Abstract
BACKGROUND
Rare mutations affecting the FOXP2 transcription factor cause a monogenic speech and language disorder. We hypothesized that neural pathways downstream of FOXP2 influence more common phenotypes, such as specific language impairment.
METHODS
We performed genomic screening for regions bound by FOXP2 using chromatin immunoprecipitation, which led us to focus on one particular gene that was a strong candidate for involvement in language impairments. We then tested for associations between single-nucleotide polymorphisms (SNPs) in this gene and language deficits in a well-characterized set of 184 families affected with specific language impairment.
RESULTS
We found that FOXP2 binds to and dramatically down-regulates CNTNAP2, a gene that encodes a neurexin and is expressed in the developing human cortex. On analyzing CNTNAP2 polymorphisms in children with typical specific language impairment, we detected significant quantitative associations with nonsense-word repetition, a heritable behavioral marker of this disorder (peak association, P = 5.0×10-5 at SNP rs17236239). Intriguingly, this region coincides with one associated with language delays in children with autism.
CONCLUSIONS
The FOXP2-CNTNAP2 pathway provides a mechanistic link between clinically distinct syndromes involving disrupted language.
Developmental disorders of speech, language, and communication account for 40% of referrals to pediatric services.1 Although many children grow out of early language delay, others have persistent difficulties with language expression and comprehension, despite normal nonverbal ability and lack of an obvious reason. In some children, developmental speech or language impairments are part of a broader syndrome such as autism, in which these deficits are accompanied by unusual repetitive behaviors and disturbances in social interaction. More commonly, such impairments occur in the absence of autistic features.2 Longitudinal studies have indicated that when language impairments persist to school age, they are likely to be associated with enduring academic and psychiatric problems.3
Developmental speech and language disorders are highly heritable, with most cases showing complex multifactorial inheritance.4 The isolation of relevant genetic effects will yield new insights into the causes of such impairments, along with improved classification, diagnosis, and treatment. One notable success in this area was the discovery that heterozygous disruptions of the FOXP2 gene cause a rare mendelian speech and language disorder.5-9 Point mutations and chromosomal abnormalities that affect FOXP2 are associated with difficulties in the learning and production of sequences of oral movements, which impair speech (also called developmental verbal dyspraxia or childhood apraxia of speech).5-9 The affected persons also have variable levels of impairment in expressive and receptive language, extending to problems with production and comprehension of grammar.10 However, FOXP2 disruptions are rare. It has been estimated that approximately 2% of people with verbal dyspraxia carry etiologic point mutations in this gene.6
Specific language impairment is the most frequently diagnosed form of developmental language disorder, affecting up to 7% of children who are 5 or 6 years of age.11 Although there is considerable variation in the profile of linguistic deficits observed and in the functions affected (expressive, receptive, or both),12 specific language impairment often occurs without accompanying difficulties in speech articulation. For example, an epidemiologic study showed that only about 5 to 8% of children with persistent specific language impairment had a significant speech delay.13 Moreover, analyses of FOXP2 in persons with typical forms of specific language impairment have not detected etiologic mutations or evidence of association.14,15 Mutation of FOXP2 itself is therefore unlikely to be a major risk factor for common language impairments. Indeed, to date we know of no report of a gene associated with typical specific language impairment.12
Because FOXP2 encodes a neurally expressed transcription factor,16,17 we reasoned that one or more of the genes that it regulates in the brain might be implicated in common language-related phenotypes. Here we describe the isolation of a novel FOXP2-regulated target with neural functions and provide evidence of its association with language-related deficits in a large set of well-characterized families with specific language impairment.
METHODS
SCREENING FOR TARGETS OF FOXP2
We engineered the human neuroblastoma SH-SY5Y cell line to stably express FOXP218 and then, using this transfected cell line, carried out unbiased screening for genomic sites bound by FOXP2 protein. This involved the use of chromatin immunoprecipitation with anti-FOXP2 antibodies, followed by shotgun sequencing of purified DNA, a process of randomly cloning fragments of DNA and then determining their sequence (for details, see the Supplementary Appendix, available with the full text of this article at www.nejm.org). We determined the positions of DNA sequences that were isolated with chromatin immunoprecipitation, using BLAT on the University of California, Santa Cruz, Genome Server (http://genome.ucsc.edu/), which enabled identification of putative target genes.
VALIDATION OF BINDING AND REGULATION BY FOXP2
Binding of FOXP2 to target sites was independently verified and further localized with the use of semiquantitative polymerase-chain-reaction (PCR) assay of chromatin isolated from additional chromatin-immunoprecipitation experiments and electrophoretic mobility shift assays (EMSAs), according to protocols reported previously.18 Regulation of putative target genes was assessed with the use of quantitative reverse-transcriptase PCR (RT-PCR) of RNA extracted from SH-SY5Y cells expressing different FOXP2 levels, as described previously18 (see Table S1 in the Supplementary Appendix for primer sequences). In situ hybridization was performed on human fetal brains,19 as described in the Methods section in the Supplementary Appendix.
STUDY SUBJECTS
The study subjects were members of epidemiologically and clinically ascertained families identified by the Specific Language Impairment Consortium.20,21 These families were recruited from four sites in the United Kingdom: the Newcomen Centre at Guy’s Hospital,20,21 the Cambridge Language and Speech Project,22 the Child Life and Health Department at Edinburgh University,23 and the Manchester Language Study.24 Families were selected through a proband with specific language impairment whose past or current language skills were 1.5 SD or more below the normative mean for the child’s age on the Clinical Evaluation of Language Fundamentals-Revised (CELF-R) scale,25 a tool that is routinely used for diagnosis and follow-up evaluation of language disorders in school-age children. (Scores on the scale range from 50 to 150, with a mean of 100 and a standard deviation of 15 in the general population. Lower scores indicate poorer performance.) We excluded any children with a nonverbal IQ of less than 80, a clinical diagnosis of an autistic-spectrum disorder, or another known medical or developmental condition that can impair language, such as hearing loss, cleft lip, or cleft palate. Moreover, for clinically ascertained samples, children were comprehensively assessed on scales evaluating language, IQ, and behavior, and those with overt pragmatic difficulties, behavioral characteristics associated with autism, or a family history indicative of autism were also excluded.
We collected quantitative phenotypic data from probands and all available siblings. We then determined composite CELF-R scores for expressive and receptive language abilities. We also used a measure of the ability to repeat nonsense words, the Children’s Test of Nonword Repetition,26 which has been established as a robust endophenotype of specific language impairment.12 (Scores on the scale range from 46 to 141, with a mean of 100 and a standard deviation of 15 in the general population. Lower scores indicate poorer performance.) This measure is thought to provide an index of phonologic short-term memory.12 Children with specific language impairment perform particularly poorly on nonsense-word repetition, and impaired phonological short-term-memory has been proposed as a core deficit in the disorder. An impairment in the ability to repeat nonsense words is highly heritable, persists in persons with historical language problems that have otherwise resolved,27 and appears to be relatively unaffected by environmental factors.28 Additional information on the consortium families has been reported previously20,21 and is available in Table S2 in the Supplementary Appendix, which shows means, standard deviations, and intertrait correlations for language measures used in this study. Written informed consent was obtained from all subjects or their parents; assent was obtained from children of appropriate age.
SINGLE-NUCLEOTIDE-POLYMORPHISM GENOTYPING
To directly test the hypothesis that variants of the identified FOXP2 target (the CNTNAP2 gene) may increase susceptibility to common language impairments, we genotyped single-nucleotide polymorphisms (SNPs) in consortium families, followed by quantitative association analyses of measures of expressive and receptive language abilities and nonsense-word repetition. We genotyped and validated 38 SNPs from the CNTNAP2 locus on chromosome 7q35 in samples from 847 members of 184 consortium families, using Golden Gate assays on the Illumina platform (for details, see the Methods section and Table S3 in the Supplementary Appendix).
STATISTICAL ANALYSIS
For analyses of differences in gene expression in SH-SY5Y cells, we assessed statistical significance using unpaired t-tests (two-tailed). For family-based association analyses of SNP data from the consortium series, we used a quantitative transmission disequilibrium test (QTDT), adopting an orthogonal association model that considers only the within-family variance and is robust to population stratification.29 After identifying significant single SNP associations, we used the Merlin package30 to generate haplotypes for the cluster of nine associated SNPs, which were similarly analyzed with QTDT. Finally, we investigated the possibility of an effect of sex or imprinting within QTDT, using these nine SNP-tag haplotypes.
RESULTS
CNTNAP2 AS A TARGET OF FOXP2
To identify candidate genes that might be involved in typical specific language impairment, we used an unbiased screening method to isolate genomic fragments bound by the FOXP2 protein in chromatin of human neuronlike cells. We thereby discovered a FOXP2-bound fragment that was of particular interest, because it was located within intron 1 of CNTNAP2 (Fig. 1A). This gene encodes CASPR2, a member of the neurexin superfamily of transmembrane proteins, found at the nodes of Ranvier in myelinated nerve fibers. In mice, Caspr2 is important for the regulation of the localization and maintenance of Shaker-type voltage-activated potassium channels31 and is implicated in neuronal recognition and cell adhesion.32 In humans, it has been suggested that CASPR2 is involved in cortical development, possibly mediating intercellular interactions during neuroblast migration and laminar organization.33
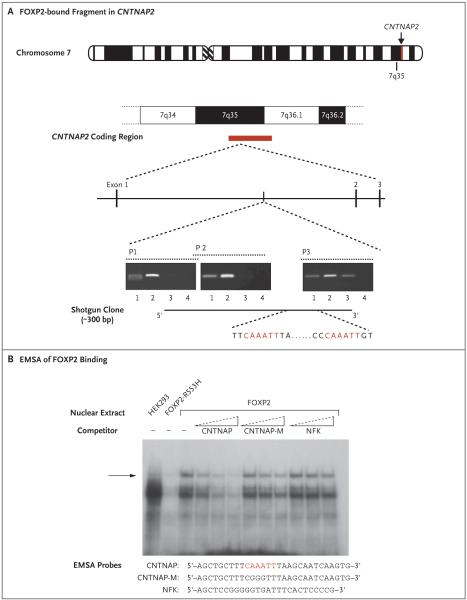
Figure 1 (facing page). Identification of CNTNAP2 as a Direct Neural Target Bound by Human FOXP2.
In Panel A, a 300-bp clone was identified through shotgun cloning of gene fragments identified by FOXP2-chromatin immunoprecipitation and localized to intron 1 of the human CNTNAP2 gene in 7q35. Semiquantitative PCR analysis indicated consistent enrichment of this region in multiple independent experiments in a neuronlike cell line immunoprecipitated with an N-terminal FOXP2 antibody (lane 2), as compared with a control sample without the antibody (lane 3) and input DNA samples (lane 1). Lane 4 shows the water control sample. Two FOXP2 consensus binding sites were identified (highlighted in red). In Panel B, electrophoretic mobility shift assays (EMSAs) using nuclear extracts from transfected HEK293T cells assessed the ability of FOXP2 protein to bind to the 5′ consensus binding site (highlighted in red). Efficient binding to the CNTNAP probe was observed when FOXP2 was present but not when either un-transfected cells or cells expressing a mutant form of FOXP2 (R553H) were used.17 Binding to the labeled probe was efficiently reduced by competition with an un-labeled probe (CNTNAP) but not by a mutant form of the probe (CNTNAP-M) or an irrelevant binding site (NFK). The arrow shows the position of the shift caused by FOXP2 binding to the CNTNAP probe.
We used PCR to amplify sequences spanning the FOXP2-bound fragment of CNTNAP2 in independent FOXP2 chromatin-immunoprecipitation samples and in control samples in which no antibodies were used and observed evidence of enrichment only when FOXP2-specific antibodies had been used to isolate the chromatin (Fig. 1A). Primers amplifying regions of 1000 bp or more away from the bound fragment did not display FOXP2-chromatin immunoprecipitation enrichment. FOXP2 is thought to bind chromatin as a dimer, and our in silico analyses of the chromatin immunoprecipitation-enriched fragment identified two adjacent sites, separated by 48 bases, matching a known consensus sequence for FOXP2 binding (CAAATT). EMSA analyses indicated that FOXP2 could bind both sites (data not shown). At each site, binding could be disrupted by the mutation of three core nucleotides of the recognition sequences (CAAATT→CGGGTT), with more dramatic effects observed for the 5′ site. Full competition assays for this site showed highly efficient and specific binding by FOXP2 (Fig. 1B).
We then used quantitative RT-PCR to directly test whether modulation of FOXP2 protein levels would yield altered CNTNAP2 expression. Indeed, CNTNAP2 messenger RNA levels were consistently and significantly reduced in neuronlike cells that were stably transfected with FOXP2, as compared with sham-transfected control samples (Fig. 2A).
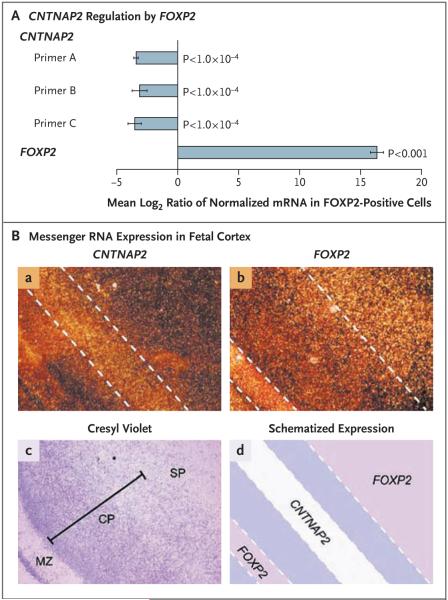
Figure 2. Analyses of the Effects of FOXP2 on Neural Expression of CNTNAP2.
Panel A shows the regulation of CNTNAP2 expression by FOXP2 in human neuronlike cells. The expression of messenger RNA (mRNA) was assessed with quantitative reverse-transcriptase PCR in SH-SY5Y cells. The cells were stably transfected either with a construct expressing FOXP2 (FOXP2-positive cells) or with an empty control vector that does not contain a gene insert (control cells). Levels of CNTNAP2 mRNA in these cells were inversely proportional to that of FOXP2. Findings were consistent for three sets of primer pairs recognizing distinct combinations of CNTNAP2 exons (primers A to C). Expression changes are given as the mean log2 expression ratios in FOXP2-positive cells, as compared with empty controls, normalized for equal expression of the internal control, GAPDH. The I bars represent standard errors. The P values were calculated with the use of two-tailed unpaired t-tests. Panel B shows nonoverlapping mRNA expression for CNTNAP2 and FOXP2 in human fetal cortex. Adjacent sections from human fetal brain (at 18 to 22 weeks’ gestation) were processed for in situ hybridization, dipped in film emulsion, and visualized by dark-field microscopy. Within the cerebral cortex, the highest levels of CNTNAP2 mRNA are observed between bands of FOXP2 expression, putatively within layers II and III of the cortical plate (subpanel a). In contrast, FOXP2 is present at high levels in the molecular zone, deep layers of the cortical plate, and subplate (subpanel b). Cortical lamination is highlighted in a bright-field image of a section stained with cresyl violet (subpanel c). Nonoverlapping expression patterns for CNTNAP2 and FOXP2 are schematized in subpanel d. CP denotes cortical plate, MZ molecular zone, and SP subplate.
A recent genomewide analysis of differential gene expression in the developing human cerebral cortex independently highlighted CNTNAP2 as a gene with substantial enrichment in frontal gray matter, which is primarily restricted to the region between the orbital gyrus and superior frontal anlage, spanning the inferior and middle frontal gyri.34 Because FOXP2 is also expressed in the developing human cortex,16,34,35 we carried out expression analyses of this structure in fetal tissue (18 to 22 weeks’ gestation) through in situ hybridization. We observed complementary patterns with respect to cortical lamination: CNTNAP2 expression was lowest in layers that showed the highest levels of FOXP2 (Fig. 2B). These in vivo findings are consistent with our data from neuronal models, supporting negative regulation of human CNTNAP2 expression by FOXP2.
ASSOCIATION ANALYSES OF CNTNAP2
Several studies underscored CNTNAP2 as a particularly compelling candidate gene to test for association with specific language impairment. In addition to our identification of it as a direct neural target of FOXP2, it has known neuronal functions,31,32 and its expression is enriched in human language-related circuitry.34 Furthermore, the gene is disrupted in a family with Tourette’s syndrome,36 and a rare point mutation causes a severe recessive disorder involving cortical dysplasia focal epilepsy, associated with language regression and autistic characteristics.33 Recent independent studies have implicated variants at the CNTNAP2 locus in autistic-spectrum disorders,37-39 with one study showing association with a measure of language delay (the age at the first spoken word) in multiplex autism families.37
We therefore went on to assess CNTNAP2 involvement in specific language impairment by genotyping polymorphisms across the locus in the large series of consortium families and testing for marker-trait association, using a family-based association method. Using an approach that was consistent with previous studies of this series,21,22 we analyzed quantitative phenotypes from probands and all available siblings, regardless of the diagnosis of specific language impairment, and focused on composite diagnostic measures of expressive and receptive language abilities, as well as a test of nonsense-word repetition, which was previously established as a robust endophenotype.12
We observed significant associations (with P values from 0.01 to 5.0×10-5) between nonsense-word repetition and nine intronic SNPs (rs851715, rs10246256, rs2710102, rs759178, rs1922892, rs2538991, rs17236239, rs2538976, and rs2710117), all mapping between exons 13 and 15. The most strongly associated SNP was rs17236239 (P = 5.0×10-5) (Fig. 3A, and Table S3 in the Supplementary Appendix). Even after an overly conservative Bonferroni correction for testing of multiple SNPs was made, this P value remained significant (P = 0.002). The rs17236239 SNP was also the marker showing strongest evidence of association with expressive language abilities (P = 0.008). The exon 13-15 region was similarly implicated in analyses of receptive language abilities, but in this case the strongest association was observed for a different SNP, rs4431523 (P = 0.003).
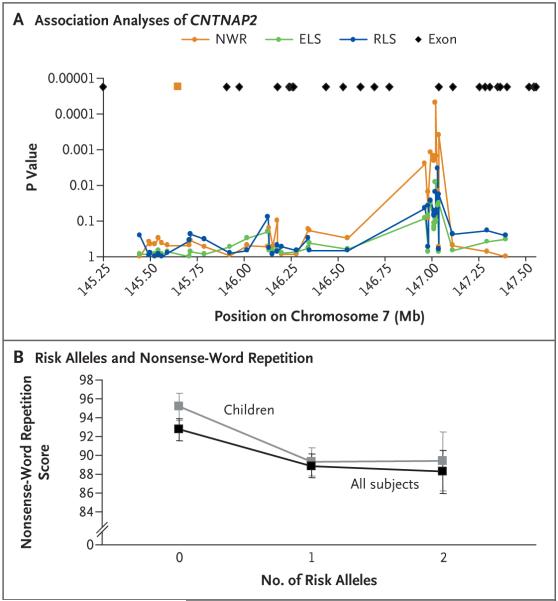
Figure 3. Association between CNTNAP2 and Language Deficits in Families with Specific Language Impairment.
Panel A shows the results of analyses with a quantitative transmission disequilibrium test (QTDT) of SNPs from the CNTNAP2 locus and their association with measures of an ability to repeat nonsense words (orange), to express language (green), and to understand language (blue). The circles show the positions of individual SNPs, and the black diamonds at the top of the graph indicate the relative positions of exons, according to the National Center for Biotechnology Information genetic sequence (build 35). The orange square denotes the position of the FOXP2-bound fragment from the shotgun cloning of gene fragments isolated by chromatin-immunoprecipitation screening. (Details regarding SNP locations, allele frequencies, QTDT results, and effect sizes are available in Table S3 in the Supplementary Appendix.) Panel B shows the effect of the multimarker haplotype ht1 on scores evaluating the ability to repeat nonsense words on the Children’s Test of Nonword Repetition.26 Scores on the scale range from 46 to 141, with a mean of 100 and a standard deviation of 15 in the general population. Lower scores indicate poorer performance. Children (gray) were divided into three groups on the basis of the numbers of copies of the putative risk allele ht1 that they carried. The mean score on nonsense-word repetition dropped by about 6 points as a consequence of carrying one or more copies of ht1. Similar results were seen with a larger sample that also included parents with available phenotypic data (black). The I bars represent standard errors. QTDT analyses of the multimarker haplotypes yielded a P value of 6.0×10-4 for the association between ht1 and measures of nonsense-word repetition (Table S4 in the Supplementary Appendix).
We constructed multimarker haplotypes with the 9 SNPs implicated in the ability to repeat nonsense words and observed 11 different combinations. Four haplotypes represented 94% of subjects (Table S4 in the Supplementary Appendix). The most common haplotype, ht1, negatively influenced the ability to repeat nonsense words; it was more often present in family members with poor scores (a frequency of 40% among those with scores of >2 SD below the population mean) than in those with good scores (a frequency of 29% among those with scores of >0.5 SD above the population mean). We classified probands and siblings according to the number of copies (none, one, or two) of this putative risk haplotype they had and calculated the mean score for nonsense-word repetition for each group (Fig. 3B). There was a decrease of approximately 6 points (0.4 SD) between the mean scores for nonsense-word repetition of children carrying no copies of ht1 (mean, 95.2) and those carrying two copies (mean, 89.4). We observed a difference of similar magnitude in scores between children carrying no copies of ht1 and those carrying one copy of ht1 (mean, 89.7), a finding that suggests a dominant effect. Although only 49 children carried two copies of ht1, as compared with 176 with no copy and 174 with one copy, we observed very similar results in an augmented data set incorporating all available parental scores (Fig. 3B). Moreover, family-based quantitative association analyses of the nine-marker haplotypes and scores on nonsense-word repetition yielded a P value of 6.0×10-4 for ht1, again indicating that this haplotype is significantly associated with impaired language performance (Table S4 in the Supplementary Appendix).
Investigations of CNTNAP2 in patients with autism showed an increased association in families with affected males37 and also suggested the possibility of imprinting.38 We therefore repeated our QTDT analysis of the haplotype associated with impaired nonsense-word repetition in the families with specific language impairment, using sex as a covariate and testing for differences in transmission of paternal and maternal alleles. We found no evidence of a role of sex (P = 6.0×10-4 with no adjustment for sex, P =8.0×10-4 with adjustment for sex) or of imprinting (P = 0.27) at this locus.
DISCUSSION
We have shown that FOXP2, which is mutant in people with a rare speech and language disorder, directly regulates expression of the CNTNAP2 gene. We went on to demonstrate that variants of CNTNAP2 are associated with deficits in common forms of language impairment. In so doing, we provide an example of how knowledge of the genetic cause of a rare single-gene disorder provides an entry point into the causes of a more complex phenotype. Further analyses of the relevant regulatory networks — including the FOXP2-CNTNAP2 pathway identified here — may lead to a better understanding of neurogenetic mechanisms involved in typical language disorders.
The FOXP2-bound fragment of CNTNAP2 lies outside the classically defined regulatory regions of the genome represented on available promoter-based microarrays. It therefore escaped detection in recently published efforts in which chromatin immunoprecipitation with FOXP2 antibodies was coupled to screening of such microarrays.18,40 Indeed, large-scale surveys of transcription-factor binding have indicated that functional regulatory sequences often lie far from known promoters, with many of such sequences mapping within introns.41
Thus far, CNTNAP2 is the only FOXP2 target that we have tested for association in specific language impairment. Of all FOXP2 targets identified to date,18,40 we prioritized CNTNAP2 for association testing in common language disorders because it represented the most compelling of candidates, with converging support from multiple lines of independent investigation. That these first association analyses were positive illustrates the promise of our function-based approach. In future work, we will go on to assess other neural targets of FOXP2 in a similar manner.
There is considerable debate over the existence of shared causes of neurodevelopmental syndromes that are treated as diagnostically distinct, such as autism and specific language impairment.2,42,43 In this study, we observed an association between the endophenotype of nonsense-word repetition and polymorphisms in the exon 13-15 region of CNTNAP2 in children with specific language impairment. A study of the gene in children with autism37 showed an association between polymorphisms in the exon 13-15 region (similarly centered on tagging-SNP rs2710102) and the age at the first spoken word. The same SNP alleles were associated with susceptibility in both studies (Table S3 in the Supplementary Appendix). Therefore, similar CNTNAP2 variants may represent susceptibility factors for language-related deficits in both specific language impairment and autism.
The CNTNAP2 associations we describe here for specific language impairment are not simply a replication of those reported previously for patients with autistic-spectrum disorders. We made a rigorous effort to exclude persons with autistic-spectrum disorders from our analysis. Although diagnostic boundaries are not always clear, it is unlikely that persons who received a misdiagnosis of autistic-spectrum disorder remained in such numbers that they accounted for the strength of the association that we observed. Moreover, we used a quantitatively defined endophenotype previously proposed to underlie typical specific language impairment, rather than a categorical designation of affected status.
Instead, our findings are compatible with the idea that different components of autistic-spectrum disorders (communication deficits, impaired social interaction, and rigid or repetitive behaviors) may be under different genetic influences.44 In this view, language impairments are observed in relatively pure form in specific language impairment, but when they occur in association with other social and behavioral deficits, such impairments can result in a diagnosis of autism.45 Thus, altered CNTNAP2 function or regulation could represent a shared mechanism contributing to language-related endophenotypes in both specific language impairment and autism. These findings illustrate the value of using endophenotypes for the genetic dissection of such disorders.
In conclusion, by integrating functional genomics and quantitative trait analyses, we have identified a shared neurogenetic pathway that is disturbed in distinct forms of language impairment. This work represents a move away from isolated studies of individual genes and toward an understanding of molecular networks that may go awry in neurodevelopmental disorders affecting language.
Supplementary Material
Acknowledgments
Supported by project and program grants from the Wellcome Trust (to Drs. Fisher and Monaco, respectively), a pilot grant from Autism Speaks (to Dr. Fisher), awards (MH75028-R21, MH081754-RO1, MH60233-R37, and HD055784-P50) from the National Institute of Mental Health (to Dr. Geschwind), a Christopher Welch Biological Sciences Scholarship from the University of Oxford (to Dr. Vernes), a postdoctoral fellowship from the Tourette Syndrome Association (to Dr. Abrahams), and a Marie Curie Intra-European Fellowship (to Dr. Nicod). Dr. Monaco is a Wellcome Trust Principal Research Fellow, and Dr. Fisher is a Royal Society Research Fellow. Drs. Newbury, Monaco, and Fisher and Ms. Winchester are members of the Specific Language Impairment Consortium.
We thank all the families who participated in this study through the Specific Language Impairment Consortium and other consortium members for their contributions to this work: L. Addis, Wellcome Trust Centre for Human Genetics, University of Oxford; I.M. Goodyer, Cambridge Language and Speech Project; E. Simonoff and P.F. Bolton, Child and Adolescent Psychiatry Department and Medical Research Council Centre for Social, Developmental and Genetic Psychiatry, Institute of Psychiatry; V. Slonims and G. Baird, Newcomen Centre, Guy’s Hospital; A. Everitt, E. Hennessy, M. Main, D. Shaw, and P.J. Helms, Department of Child Health, University of Aberdeen; A.D. Kindley, Raeden Centre and Grampian University Hospitals Trust; A. Clark and J. Watson, Speech and Hearing Sciences, Queen Margaret University; A. O’Hare, Department of Reproductive and Developmental Sciences, University of Edinburgh; J. Seckl, Molecular Medicine Centre, University of Edinburgh; H. Cowie, Department of Speech and Language Therapy, Royal Hospital for Sick Children, Edinburgh; W. Cohen, Department of Educational and Professional Studies, University of Strathclyde; J. Nasir, Clinical Developmental Sciences, St. George’s University of London; D.V.M. Bishop, Department of Experimental Psychology, University of Oxford; Z. Simkin and G. Conti-Ramsden, Human Communication and Deafness, School of Psychological Sciences, University of Manchester; and M. Falcaro and A. Pickles, Biostatistics Group, School of Epidemiology and Health Science, University of Manchester; as well as D.V.M. Bishop, G. Baird, G. Conti-Ramsden, and A. O’Hare for their assistance with the preparation of the manuscript.
Footnotes
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Harel S, Greenstein Y, Kramer U, et al. Clinical characteristics of children referred to a child development center for evaluation of speech, language, and communication disorders. Pediatr Neurol. 1996;15:305–11. doi: 10.1016/s0887-8994(96)00222-6. [DOI] [PubMed] [Google Scholar]
- 2.Conti-Ramsden G, Simkin Z, Botting N. The prevalence of autistic spectrum disorders in adolescents with a history of specific language impairment (SLI) J Child Psychol Psychiatry. 2006;47:621–8. doi: 10.1111/j.1469-7610.2005.01584.x. [DOI] [PubMed] [Google Scholar]
- 3.Cohen NJ. Language impairment and psychopathology in infants, children, and adolescents. Sage; Thousand Oaks, CA: 2001. [Google Scholar]
- 4.Fisher SE, Lai CS, Monaco AP. Deciphering the genetic basis of speech and language disorders. Annu Rev Neurosci. 2003;26:57–80. doi: 10.1146/annurev.neuro.26.041002.131144. [DOI] [PubMed] [Google Scholar]
- 5.Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–23. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 6.MacDermot KD, Bonora E, Sykes N, et al. Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am J Hum Genet. 2005;76:1074–80. doi: 10.1086/430841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feuk L, Kalervo A, Lipsanen-Nyman M, et al. Absence of a paternally inherited FOXP2 gene in developmental verbal dyspraxia. Am J Hum Genet. 2006;79:965–72. doi: 10.1086/508902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shriberg LD, Ballard KJ, Tomblin JB, Duffy JR, Odell KH, Williams CA. Speech, prosody, and voice characteristics of a mother and daughter with a 7;13 translocation affecting FOXP2. J Speech Lang Hear Res. 2006;49:500–25. doi: 10.1044/1092-4388(2006/038). [DOI] [PubMed] [Google Scholar]
- 9.Zeesman S, Nowaczyk MJ, Teshima I, et al. Speech and language impairment and oromotor dyspraxia due to deletion of 7q31 that involves FOXP2. Am J Med Genet A. 2006;140:509–14. doi: 10.1002/ajmg.a.31110. [DOI] [PubMed] [Google Scholar]
- 10.Watkins KE, Dronkers NF, Vargha-Khadem F. Behavioural analysis of an inherited speech and language disorder: comparison with acquired aphasia. Brain. 2002;125:452–64. doi: 10.1093/brain/awf058. [DOI] [PubMed] [Google Scholar]
- 11.Tomblin JB, Records NL, Buckwalter P, Zhang X, Smith E, O’Brien M. Prevalence of specific language impairment in kindergarten children. J Speech Lang Hear Res. 1997;40:1245–60. doi: 10.1044/jslhr.4006.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newbury DF, Bishop DV, Monaco AP. Genetic influences on language impairment and phonological short-term memory. Trends Cogn Sci. 2005;9:528–34. doi: 10.1016/j.tics.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Shriberg LD, Tomblin JB, McSweeny JL. Prevalence of speech delay in 6-year-old children and comorbidity with language impairment. J Speech Lang Hear Res. 1999;42:1461–81. doi: 10.1044/jslhr.4206.1461. [DOI] [PubMed] [Google Scholar]
- 14.Newbury DF, Bonora E, Lamb JA, et al. FOXP2 is not a major susceptibility gene for autism or specific language impairment. Am J Hum Genet. 2002;70:1318–27. doi: 10.1086/339931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Brien EK, Zhang X, Nishimura C, Tomblin JB, Murray JC. Association of specific language impairment (SLI) to the region of 7q31. Am J Hum Genet. 2003;72:1536–43. doi: 10.1086/375403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai CS, Gerrelli D, Monaco AP, Fisher SE, Copp AJ. FOXP2 expression during brain development coincides with adult sites of pathology in a severe speech and language disorder. Brain. 2003;126:2455–62. doi: 10.1093/brain/awg247. [DOI] [PubMed] [Google Scholar]
- 17.Vernes SC, Nicod J, Elahi FM, et al. Functional genetic analysis of mutations implicated in a human speech and language disorder. Hum Mol Genet. 2006;15:3154–67. doi: 10.1093/hmg/ddl392. [DOI] [PubMed] [Google Scholar]
- 18.Vernes SC, Spiteri E, Nicod J, et al. High-throughput analysis of promoter occupancy reveals direct neural targets of FOXP2, a gene mutated in speech and language disorders. Am J Hum Genet. 2007;81:1232–50. doi: 10.1086/522238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abu-Khalil A, Fu L, Grove EA, Zecevic N, Geschwind DH. Wnt genes define distinct boundaries in the developing human brain: implications for human forebrain patterning. J Comp Neurol. 2004;474:276–88. doi: 10.1002/cne.20112. [DOI] [PubMed] [Google Scholar]
- 20.The SLI Consortium A genomewide scan identifies two novel loci involved in specific language impairment. Am J Hum Genet. 2002;70:384–98. doi: 10.1086/338649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Idem Highly significant linkage to the SLI1 locus in an expanded sample of individuals affected by specific language impairment. Am J Hum Genet. 2004;74:1225–38. doi: 10.1086/421529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burden V, Stott CM, Forge J, Goodyer I, The Cambridge Language and Speech Project (CLASP) I. Detection of language difficulties at 36 to 39 months. Dev Med Child Neurol. 1996;38:613–31. doi: 10.1111/j.1469-8749.1996.tb12126.x. [DOI] [PubMed] [Google Scholar]
- 23.Clark A, O’Hare A, Watson J, et al. Severe receptive language disorder in childhood — familial aspects and long-term outcomes: results from a Scottish study. Arch Dis Child. 2007;92:614–9. doi: 10.1136/adc.2006.101758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conti-Ramsden G, Botting N. Characteristics of children attending language units in England: a national study of 7-year-olds. Int J Lang Commun Disord. 1999;34:359–66. doi: 10.1080/136828299247333. [DOI] [PubMed] [Google Scholar]
- 25.Semel EM, Wiig EH, Secord W. Clinical evaluation of language fundamentals — revised. Psychological Corporation; San Antonio, TX: 1992. [Google Scholar]
- 26.Gathercole SE, Willis CS, Baddeley AD, Emslie H. The Children’s Test of Nonword Repetition: a test of phonological working memory. Memory. 1994;2:103–27. doi: 10.1080/09658219408258940. [DOI] [PubMed] [Google Scholar]
- 27.Bishop DV, North T, Donlan C. Nonword repetition as a behavioural marker for inherited language impairment: evidence from a twin study. J Child Psychol Psychiatry. 1996;37:391–403. doi: 10.1111/j.1469-7610.1996.tb01420.x. [DOI] [PubMed] [Google Scholar]
- 28.Gathercole SE. Nonword repetition and word learning: the nature of the relationship. App Psycholinguist. 2006;27:513–43. [Google Scholar]
- 29.Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–92. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin — rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 31.Poliak S, Salomon D, Elhanany H, et al. Juxtaparanodal clustering of Shaker-like K+ channels in myelinated axons depends on Caspr2 and TAG-1. J Cell Biol. 2003;162:1149–60. doi: 10.1083/jcb.200305018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasband MN. It’s “juxta” potassium channel! J Neurosci Res. 2004;76:749–57. doi: 10.1002/jnr.20073. [DOI] [PubMed] [Google Scholar]
- 33.Strauss KA, Puffenberger EG, Huentelman MJ, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354:1370–7. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 34.Abrahams BS, Tentler D, Perederiy JV, Oldham MC, Coppola G, Geschwind DH. Genome-wide analyses of human perisylvian cerebral cortical patterning. Proc Natl Acad Sci U S A. 2007;104:17849–54. doi: 10.1073/pnas.0706128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teramitsu I, Kudo LC, London SE, Geschwind DH, White SA. Parallel FoxP1 and FoxP2 expression in songbird and human brain predicts functional interaction. J Neurosci. 2004;24:3152–63. doi: 10.1523/JNEUROSCI.5589-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verkerk AJ, Mathews CA, Joosse M, Eussen BH, Heutink P, Oostra BA. CNTNAP2 is disrupted in a family with Gilles de la Tourette syndrome and obsessive compulsive disorder. Genomics. 2003;82:1–9. doi: 10.1016/s0888-7543(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 37.Alarcón M, Abrahams BS, Stone JL, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82:150–9. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arking DE, Cutler DJ, Brune CW, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82:160–4. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bakkaloglu B, O’Roak BJ, Louvi A, et al. Molecular cytogenetic analysis and re-sequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82:165–73. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spiteri E, Konopka G, Coppola G, et al. Identification of the transcriptional targets of FOXP2, a gene linked to speech and language, in developing human brain. Am J Hum Genet. 2007;81:1144–57. doi: 10.1086/522237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cawley S, Bekiranov S, Ng HH, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 42.De Fossé L, Hodge SM, Makris N, et al. Language-association cortex asymmetry in autism and specific language impairment. Ann Neurol. 2004;56:757–66. doi: 10.1002/ana.20275. [DOI] [PubMed] [Google Scholar]
- 43.Whitehouse AJ, Barry JG, Bishop DV. The broader language phenotype of autism: a comparison with specific language impairment. J Child Psychol Psychiatry. 2007;48:822–30. doi: 10.1111/j.1469-7610.2007.01765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Happé F, Ronald A, Plomin R. Time to give up on a single explanation for autism. Nat Neurosci. 2006;9:1218–20. doi: 10.1038/nn1770. [DOI] [PubMed] [Google Scholar]
- 45.Bishop DV. Autism and specific language impairment: categorical distinction or continuum? Novartis Found Symp. 2003;251:213–26. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.