 2006).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We are grateful to Prof. Dr. M. Hasebe (NIBB, Okazaki, Japan) for the kind gift of the ppcry1a/1b double disruptant line and Dr. B. Reiss and Prof. Dr. G. Coupland (MPIZ, Cologne, Germany) for providing the P. patens RNA samples used for circadian and light quality dependent expression analysis. We thank our colleagues in the lab for helpful discussions.
Abbreviations
- SD
Short day
- LD
Long day
- CRY
Cryptochrome
- SBP
SQUAMOSA PROMOTER BINDING PROTEIN
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s00425-007-0661-5) contains supplementary material, which is available to authorized users.
References
- Ahmad M, Cashmore AR (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366:62–166 [DOI] [PubMed]
- Ahmad M, Lin C, Cashmore AR (1995) Mutations throughout an Arabidopsis blue-light photoreceptor impair blue light-responsive anthocyanin accumulation and inhibition of hypocotyl elongation. Plant J 8:653–658 [DOI] [PubMed]
- Birkenbihl RP, Jach G, Saedler H, Huijser P (2005) Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J Mol Biol 352:585–596 [DOI] [PubMed]
- Cardon GH, Höhmann S, Nettesheim K, Saedler H, Huijser P (1997) Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: a novel gene involved in floral transition. Plant J 12:367–377 [DOI] [PubMed]
- Cardon G, Höhmann S, Klein J, Nettesheim K, Saedler H, Huijser P (1999) Molecular characterization of the Arabidopsis SBP-box genes. Gene 237:91–140 [DOI] [PubMed]
- Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher pPlants. Annu Rev Genet 38:87–117 [DOI] [PubMed]
- Cove DJ, Schild A, Ashton NW, Hartmann E (1978) Genetic and physiological studies of the effect of light on the development of the moss Physcomitrella patens. Photochem Photobiol 27:249–254
- Cove DJ, Bezanilla M, Harrues P, Quatrano R (2006) Mosses as model system for study of metabolism and development. Annu Rev Plant Biol 57:497–520 [DOI] [PubMed]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190 [DOI] [PMC free article] [PubMed]
- Devlin PF, Kay SK (2000) Cryptochromes are required for phytochrome signalling to the circadian clock but not for rhythmicity. Plant Cell 12:2499–2509 [DOI] [PMC free article] [PubMed]
- Folta KM, Pontin MA, Karlin-Neumann G, Bottini R, Spalding EP (2003) Genomic and physiological studies of early cryptochrome 1 action demonstrate role for auxin and gibberellin in the control of hypocotyl growth by blue light. Plant J 36:203–214 [DOI] [PubMed]
- Gandikota M, Birkenbihl RP, Höhmann S, Cardon GH, Saedler H, Huijser P (2007) The miRNA156/157 recognition element in the 3’ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J 49:683–693 [DOI] [PubMed]
- Guo H, Yang H, Mockler TC, Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279:1360–1363 [DOI] [PubMed]
- Gyula P, Schäfer E, Nagy F (2003) Light perception and signalling in higher plants. Curr Opin Plant Biol 6:446–452 [DOI] [PubMed]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27:297–300 [DOI] [PMC free article] [PubMed]
- Ichikawa K, Sugita M, Imaizumi T, Wada M, Aoki S (2004) Differential expression on daily basis of plastid sigma factor genes from the moss Physcomitrella patens. Circadian clock, and blue light signalling mediated by cryptochromes. Plant Physiol 136:4285–4298 [DOI] [PMC free article] [PubMed]
- Imaizumi T, Kanegae T, Wada M (2000) Cryptochrome nucleoplasmic distribution and gene expression are regulated by light quality in the fern Adiantum capilus-veneris. Plant Cell 12:81–95 [DOI] [PMC free article] [PubMed]
- Imaizumi T, Kadota A, Hasebe M, Wada M (2002) Cryptochrome light signals control development to suppress auxin sensitivity in the moss Physcomitrella patens. Plant Cell 14:373–386 [DOI] [PMC free article] [PubMed]
- Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8:217–230 [DOI] [PubMed]
- Klein J, Saedler H, Huijser P (1996) A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol Gen Genet 250:7–16 [DOI] [PubMed]
- Kropat J, Tootey S, Birkenbihl RP, Depège N, Huijser P, Merchant S (2005) A regulator of nutritional copper signalling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc Natl Acad Sci USA 102:18730–18735 [DOI] [PMC free article] [PubMed]
- Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR (1998) Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA 95:2686–2690 [DOI] [PMC free article] [PubMed]
- Manning K, Tör M, Poole M; Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Syemour GB (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38:948–952 [DOI] [PubMed]
- Markmann-Mulisch U, Reiss B, Mulisch M (1999) Cell type-specific gene expression in the cell cycle of the dimorphic ciliate Eufolliculina uhligi. Mol Gen Genet 262:390–399 [DOI] [PubMed]
- Mathews S (2006) Phytochrome-mediated development in land plants: red light sensing evolves to meet the challenges of changing light environments. Mol Ecol 15:83–3503 [DOI] [PubMed]
- Mockler TC, Guo H, Yang H, Duong H, Lin C (1999) Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126:2073–2082 [DOI] [PubMed]
- Moreno A, Harper LC, Krueger RW, Dellaporta SL, Freeling M (1997) Liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev 11:616–628 [DOI] [PubMed]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326 [DOI] [PMC free article] [PubMed]
- Riese M, Höhmann S, Saedler H, Münster T, Huijser P (2007) Comparative analysis of the SBP-Box gene families in P. patens and seed plants. Gene 401:28–37. doi:10.1016/j.gene.2007.06.018 [DOI] [PubMed]
- Russell AJ, Cove DJ, Trewavas AJ, Wang TL (1998) Blue light but not red light induces a calcium transient in the moss Physcomitrella patens (Hedw.) B., S. & G. Planta 206:278–283 [DOI]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
- Schaefer DG, Zryd JP, Knight CD, Cove DJ (1991) Stable transformation of the moss Physcomitrella patens. Mol Gen Genet 226:418–424 [DOI] [PubMed]
- Schumaker KS, Dietrich MA (1997) Programmed changes in form during moss development. Plant Cell 9:1099–1107 [DOI] [PMC free article] [PubMed]
- Somers DE, Devlin PF, Kay AK (1998) Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282:1488–1490 [DOI] [PubMed]
- Stone JM, Liang X, Nekl E, Stiers J (2005) ArabidopsisAtSPL14, a plant specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J 41:744–754 [DOI] [PubMed]
- Tichopad A, Dilger M, Schwarz G, Pfaffl MW (2003) Standardized determination of real-time PCR efficiency from a single reaction setup. Nucleic Acids Res 31:e122 [DOI] [PMC free article] [PubMed]
- Thijs G, Marchal K, Lescot M, Rombauts S, De Moor B, Rouzé P, Moreau Y (2002) A Gibbs Sampling method to detect over-represented motifs in upstream regions of coexpressed genes. J Comput Biol 9:447–464 [DOI] [PubMed]
- Uneaka H, Wada M, Kadota A (2005) Four distinct photoreceptors contribute to light-induced side branch formation in the moss Physcomitrella patens. Planta 222:623–631 [DOI] [PubMed]
- Unte US, Sorensen AM, Pesaresi P, Gandikota M, Leister D, Saedler H, Huijser P (2003) SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell 15:1009–1019 [DOI] [PMC free article] [PubMed]
- Vandenbussche F, Habricot Y, Condiff AS, Maldiney R, Van der Straeten D, Ahmad M (2007) HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsisthaliana. Plant J 49:428–441 [DOI] [PubMed]
- Wang H, Nussbaum-Wagler T, Li B, Zhao Q, Vigouroux Y, Faller M, Bomblies K, Lukens L, Doebley JF (2005) The origin of naked grains of maize. Nature 436:714–719 [DOI] [PMC free article] [PubMed]
- Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133:3539–3547 [DOI] [PMC free article] [PubMed]
- Yamasaki K, Kigawa T, Inoue M, Tateno M, Yamasaki T, Yabuki T, Aoki M, Seki E, Matsuda T, Nunokawa E, Ishizuka Y, Terada T, Shirouzu M, Osanai T, Tanaka A, Seki M, Shinozaki K, Yokoyama S (2004) A novel zinc-binding motif revealed by solution structures of DNA-DNA-binding domains of Arabidopsis SBP-family transcription factors. J Mol Biol 337:49–63 [DOI] [PubMed]
- Zhang Y, Schwarz S, Saedler H, Huijser P (2007) SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol Biol 63:429–439 [DOI] [PubMed]
- Zobell O, Coupland G, Reiss B (2005) The family of CONSTANS-like genes in Physcomitrella patens. Plant Biol 7:266–275 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.