Abstract
We have determined the complex sequence of events from the point of injury until reepithelialization in axolotl skin explant model and shown that cell layers move coherently driven by cell swelling after injury. We quantified three-dimensional cell migration using correlation spectroscopy and resolved complex dynamics such as the formation of dislocation points and concerted cell motion. We quantified relative behavior such as velocities and swelling of cells as a function of cell layer during healing. We propose that increased cell volume (∼37% at the basal layer) is the driving impetus for the start of cell migration after injury where the enlarged cells produce a point of dislocation that foreshadows and dictates the initial direction of the migrating cells. Globally, the cells follow a concerted vortex motion that is maintained after wound closure. Our results suggest that cell volume changes the migration of the cells after injury.
Introduction
The skin provides a barrier that protects the body from physical, chemical, and biological stress of the outside world. Its position, at the interface between the internal and external environments, makes the skin particularly prone to injury. Failure to repair damaged skin can have serious consequences, such as increasing the risk of infection. It is important that injuries be repaired as quickly as possible. Re-establishment of the epidermal barrier is an essential part of wound repair. Wound healing involves a complex sequence of events including keratinocyte migration, proliferation, and differentiation (1–3). In this study, we focus on the early events in wound healing; specifically, the dynamics of re-epithelialization of the wound bed, which precedes the aforementioned processes. Re-epithelialization is almost entirely the result of cell migration from the wound edge with minimal cell proliferation within the wound bed (4,5). Mobilization of keratinocytes from a stationary to a migratory state requires alterations in cell-cell and cell-matrix attachments, as well as intracellular cytoskeletal changes. Although much is known about the cellular mechanisms involved in keratinocyte migration, there are still unresolved questions about which cells move into the wound bed and the overall tissue dynamics of re-epithelialization. In particular, do keratinocytes migrate as a single sheet of cells or is re-epithelialization a result of cells “rolling or sliding” over each other (6,7). To answer this question, it is necessary to distinguish between cell layers. However, spatial sectioning of thick tissue is problematic in vivo. Therefore, we developed a model system that readily mimics in vivo responses after injury. We used real-time live cell imaging capabilities with sufficient resolution to follow the complex migration dynamics, and globally discern the concerted motion of cells within a tissue sample. Our data analysis techniques based on macroscopic correlations of cell motion are able to distinguish correlated cellular motion that can be associated to different layers and quantify dynamical quantities such as velocities, displacements, and changes in volume and morphology.
Coordinated cell motion is a widespread phenomenon in biology. During embryogenesis cell migration is essential for normal development. For example, the formation of the primitive streak during embryogenesis involves the establishment of two counter-rotating cell flow vortices in the epiblast that leads to the accumulation of cells at the site initiation of primitive streak formation (8). Although much is known about the mechanisms underlying individual cell migration, the factors driving coordinated cell motion are still the subject of debate. Orientated cell division, cell-cell intercalation, and chemotaxis have been proposed as mediators but none can fully account for the initiation of the vortex motion observed during primitive streak formation (9). We have observed similar vortex motion during wound healing in the skin explant model used in this work. We have used this model to investigate the initiation of the cell motion and have identified a marked increase in cell volume preceding cell migration. We propose that this increased cell volume provides the driving impetus for the start of cell migration after injury.
Axolotl wound healing model
Urodele amphibians, including newts and the axolotl (Ambystoma mexicanum), form a special subset of vertebrates that possess the ability to fully regenerate complex tissues including their limbs, tail, spinal cord, and other structures. They are a well-established biological model for studying regeneration and wound healing, particularly limb regeneration after amputation (10,11). Wound healing in urodeles is rapid; re-epithelialization of an amputated limb typically occurs in <10 h (12,13). In contrast, a similarly sized wound in mammals takes 2–3 days to re-epithelialize (7,14,15). Understanding the mechanisms that underlie these differences may lead to novel strategies for improving wound healing in humans. Recently, Ferris et al. (16) developed an in vitro system, for studying axolotl wound healing. Briefly, the system comprises tissue excised from the limb of a transgenic axolotl that ubiquitously expresses green fluorescent protein (17). A wound is produced in the explant and cell movement of epidermal cells is followed in vitro using fluorescence imaging. In this system, complete re-epithelialization of a 2-mm diameter wound typically occurs in <11 h. In this model, wound healing via the full re-epithelialization of the wound bed (closure) is highly reproducible.
Imaging techniques
Camera-based transmission microscopy was used to observe the entire area of the wound and skin from the onset of injury (Fig. 1 A). Although this approach has the advantage of a wide field view that is crucial to observe the overall motion of cells, transmission imaging cannot be used to differentiate cell layers. We used laser scanning systems in one photon confocal and two-photon (excitation) microscopy to spatially interrogate thick samples in the axial direction because they offer sufficient resolution to discern cells within the dermis and at least five cell layers into the epidermis as shown in Fig. 1 B for an intact tissue layer (Fig. 1 B, panels A–C). Hence, a true three-dimensional (3D) reconstruction of the cellular network within the tissue is obtained. The spatial resolution provided by confocal microscopy allowed us to determine cell types based on morphology in this highly heterogeneous tissue. By acquiring four-dimensional (4D) images during wound healing (3D images as a function of time), we explicitly followed the average migration of cells layers in a region of ∼40 μm deep starting at the basal layer and measured the average local velocity of the different cell layers, as well as the velocity of individual cells. This real-time dynamical information was used to quantify and better understand the global as well as single cell motion within a sample and avoid ambiguities that may arise by comparing across sections with different samples in fixed tissue.
Figure 1.
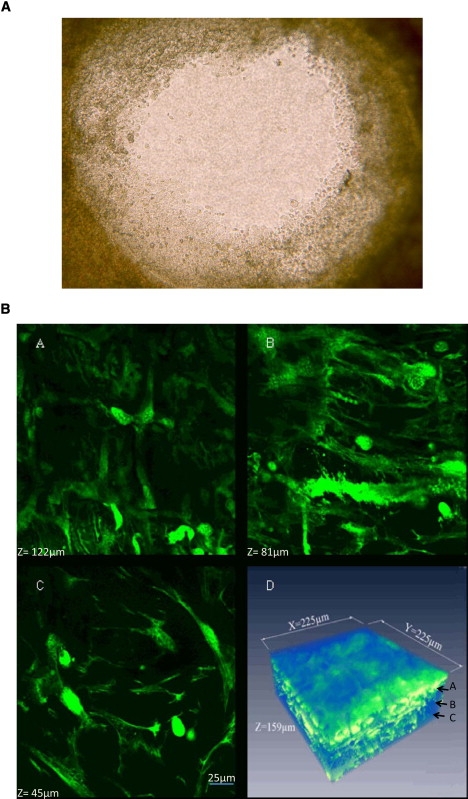
(A) Transmission image taken 5-h post injury showing the full view of the wound bed of diameter 2 mm and the cells that have migrated into the wound bed where the individual cell layers cannot be discerned. (B) Panels A–C show images obtained of a fully intact explant at a height of 45, 81, and 122 μm, respectively, above the microscope objective using two-photon microscopy where we can observe the transition from the dermis as identified by fibroblasts (height, 45 μm) to the epidermal layers as identified by keratinocytes (from >81 μm). Panel D shows the 3D reconstruction using the Amira software where the dimensions are 225 × 225 × 159 μm3.
Data analysis techniques
In our system, we have ∼105 cells involved in a concerted motion. We used techniques based on image correlation spectroscopy (ICS) and spatio-temporal image correlation spectroscopy (STICS) that were originally developed to provide a global analysis of the average size and flows of the molecular aggregates in images of cells (18,19). We use these algorithms to determine average cell movement and average cell size thereby providing dynamical information such as spatially resolved cellular velocities and displacements for entire tissue layers. Consequently, we can compare average quantities such as velocities, sizes, and cell numbers as a function of cell layer.
From the information derived from the correlation analysis that provides simultaneous tracking of multiple cells, we observe regions of concerted cell motion. Our observations have detailed the process of re-epithelialization such as the mechanism by which cells repopulate the wound bed. They suggest that physical constraints act as an impetus for the initiation of cell migration.
We examined nine different explants where the wound completely re-epithelialized and four where the wound did not fully close. We also imaged three samples, where no injury was made to the tissue, to show that we have enough spatial resolution to resolve each cell layer with sufficient penetration to observe from below the basement membrane, through the basal cell layer and into the suprabasal layers of the epidermis (Fig. 1 B, panel D). We present data that are representative of the collective behavior of the cells within the tissue explant and describe the complex dynamics associated with wound re-epithelialization.
Materials and Methods
Sample preparation
Collagen gels
A 1-mL collagen gel was adjusted to a final collagen concentration of 2.36 mg/mL of the solution by combining 721 μL of the original stock of rat tail collagen type I (BD Bioscience, San Mateo, CA, 3.27 mg/mL), 100 μL of 600% Leibovitz L-15 culture medium (Sigma, Irvine, CA), 50 μL fetal bovine serum (FBS) (Atlanta Biological, Lawrenceville, GA), 89 μL sterile H2O, and 40 μL sodium bicarbonate (NaHCO3). One hundred microliters of the resultant collagen mixture was added to a 35-mm MatTek (Ashland, MA) culture dish, and allowed to cure for 30 min at room temperature. After polymerization, collagen gels were equilibrated using one of the following conditions. In one protocol, the gels were equilibrated in 1 mL of culture medium (60% Dulbecco's modified Eagle's medium (DMEM) with 10% FBS, 0.5% gentamycin sulfate and 1.0% ITS (contains insulin, human transferrin, and selenious acid); Sigma) and incubated at 26°C, 5% CO2/95% air atmosphere. The other protocol involved equilibration with 60% Leibovitz L-15 culture medium with 10% FBS, 0.05% gentamicin, and 1.0% ITS, and incubated at 26°C without CO2.
Explant preparation and surgical procedures
Explants were prepared in a tissue culture hood by removing the forelimbs at mid humerus, sterilized in 70% EtOH for 7–10 s and rinsed in three changes of calcium- magnesium free Hank's (Sigma). Forelimbs were placed in a petri dish and labeled right or left to establish dorsal ventral orientation. After surgically removing the autopod, we made a full-length lateral incision along the posterior margin of the limb. Full thickness skin (epidermis and dermis) was peeled away with forceps and laid out flat (dermal surface up) in a petri dish. A 6-mm sterile biopsy punch (Miltex, York, PA) was used to remove a single explant, and a concentric 2-mm hole was punched in the center of the 6-mm disc to form a “donut” shaped explant). Explants were transferred to glass bottom 35-mm tissue culture dish (MatTek, Ashland, MA) dermal surface down on collagen gel; 1 ml of culture medium was added to the culture dish.
Imaging parameters
Samples were imaged using three instruments; an inverted Olympus FV 1000 Confocal microscope (San Diego, CA), an inverted Olympus IMT-2 microscope, and an inverted Zeiss LSM 510Meta (Jena, Germany). One photon confocal 3D images of 256 × 256 pixels (lateral dimensions) were acquired on the Olympus Fluoview 1000 using a 20× air objective with a NA of 0.75 using a zoom corresponding to an area of 452.6 × 452.6 μm2 and a total axial displacement of 50 μm using a step size of 0.5 μm. We excited our sample with the 488 nm line from an argon ion laser with a power of <3% while the bandpass filters were set to 505–525 nm and a gain of 700 on the amplifier in the emission pathway. Sequential transmission images were acquired every minute using a 4× phase objective on the Olympus IMT-2 microscope where the gain was set to 2 on the camera (detector). The sample was illuminated using a halogen lamp. The area of exposure corresponded to 1600 × 1200 pixels (2.34 mm × 1.76 mm) as detected by Spot Insight 11.2 Color Mosaic digital camera (Diagnostic Instrument, Sterling Heights, MI). The camera features a high quantum efficiency CCD capable of a sampling rate of 40 MHz (single channel) for real-time imaging. Two-photon fluorescence images were obtained on the Zeiss LSM 510Meta using a 10× dry, 20× dry, and 40× water objectives corresponding to areas of 900, 450, and 225 μm in the x and y directions, respectively. (1024 × 1024 pixels). The maximum axial displacement measured was 200 μm. We acquire images in incremental steps of 0.5–2 μm in the axial direction. Fig. 5 B, inset, shows the schematic of the intact sample as imaged where the dermal layer was closest to the objective and increasing in the axial level until we reach a limit in penetration depth of ∼200 μm. This allowed us to image a few cell layers into the epidermis (depending on sample thickness). The time to take one complete Z stack was ∼7 min for the Zeiss 510Meta. The excitation wavelength was 910 nm with the bandpass filters set to 505–525 nm in the emission pathway.
Figure 5.
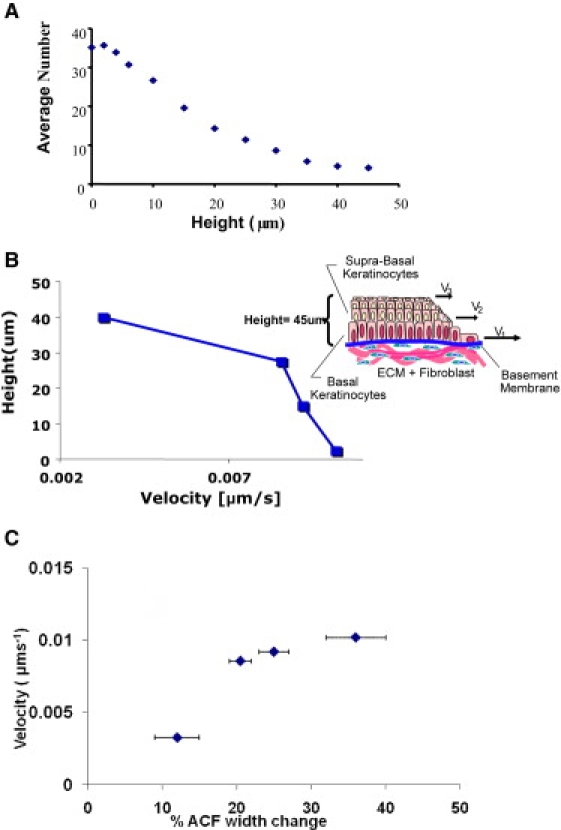
(A) Average number of cells calculated using ICS that move out for the total time (11 h) of wound re-epithelialization. The average number of basal keratinocytes for the cell layer at height = 5 μm is sevenfold greater than that of the average number of the suprabasal keratinocytes at height = 45 μm. (B) Representative data where the velocity of the individual cell layers at the leading edge of migration into the wound bed varies as a function of height where the basal layer (height = 0 μm) moves three times faster than the suprabasal layers (height = 15-45 μm). The inset shows a schematic that depicts the geometry with respect to our optical setup. (C) Correlation between changes in the width of the ACF and average velocity of a given layer. The changes in width are from Fig. 2B. The uncertainty in the width is obtained by estimating the changes in width occurring for an axial displacement of ∼2 μm from the shaded areas in Fig. 2. The error in the velocity as very small for a given layer because the average velocity is obtained by analyzing a region containing ∼10,000 cells.
Osmolyte treatment of explants
Axolotl skin explants were prepared as described above with the exception that 100 mM sorbitol (D-sorbitol) was added to the culture medium. The tissue culture dish containing the skin explant and culture medium was placed on the stage of the Olympus IMT-2 inverted microscope. Time-lapse microscopy was used to observe wound closure in explants treated with 100 mM sorbitol from the onset of injury. Sequential images (4× magnification) were collected at 1-min intervals for 6 h using the Spot 11.2 Color Mosaic digital camera. At 6 h explants were washed with PBS and fresh culture medium, without sorbitol, was added. Sequential imaging of wound closure was continued for a further 6 h. Images were exported using Spot software, as either AVI or Quick Time movies. The area of re-epithelialization was calculated from images taken at various time points using ImageJ software.
Data analysis
We use two approaches; one where we obtain a global analysis of the system, and another where we examine subpopulations such as determining the motion and volume changes of individual cells. We use techniques based on fluctuation spectroscopy to determine average changes in size, cell number, and velocities. The correlation techniques are robust in that they provide statistically weighted average quantities that describe the global behavior of the system. ICS as described by Petersen et al. (19) allows the determination of the average size and number of protein aggregates by calculating the spatial auto-correlation function:
| (1) |
Conceptually, if we have objects (i.e., cells) in the volume of observation, the width of the spatial autocorrelation function is proportional to the average size of the objects if the objects are sufficiently large (larger than the point spread function). The amplitude of the autocorrelation function is inversely related to the number of objects in the observation volume. STICS as described by Herbert et al. (18) is used to extract the average velocity by tracking the displacement of the spatial autocorrelation function as a function of time, in our case as a function of individual frames:
| (2) |
In short, any directed motion will result in the displacement of the spatial autocorrelation function as a function of time. By dividing the displacement per frame by the frame time, we obtain the average velocity.
We use the 2D ICS formulation to determine the size and shape changes for each focal plane. This approach allows us to maintain the spatial resolution in the axial direction so that we can quantify the geometric changes as a function of cell layer. In the second approach, we determine size changes by using 3D reconstruction software, Amira (Carlsbad, CA) for selected cells. By composing all of the 2D (x and y) images taken as a function of both time and depth (t and z), we can then truly follow the entire volume of tissue as a function of time. Using the volume statistics option of the Amira software, we can then determine the volume changes of individual cells. We carry out ICS analysis to glean information of the entire area for each focal plane whereas the Amira analysis is restricted to single cell that may or may not be coincidental with the global analysis as the single cell analysis may not be necessarily representative of the average. The procedure is repeated at different times to track volume changes and the results displayed to show the volume-time relationship. We note that the volume of the point spread function is small compared to the dimension of the cell so that correction due to the change of the point spread function as a function of the cell layer is not needed.
Additionally, the cell velocity at individual cells can be determined by tracking the displacement of the cell as a function of the number of 3D frames.
Results and Discussion
Cell volume and implications on wound re-epithelialization
In our model system, we address the collective cellular response to injury, namely if there are any physical changes such as cell swelling, cell shrinkage, or simply morphological changes. By calculating the width of the spatial autocorrelation functions (spatial ACF), we determine the average size of the ensemble of cells at different depths as this function is directly proportional to this parameter. In Fig. 2 we observe a size increase of ∼37%, when we compare the widths of spatial ACF at 11 min and 51 min post injury (Fig. 2). In Table 1, we compare the widths of the spatial autocorrelation function at different heights with respect to the microscope objective and the intact explant (from basal (0 μm) to suprabasal layers (40 μm)) where the widths were obtained by a Gaussian fit of the spatial ACF. An increase in the width of the spatial ACF is seen at all focal planes (all points are positive in Table 1) although there is an oscillation of the width parameter that matches with the position of the cell layers. There is also a clear trend of the overall width (Fig. 2 B, dotted line) with the basal layer showing the larger change. This result is consistent across nine samples where the average size increase ranges from 14% to 50%.
Figure 2.
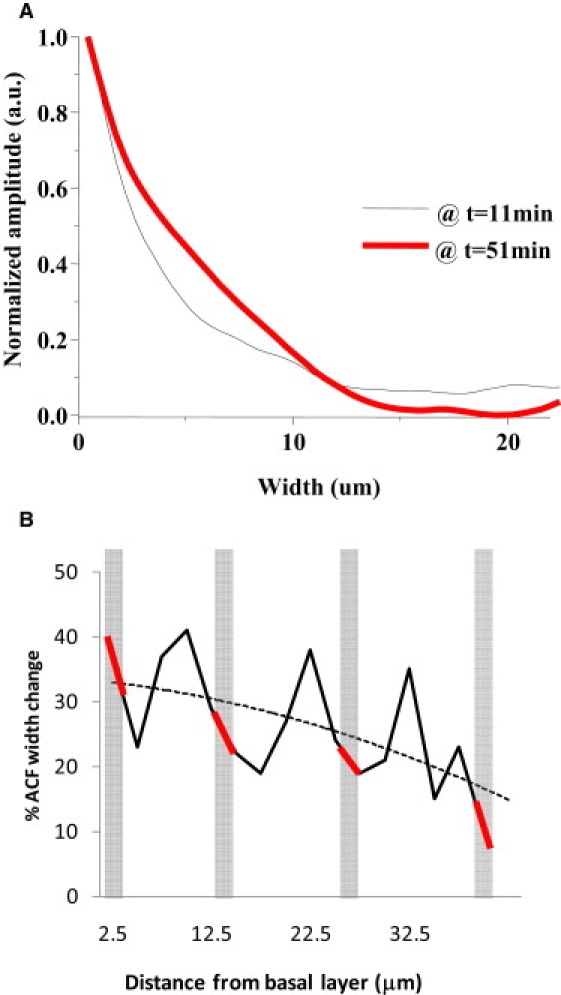
(A) Normalized horizontal section of the spatial autocorrelation function (spatial ACF) as a function of pixel shift (in μm). The width of the spatial ACF is proportional to the average size of cells in the observed region and in the observed plane. Using one-photon microscopy, the observed area at the leading edge of migration was ∼0.25 mm2. The spatial ACF were calculated using the ICS algorithm at 11 and 51 min post injury. (B) Data from Table 1. The oscillations in the width of the ACF correspond to the different cell layers. There is an overall trend (dotted line) of the width change as a function of the height above the basal layer. The shaded rectangles indicate the height at which the velocity of the cell layer is calculated.
Table 1.
Comparison of the average widths of the cells at the leading edge of migration for a representative experiment
| Axial position Z (μm) | Width at time of injury (μm) | Width 1-h post injury (μm) | Increase (%) |
|---|---|---|---|
| 2.5 | 13.1 | 17.9 | 37 |
| 5.0 | 16.7 | 20.6 | 23 |
| 7.5 | 8.8 | 13.0 | 37 |
| 10.0 | 14.4 | 20.3 | 41 |
| 12.5 | 25.2 | 32.5 | 29 |
| 15.0 | 13.9 | 16.9 | 22 |
| 17.5 | 11.78 | 14 | 19 |
| 20.0 | 12.8 | 16.2 | 27 |
| 22.5 | 11.1 | 15.3 | 38 |
| 25.0 | 12.6 | 15.6 | 24 |
| 27.5 | 11.3 | 13.4 | 19 |
| 30.0 | 13.9 | 16.8 | 21 |
| 32.5 | 12.1 | 16.3 | 35 |
| 35.0 | 11.3 | 13.0 | 15 |
| 37.5 | 10.5 | 12.9 | 23 |
| 40.0 | 11.9 | 13.1 | 10 |
Widths at the time of injury and 1-h post injury are compared as calculated using a Gaussian fit of the horizontal spatial ACF as a function of cell layer where 2.5 μm refers to the basal layer and 40 μm refers to the suprabasal layer. The error in the determination of the average spatial width is very small for a given sample because the average arises from many (>10,000 cells). However, different samples show different changes in the width of the ACF per equal time after injury.
Whereas the spatial ACF will give a global analysis, we carried out volume analysis using the Amira software (Carlsbad, CA) on 30 cells to determine the size changes of individual cells. Fig. 3 A shows that some cells undergo as much as a 50% increase in volume, whereas other cells show a negligible change. Fig. 3 B shows that there is variability in the rate and duration of the cellular volume changes when we carry out single cell analysis as a function of time. However, an initial increase in volume is seen in all cells studied (Fig. 3 B). There is no reason for the average number attained by the determination of the widths from the spatial ACF to be the same as that obtained from the single cell volume analysis as the single cell analysis provides a “snapshot” of cells rather than global information. Nevertheless, we do expect the trend, i.e., the initial average increase of volume to be similar (compare Table 1 and Fig. 3 B). If we used only techniques restricted to the selection of (few) individual cells, such as examination of cells across fixed histological samples or where we only perform single cell tracking, we could be biased on the selection of the region, type of cell and cell layer. The single cell analysis gives inconsistent results and it is difficult to extract the general trend. Instead the correlation methods used to determine the average size changes of a large number of cells are robust. It has been known for some time that cell swelling (cell hypertrophy) occurs during the post wound period and before cell migration. Viziam et al. (15) reported cell hypertrophy at the wound edge ∼18–21 h after wounding in a rabbit wound healing model. The hypertrophy preceded migration by at least 3 h. Odland and Ross (14) also noted increased cell volume in keratinocytes at the wound edge in humans during the premigratory phase. Using spatial correlation methods, we were able to identify global increases in cell volume in our explant model. Although these changes can be observed by more conventional volume reconstruction techniques, the nature of the individual cell measurements does not fully describe the state of the tissue at any given time point (Fig. 3 B). A major consequence of increasing cell volume is that it causes stresses within the tissue, which must be accommodated.
Figure 3.
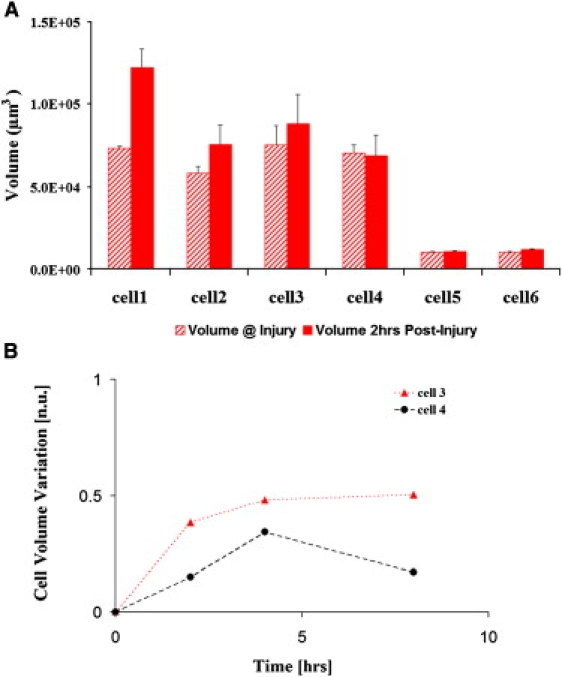
(A) Representative data of volume changes from multiple experiments where the wound was fully re-epithelialized. Volume comparison at point of injury and 2-h post injury using the Amira single cell analysis shows that some cells undergo a significant increase whereas others do not. The volume error is obtained by carrying out repeated measurements within the sample. (B) Temporal evolution of the fractional change in volume of a subset of cells. Cell 3 and 4 were tracked for 8 h. Starting from the onset of injury, some cells can show a continuous increase or a decrease with respect to the achieved maximum volume. However, all cells examined undergo an initial volume increase. The error was estimated by repeated measurement of the volume of the same cell.
If we look at the early stages of wound closure, when ∼10% of the wound bed has been re-epithelialized, we observe a point of dislocation where a sheet of cells migrate in a tongue-like fashion to indicate the beginning of closure of the wound as indicated by the red arrows (Fig. 4, top right panel). Fig. 4, top right panel, also shows the velocities of the adjacent cells (black arrows) near the line of dislocation. In each of the explants, we observe this dislocation as the origin for the vortex movement of the inward closure. Continued observation of re-epithelialization by transmission microscopy shows a concerted movement of the cells that produces a vortex-like motion (data not shown). Fig. 4, bottom right panel, shows the velocity map of the cells 5-h post injury where 60% of the wound has been repopulated. This edge centric-vortex motion is maintained even after the wound is completely closed; the cells still continue to migrate.
Figure 4.
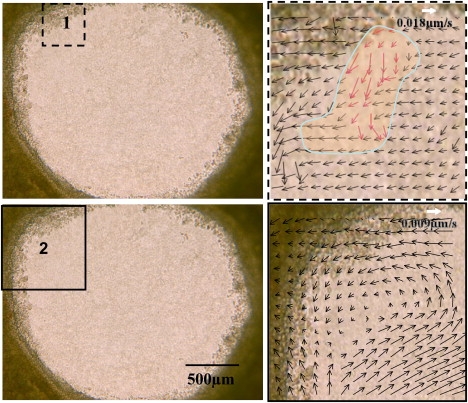
Left panels show the original transmission image of the explant corresponding to an area of 2.34 mm × 1.76 mm at the time and point of injury with the square (black dashes and solid lines, respectively) indicating the regions of the velocity map analysis. The right upper panel shows the formation of the initial dislocation of cells at the wound border shown by the blue line-shaded region. The velocity map was obtained using the STICS analysis averaging over a region of 32 × 32 pixels for a time of 64 min from the initial injury. The pixel resolution of this image is the same as that of the original image. The right bottom panel shows the coordinated motions of the cell in a vortex like manner. The velocity map was obtained using a lower resolution image (binning by 2 the original image) analyzed over a region of 32 × 32 pixels for a period of 200 min, 5-h post injury. In each of the velocity maps, the scale of the velocity is shown by the white arrow.
We propose that the dislocation and resultant vortex motion is a direct result of cell swelling and the nearest neighbor forces that each cell experiences given the geometry of the wound. This effect is seen when we observe the velocities of the cells from the time of injury to complete wound re-epithelialization (Fig. 4, panel 2). As in mammalian cells, the volume change precedes cell migration in the axolotl. However, in the case of the axolotl these changes occur within minutes rather than hours.
The mechanism by which cell volume is increased during wound healing is unknown, but the rapidity of the volume increase in the axolotl explant suggests that it is unlikely to involve changes in protein synthesis. Recently, it has been shown that downregulation of the aquaporin 3 channels, which facilitate water transport in keratinocytes, leads to decreased cell migration and impaired wound healing (20,21). We speculate that differences in volume regulation may account for the differences in the timing of the cell volume changes between mammals and salamanders. To alter the volume changes effect we treated the explant with osmolytes at different times during the healing process. Although we did experiments inhibiting aquaporin 3 channels and we found that this inhibition has a large effect on the rate of wound closure, we prefer to report here the effect of osmolytes that we assume have a different, more physical effect than channel inhibition. Fig. 6 A shows a typical example for one explant and Fig. 6 B shows the results for nine different explants treated with the osmolytes as described in Materials and Methods. It seems that changing the rate of swelling of the cells directly affects the rate of wound closure.
Figure 6.
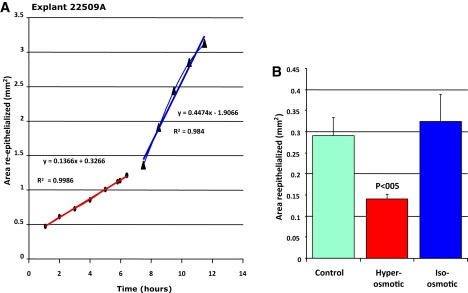
Changes of rate of re-epithelialization on application of osmolytes. (A) Typical behavior of an explant treated with the osmolyte. The osmolyte is washed away after ∼6 h. (B) Comparison of control (untreated), treated explant with an hyper-osmotic solution and with an iso-osmotic solution. Nine explants were measured in each group.
Keratinocyte migration in axolotl skin explants
Two models of wound healing in stratified epithelia have been described and remain the subject of debate. In short, each model predicts that either the basal or suprabasal cell layers move out first after injury. We applied algorithms to determine the relative velocities as well as quantify the number of migrating cells as a function of cell layer, thus addressing this source of controversy. Fig. 5 A shows that the average number of cells migrating into the wound bed decreases as a function of height from the basement membrane. The number of basal keratinocytes at the leading edge (at height 5 μm) that migrate is ∼7-fold greater than cells in the suprabasal layers (at height 40 μm). This average number is shown for the cells at the leading edge moving into the wound bed at the onset of injury until the wound has fully closed. This decrease as a function of height was consistently observed for nine samples of circular wounds. Specifically, the ratio of the average number of cells, comparing the basal and suprabasal cells, ranged from 5 to 10 depending on the sample. Velocity analysis of 4D fluorescence images shows that the basal keratinocytes at the leading edge (bottom layers) migrate a factor of 3 greater than that of the suprabasal keratinocytes (upper layers) as shown in Fig. 5 B. Expanding on the physical manifestations of each model, the first model described by Krawczyk (7) is based on light and electron microscopy of subepidermal blister wound healing. It suggests that keratinocytes near the wound edge crawl or “leapfrog” over one another and a number of studies have provided further evidence to support this model of wound healing (7,22–25). The “Leap-frog” model implies that cells in the suprabasal layers will move into the basal layer. If wound healing conforms to this model then the velocity of the upper layers should be faster than the basal layer and we should observe a migration of the cells in the axial direction (Z direction) signifying the descent of the suprabasal cells into the wound bed. In the second model, the cells migrate as a coherent sheet while maintaining their position within each layer (6,26–28). Thus, the “Tractor-tread” model, predicts the cells in the bottom layer of the epidermis will move out first. In our explant system, we observe cells migrating from the basal layer at the onset of injury and that the velocity of the layers decreased with distance from the basement membrane. We see no evidence to support the Leap-frog model in our system. However, we note that in our model system the basal keratinocytes seem to adhere to the substrate loosely that may facilitate their relative rapid motion during the re-epithelialization. The differential velocities of the cell layers imply that there is a driving impetus for wound closure as well as the breakdown of cell adhesion between the layers. One mechanism proposes that the cells in the basal compartment migrate into the suprabasal layer resulting in an apparent loss of number of cells in the basal layer (22). Our observations suggest an alternative hypothesis whereby the continuous spiral motion allows for the conservation of cells in the basal cell layer whereas at the same time reducing the number of cells at the wound edge.
In Fig. 5 C we show that there is correlation between the velocities of the layers as a function of the height and the width change of the spatial autocorrelation function as reported in Table 1. We identified the increase with respect to the initial time just after injury of width of the correlation function with the average swelling of the cells. The graph in Fig. 5 C shows that an increase in swelling at the different height correlates with an increase in layer velocity.
Wound re-epithelialization summary
In summary, we are able to follow the chronological events including the global migration of sheets of cells as well as the cell volume changes after injury. We observe a sequence whereby at the point of injury, cells swell. Swelling causes a dislocation at the wound edge that provides the impetus for the re-epithelialization to begin (Fig. 7, right panel). Cells continue to move in a spiral motion even after full re-epithelialization is completed. We propose that cell swelling is the driving impetus for the start of cell migration after injury. The constant nearest neighbor forces result in a dislocation. Only then do the cells begin to migrate in a vortex motion. This spiral motion easily accommodates the packing distribution of cells.
Figure 7.
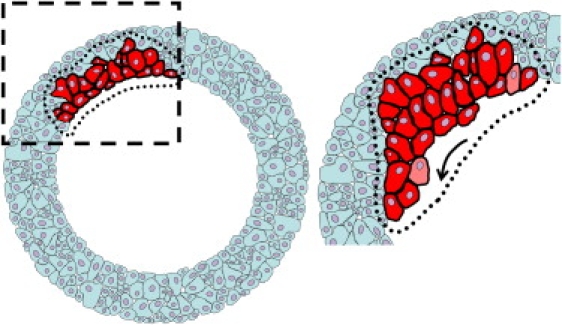
Schematic of the formation of the dislocation that occurs after ∼1-h post injury. Left panel shows the explant at the time and point of injury with a highlighted region of cells. All cells undergo cell hypertrophy (not represented). Right panel show the highlighted cells undergoing hypertrophy and the consequential dislocation. The arrow indicates the direction of the inward re-epithelialization of the wound bed.
We used camera-based, one- and two-photon microscopy to follow real-time dynamics of epithelial cell migration in a 3D axolotl wound healing model. This approach yielded information that cannot be gleaned easily from static images of fixed tissue. We were able to discern the movements of individual cell layers within the tissue, which allows us to determine the origin of cells that migrate into the wound bed. In addition, global analyses using ICS and STICS (18,19) show properties of the tissue, which may not be apparent at the level of the individual cell (Figs. 4 and 5). We show that the image correlation global analysis in conjunction with 4D optical sectioning can provide overall dynamics, concerted motion, and information about cell volume where the integrity of the tissue is maintained due to the noninvasive nature of the technique. This allows a true dynamical examination of migratory cells within tissue.
Acknowledgments
This work was supported jointly by the National Institutes of Health (PHS 5 P41-RR003155 to E.G, L.L. W.W.M and K.T., and P50 GM076516-029001 to E.G.), the Defense Advance Research Projects Agency (D.M.G., E.L.R), the Laser Microbeam and Medical Program (LAMMP) National Institutes of Health (P41-RR01192), and the National Science Foundation through its support of the Ambystoma Genetic Stock Center at the University of Kentucky, Lexington.
Footnotes
K. Tanner and D.R. Ferris contributed equally to this work.
References
- 1.Clark R.A. Plenum Press; New York: 1996. The Molecular and Cellular Biology of Wound Repair. [Google Scholar]
- 2.Paladini R.D., Takahashi K., Bravo N.S., Coulombe P.A. Onset of re-epithelialization after skin injury correlates with a reorganization of keratin filaments in wound edge keratinocytes: defining a potential role for keratin 16. J. Cell Biol. 1996;132:381–397. doi: 10.1083/jcb.132.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singer A.J., Clark R.A. Cutaneous wound healing. N. Engl. J. Med. 1999;341:738–746. doi: 10.1056/NEJM199909023411006. [DOI] [PubMed] [Google Scholar]
- 4.Bereiter-Hahn, J. 1986. Epidermal cell migration and wound repair. In Biology of the Integument, Vol. 2. Vertebrates. Springer-Verlag, Berlin. 443–471.
- 5.Patel G.K., Wilson C.H., Harding K.G., Finlay A.Y., Bowden P.E. Numerous keratinocyte subtypes involved in wound re-epithelialization. J. Invest. Dermatol. 2006;126:497–502. doi: 10.1038/sj.jid.5700101. [DOI] [PubMed] [Google Scholar]
- 6.Hanna C., Keatts H.C. Cell migration in the adult rat and rabbit lens. Exp. Eye Res. 1968;7:244–246. doi: 10.1016/s0014-4835(68)80074-0. [DOI] [PubMed] [Google Scholar]
- 7.Krawczyk W.S. A pattern of epidermal cell migration during wound healing. J. Cell. Biochem. 1971;49:247–263. doi: 10.1083/jcb.49.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui C., Yang X., Chuai M., Glazier J.A., Weijer C.J. Analysis of tissue flow patterns during primitive streak formation in the chick embryo. Dev. Biol. 2005;284:37–47. doi: 10.1016/j.ydbio.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 9.Chuai M., Zeng W., Yang X., Boychenko V., Glazier J.A. Cell movement during chick primitive streak formation. Dev. Biol. 2006;296:137–149. doi: 10.1016/j.ydbio.2006.04.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardiner D.M., Bryant S.V. John Wiley & Sons; Chichester, UK: 1998. The Tetrapod Limb. 187–205. [Google Scholar]
- 11.Wallace H. John Wiley & Sons; Chichester, UK: 1981. Vertebrate Limb Regeneration. [Google Scholar]
- 12.Endo T., Bryant S.V., Gardiner D.M. A stepwise model system for limb regeneration. Dev. Biol. 2004;270:135–145. doi: 10.1016/j.ydbio.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 13.Repesh L.A., Oberpriller J.C. Ultrastructural studies on migrating epidermal cells during the wound healing stage of regeneration in the adult newt, Notophthalmus viridescens. Am. J. Anat. 1980;159:187–208. doi: 10.1002/aja.1001590207. [DOI] [PubMed] [Google Scholar]
- 14.Odland G., Ross R. Human wound repair. I. Epidermal regeneration. J. Cell Biol. 1968;39:135–151. doi: 10.1083/jcb.39.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viziam C.B., Matoltsy A.G., Mescon H. Epithelialization of small wounds. J. Invest. Dermatol. 1964;43:499–507. [PubMed] [Google Scholar]
- 16.Ferris D.R., Satoh A., Mandefro B., Bryant S.B., Gardiner D.M., Rugg E.L. An in vitro model for studying axolotl wound healing. 2008 International Investigative Dermatology Meeting Abstracts. J. Invest. Dermatol. 2008;128:S99. [Google Scholar]
- 17.Sobkow L., Epperlein H.H., Herklotz S., Straube W.L., Tanaka E.M. A germline GFP transgenic axolotl and its use to track cell fate: dual origin of the fin mesenchyme during development and the fate of blood cells during regeneration. Dev. Biol. 2006;290:386–397. doi: 10.1016/j.ydbio.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 18.Hebert B., Costantino S., Wiseman P.W. Spatiotemporal image correlation spectroscopy (STICS) theory, verification, and application to protein velocity mapping in living CHO cells. Biophys. J. 2005;88:3601–3614. doi: 10.1529/biophysj.104.054874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersen N.O., Hoddelius P.L., Wiseman P.W., Seger O., Magnusson K.E. Quantitation of membrane receptor distributions by image correlation spectroscopy: concept and application. Biophys. J. 1993;65:1135–1146. doi: 10.1016/S0006-3495(93)81173-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hara-Chikuma M., Verkman A.S. Aquaporin-3 facilitates epidermal cell migration and proliferation during wound healing. J. Mol. Med. 2008;86:221–231. doi: 10.1007/s00109-007-0272-4. [DOI] [PubMed] [Google Scholar]
- 21.Papadopoulos M.C., Saadoun S., Verkman A.S. Aquaporins and cell migration. Pflugers Arch. 2008;456:693–700. doi: 10.1007/s00424-007-0357-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danjo Y., Gipson I.K. Specific transduction of the leading edge cells of migrating epithelia demonstrates that they are replaced during healing. Exp. Eye Res. 2002;74:199–204. doi: 10.1006/exer.2001.1115. [DOI] [PubMed] [Google Scholar]
- 23.Garlick J.A., Taichman L.B. Fate of human keratinocytes during reepithelialization in an organotypic culture model. Lab. Invest. 1994;70:916–924. [PubMed] [Google Scholar]
- 24.Gibbins J.R. Epithelial migration in organ culture. A morphological and time lapse cinematographic analysis of migrating stratified squamous epithelium. Pathology. 1978;10:207–218. doi: 10.3109/00313027809063503. [DOI] [PubMed] [Google Scholar]
- 25.Repesh L.A., Oberpriller J.C. Scanning electron microscopy of epidermal cell migration in wound healing during limb regeneration in the adult newt, Notophthalmus viridescens. Am. J. Anat. 1978;151:539–555. doi: 10.1002/aja.1001510408. [DOI] [PubMed] [Google Scholar]
- 26.Buck R.C. Cell migration in repair of mouse corneal epithelium. Invest. Ophthalmol. Vis. Sci. 1979;18:767–784. [PubMed] [Google Scholar]
- 27.Mahan J.T., Donaldson D.J. Events in the movement of newt epidermal cells across implanted substrates. J. Exp. Zool. 1986;237:35–44. doi: 10.1002/jez.1402370107. [DOI] [PubMed] [Google Scholar]
- 28.Zhao M., Song B., Pu J., Forrester J.V., McCaig C.D. Direct visualization of a stratified epithelium reveals that wounds heal by unified sliding of cell sheets. FASEB J. 2003;17:397–406. doi: 10.1096/fj.02-0610com. [DOI] [PMC free article] [PubMed] [Google Scholar]


