Abstract
Epilepsy and mental retardation limited to females (EFMR) is a disorder with an X-linked mode of inheritance and an unusual expression pattern. Disorders arising from mutations on the X chromosome are typically characterized by affected males and unaffected carrier females. In contrast, EFMR spares transmitting males and affects only carrier females. Aided by systematic resequencing of 737 X chromosome genes, we identified different protocadherin 19 (PCDH19) gene mutations in seven families with EFMR. Five mutations resulted in the introduction of a premature termination codon. Study of two of these demonstrated nonsense-mediated decay of PCDH19 mRNA. The two missense mutations were predicted to affect adhesiveness of PCDH19 through impaired calcium binding. PCDH19 is expressed in developing brains of human and mouse and is the first member of the cadherin superfamily to be directly implicated in epilepsy or mental retardation.
EFMR (MIM 300088) is a condition characterized by seizure onset in infancy or early childhood (6-36 months) and cognitive impairment. The disorder is sex-limited, with the phenotype being restricted to females. Males are apparently spared, with normal cognitive function and absence of seizures. Evidence for the trait being due to a gene on the X chromosome derives from the pattern of inheritance and from genetic linkage to Xq22 (ref. 1). EFMR is recognizable in multi-generational pedigrees by the unique sex-limited expression pattern, wherein affected females are connected through unaffected transmitting males1-3.
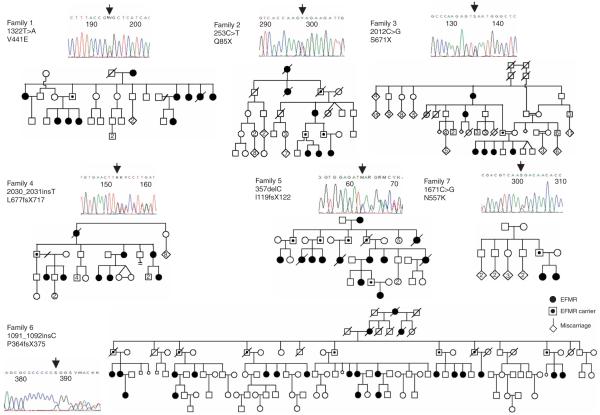
We ascertained five new families carrying EFMR on the basis of their inheritance pattern, electroclinical features and gene localization to Xq22 (ref. 4 and S.M., unpublished data). To identify the underlying genetic defect, we resequenced 737 genes from the Vertebrate Genome Annotation (VEGA) database in probands from three families. We identified a single nonsense nucleotide change 2012C>G (S671X) in the PCDH19 gene in family 3 (Fig. 1). Families 1 and 2 initially did not show any changes.
Figure 1.
Mutations in PCDH19 cause EFMR. Pedigrees of the seven EFMR families showing the characteristic inheritance pattern of affected females and transmitting males. Each of the X chromosome-encoded PCDH19 mutations segregated with the EFMR clinical phenotype. An example of a sequence chromatogram of a PCDH19 mutation as detected in an affected female is shown for each family.
Subsequent comparative sequence analysis of the annotated PCDH19 ORF revealed that it was incomplete, consistent with a previous report5. We annotated the complete 3,447-bp ORF of PCDH19, which consists of six exons. The full-length, processed PCDH19 mRNA is 9,765 nt long, with exon 2 alternatively spliced (Supplementary Fig. 1 online). Sequencing of the entire PCDH19 ORF identified a missense change 1322T>A (V441E) in family 1, a nonsense change 253C>T (Q85X) in family 2, a frameshift change 2030_2031 insT (L677fsX717) in family 4 and a frameshift change 357delC (I119fsX122) in family 5. Finally, we identified a frameshift change 1091_1092insC (P364fsX375) in the original EFMR-bearing family (family 6, ref. 1) (Fig. 1). In summary, we identified PCDH19 nucleotide changes in all six families bearing EFMR that we studied. Another unique nucleotide change (1671C>G, N557K) was identified by screening a cohort of 87 females that included both isolated and familial cases with epilepsy and varying degrees of mental impairment. We found that the nucleotide change in this small family, with two affected girls with epilepsy and autism spectrum disorder, arose de novo in their carrier father on the grandmaternal X chromosome (Fig. 1, family 7).
All seven nucleotide changes (Fig. 2a) segregated with the clinical phenotype in the respective families (Fig. 1) and were not present in 250 male probands from families with putative X-linked mental retardation or in 750 control (350 male and 400 female) X chromosomes. Further, we did not detect any other potentially deleterious nucleotide changes in probands from the three families in any of the other 736 X-chromosome genes sequenced (data not shown). Taken together, the predicted loss of function of all seven PCDH19 changes (see below), the high degree of conservation of the residues affected by the two missense changes (Fig. 2a,b) and our PCDH19 mRNA studies (see below) strongly support PCDH19 as the gene whose mutation causes EFMR. Moreover, PCDH19 is located at Xq22 (Ensembl database) within the original linkage region1. EFMR locus homogeneity in these families has now been molecularly defined, and the combined lod score with markers in the Xq22 region reached 11.9 (refs. 1,4 and S.M., unpublished data). Among the seven families bearing EFMR studied, there were two (of 68) carrier females who had been classified (at the time of the original assessment) as unaffected. Therefore, the estimated penetrance of the known EFMR mutations in females was >90%.
Figure 2.
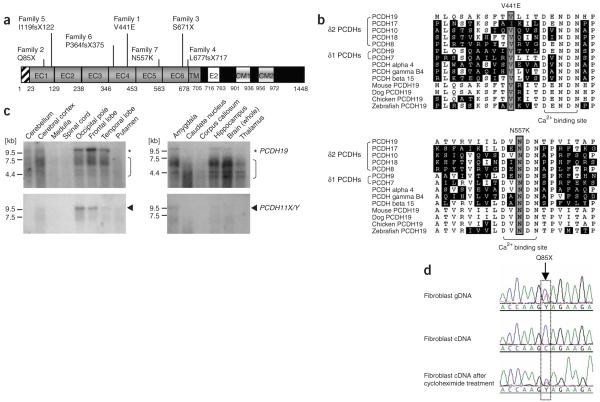
Structure and expression analysis of PCDH19. (a) Schematic diagram of the PCDH19 protein showing the signal peptide, extracellular cadherin (EC), transmembrane (TM) and cytoplasmic (CM) domains. The positions of the mutations found in the families bearing EFMR are shown. (b) Partial alignment of human PCDHs and orthologs of PCDH19 from other species, showing the high conservation of the residues affected by the two missense mutations, V441E (top) and N557K (bottom). Asn557 is invariant and one of the essential residues for calcium ion binding13,14. Val441 is highly conserved and in close proximity to the calcium-binding acidic residues (bracket). (c) RNA blot analyses of PCDH19 and PCDH11X/Y in various human tissues. Asterisks, the ~9.8 kb PCDH19 transcripts; arrowheads, the smaller, ~9.5 kb PCDH11X/Y mRNAs. Brackets, either nonspecific binding of the PCDH19 probe or PCDH19 degradation products, as there is no evidence for such alternative isoforms of PCDH19 available from our data (data not shown) or from published data. (d) Nonsense-mediated RNA decay of mutant PCDH19 transcripts. Sequence chromatogram from EFMR-affected female from family 2 showing the detection of the mutation 253C>T in genomic DNA (gDNA) (top), the absence of the mutant sequence in fibroblast cDNA (middle) and the presence of the both mutant and wild-type cDNA after the treatment of fibroblasts with cycloheximide (bottom), which inhibits the pioneer round of translation and thus NMD. The position of the mutation is boxed.
PCDH19 was expressed predominantly in neural tissues and at different developmental stages. By RNA blot analysis using a PCDH19-specific probe, we detected a transcript size of approximately 9.8 kb from various areas of the male and female adult human brain (Fig. 2c and below). We also identified PCDH19 mRNA expression in primary skin fibroblasts. This allowed us to examine the consequences of the PCDH19 mutations 253C>T, Q85X (family 2) and 2012C>G, S671X (family 3) on the stability of their respective mRNAs. Both mutations introduce a premature termination codon (PTC) into the PCDH19 mRNA. Such PTC-containing mRNAs are usually recognized by the nonsense-mediated decay (NMD) surveillance complexes and efficiently degraded6. We found that the PTC mutations in family 2 (Fig. 2d) and family 3 (data not shown) led to PCDH19 mRNA degradation by NMD. Inhibition of translation by cycloheximide treatment of skin fibroblast cells preserved the PTC mutation-containing mRNA (Fig. 2d). The absence of mutant PCDH19 mRNAs could not be attributed to skewing of X-inactivation. We confirmed random X-inactivation in DNA isolated from blood and skin fibroblast cultures of each affected female available (data not shown), in agreement with previous data1.
PCDH19 encodes a 1,148-amino acid protein belonging to the protocadherin δ2 subclass of the cadherin superfamily of cell-cell adhesion molecules. PCDH19 contains a signal sequence, six extracellular cadherin (EC) repeats, a transmembrane domain and a cytoplasmic region with conserved CM1 and CM2 domains (Fig. 2a). The biological role of the PCDH19 protein is not known. However, members of the protocadherin family are predominantly expressed in the nervous system7,8 and are postulated to be involved in the establishment of neuronal connections and in signal transduction at the synaptic membrane9,10. PCDH10, also a δ2 protocadherin, is required for growth of striatal axons and thalamocortical projections11. Usher syndrome type 1, involving combined hearing loss and blindness, was the first human disorder to be associated with mutations in a PCDH gene (PCDH15)12. Mutations in PCDH19 have now been associated with epilepsy and mental retardation.
All mutations identified in this study were located in the large PCDH19 extracellular domain (Fig. 2a) containing the cadherin repeats, which facilitate cell-cell interactions. We predict five of these mutations to be complete loss-of-function mutations as a consequence of NMD degradation of their respective PTC-containing mRNAs (Fig. 2d and data not shown). Given the similarity among the clinical phenotypes associated with all seven PCDH19 mutations, it is reasonable to suggest that the remaining two mutations, missense mutations leading to V441E and N557K substitutions, also lead to loss of PCDH19 function. The N557K mutation affects an invariant asparagine residue within the EC5 domain (Fig. 2a,b). The equivalent asparagine residue of EC1 of classical cadherins (for example, Asn100 of N-cadherin13) and protocadherins (for example, Asn101 of Pcdhα14) is essential for calcium binding and thus for the adhesive function of the EC1 domain13,14. The valine residue at position 441 (in EC4, or the equivalent of Val96 in EC1 of N-cadherin13 or Val97 in EC1 of Pcdhα14) is also highly conserved (Fig. 2b) and in close proximity to the calcium-binding site13,14. We speculate that both PCDH19 missense variants adversely affect PCDH19 adhesive function through impaired calcium binding.
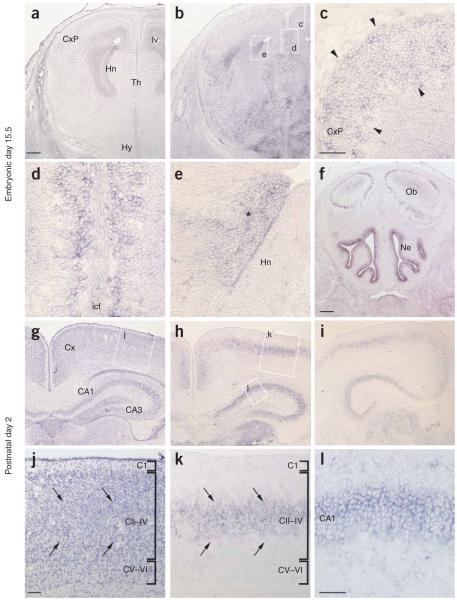
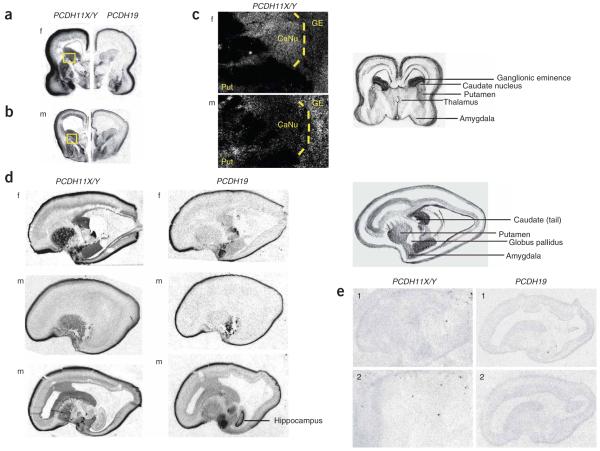
To investigate the expression of PCDH19 in the developing mouse and human central nervous system (CNS), we performed in situ hybridization analysis. In mice, Pcdh19 had widespread expression in both the embryonic and adult brain, including the developing cortex and hippocampus (Fig. 3 and Supplementary Note online), which is consistent with our finding that mutations of this gene in humans are associated with cognitive impairment. In human tissue, in addition to PCDH19, we also investigated the expression pattern of PCDH11X and PCDH11Y, two paralogous protocadherin genes, whose expression and function we speculate to be relevant to EFMR (see Fig. 4 and below). Each of the three PCDH genes was expressed in developing cortical plate, amygdala and subcortical regions and in the ganglionic eminence (Fig. 4). However, PCDH11X/Y expression showed a marked sexual dimorphism in the caudate nucleus: virtually absent expression in the male and high expression in the female (Fig. 4a-c). Additionally, closer inspection of the amygdala showed that PCDH19 was expressed in lateral nuclei, whereas PCDH11X/Y was more medial.
Figure 3.
Expression of Pcdh19 in the developing mouse CNS. (a-l) Expression at embryonic day 15.5 (a-f) and postnatal day 2 (g-l) (representative of two males and two females studied). (a,b) Adjacent coronal sections through the hippocampal region stained with hematoxylin and eosin or processed for Pcdh19 in situ, respectively. (c-e) Higher magnification images of the boxed regions in b. Arrowheads in c, Pcdh19-expressing cells within the cortex; asterisk in e, dorsolateral wall of the lateral ventricle. (f) Coronal section through the olfactory bulb highlighting Pchd19 expression in the nasal epithelium. (g,h) Adjacent coronal sections through the mid-hippocampal region stained with hematoxylin and eosin or processed for Pcdh19 in situ, respectively. (i) A brain section more posterior than that in h, highlighting Pcdh19 expression. (j-l) Higher-magnification images of the regions boxed in g and h, as indicated. Arrows in j,k, Pcdh19 expression within cortical layers II-IV. Cx, cortex; CxP, cortical plate; Hn, hippocampal neuroepithelium; lv, lateral ventricle; Th, thalamus; Hy, hypothalamus; icf, intercerebral fissure; Ob, olfactory bulbs; Ne, nasal epithelium. Scale bars in a,b,f-i, 200 μM; in c-e,j-l, 50 μM.
Figure 4.
Expression of PCDH11X/Y and PCDH19 in midgestation developing human CNS. (a,b) Autoradiography in situ hybridization of female (f; panel a) and male (m; panel b) coronal sections through the basal regions of the brain using probes for PCDH11X/Y and PCDH19, showing several areas of overlap and differences, including high expression of PCDH11X/Y in the ganglionic eminence and sexually dimorphic expression in the caudate nucleus. Dark color denotes high expression. Both genes are highly expressed in amygdala and developing cortical plate. A cartoon key to these sections is shown at the far right and the boxed regions are shown magnified in c. (c) Micrographs from photographic emulsions of magnified regions of PCDH11X/Y expression show low expression in male caudate. White denotes strong expression. GE, ganglionic eminence; Put, putamen; CaNu, caudate nucleus. (d) Sagittal photomicrographs highlight areas of differential expression between the two PCDH genes, indicating high putamen expression for PCDH11X/Y and high hippocampal expression for PCDH19. We show sections from two different planes in the male, the first more lateral and the second more medial, to show that the absence of male expression is not an artifact of section plane. (e) Sections hybridized with sense probes show absence of specific staining.
Typically, in most X-linked dominant conditions, males are more severely affected and often die prenatally. In EFMR, the transmitting males are apparently normal (that is, without epilepsy or intellectual disability), but with some obsessive traits reported anecdotally4. Previous hypotheses to explain the EFMR expression pattern1 considered either a dominant negative effect of the mutant protein in carrier females (for example, similar to that of the C-terminally truncated P-cadherin in malignant melanoma15) or the presence of a compensatory or rescue factor in males. The dominant negative effect of mutant PCDH19 is unlikely, based on our NMD results (see above), and PCDH19 does not have a Y-chromosome paralog to provide male rescue. Sexually dimorphic expression and function16,17 of PCDH19 and/or relevant compensatory genes is also a possibility.
We propose an alternative hypothesis, based on the identity of the EFMR gene, that invokes functional rescue by a related, but nonparalogous, protocadherin gene in males. The PCDH19 gene is subject to X inactivation (Supplementary Figs. 2 and 3 online). Therefore, hemizygous transmitting males will have a homogeneous population of PCDH19-negative cells, whereas affected females are likely to be mosaics comprising PCDH19-negative and PCDH19-wild type cells. Such tissue mosaicism may scramble cell-cell communication, which manifests clinically as EFMR. A similar mechanism was proposed for the craniofrontonasal syndrome CFNS (MIM 304110)18. We hypothesize that the absence of PCDH19 function in males may potentially be compensated for by the related but nonparalogous protocadherin gene PCDH11Y, a Y-chromosome gene that is expressed in human brain19,20. PCDH11Y has an X chromosome paralog, PCDH11X, that has strong sequence similarity19. However, the differences in brain expression patterns between these two genes may account for differential ability to compensate for absence of PCDH19. The potential role of PCDH11Y (and that of its paralog PCDH11X) in the EFMR phenotype will need to be addressed in the future by engineering the appropriate animal models.
Cadherins are a large family of genes with crucial functions in human brain21. Our data directly implicate the protocadherin gene family in epilepsy and intellectual disability. On the basis of our observations, molecular diagnosis will result in wider recognition of EFMR, especially in smaller families and single cases, with important consequences for counseling.
METHODS
Subjects
The clinical details of families 1-4 have been reported4. The proband in family 5 (IV.2) experienced multiple seizures in the first year of life and has severe learning disabilities, no speech, and behavioral disturbances (S.M., unpublished data). The clinical details of family 6 have been published1-3. Family 7 was ascertained on the basis of one sister having infantile seizures and Asperger’s syndrome and the other having epilepsy and mild intellectual disability (I.E.S. and S.F.B., unpublished data).
RNA blotting
Human brain MTN blots II and V (Clontech) were hybridized per the manufacturer’s instructions with a probe containing nucleotides 2884-3257 of the human PCDH19 ORF. The 374-bp PCR product (see Supplementary Table 1 online for PCR primer sequences) was labeled with radioactive [32P]dCTP (Perkin Elmer) using the Megaprime DNA labeling system (GE Healthcare).
RT-PCR analyses
Total RNA was extracted from fibroblast cells with the RNeasy Mini kit (Qiagen) and treated with DNase I (Qiagen). About 2 μg of RNA was primed with 1 μg of random hexanucleotides and subjected to reverse transcription for 90 min at 42 °C using Superscript II (Invitrogen). The efficiency of the reaction was tested by PCR using primers specific to the ubiquitously expressed ESD gene. cDNAs were amplified with Taq DNA polymerase (Roche) and specific single-stranded DNA primers for 35 cycles (denaturation, 94 °C for 30 s; annealing, at specific melting temperature for each pair of primers for 30 s; extension, 72 °C for 30 s). PCR products were separated on an agarose gel stained with 1% ethidium bromide.
Tissue culture of primary skin fibroblast lines
A 3-mm skin biopsy excised from the upper arm was cut finely and transferred to a tissue culture flask. The biopsy was cultured in RPMI-1640, 20% FCS (supplemented with 4 mM l-glutamine, 0.017 mg/ml benzylpenicillin) and grown at 37 °C, with 5% CO2. Once established, fibroblasts were cultured in RPMI-1640, 10% FCS (supplements as above).
Cycloheximide treatment of skin fibroblast cell lines
Primary fibroblast cells were seeded at 1 × 104 per cm2 in RPMI-1640, 10% FCS, and incubated with 50 μg/ml cycloheximide (Sigma) or medium alone for 6 h. Fibroblasts were harvested using a sterile cell scraper (TPP), then washed once in PBS before total RNA extraction and processing to cDNA as described above.
Human in situ hybridization analysis
Human tissue was obtained from the Brain and Tissue Bank for Developmental Disorders at the University of Maryland and was treated as previously described22. Five brains were used, ranging in gestational age from 16 to 20 weeks, with known post mortem intervals from 1 to 2 h. For probe preparation, we performed cDNA synthesis using Superscript III (Invitrogen) from RNA of the inferior frontal gyrus of a 20-gestational-week fetus (RNeasy Mini kit, Qiagen) and amplified two nonoverlapping cDNA transcripts for both PCDH11X/Y and PCDH19 using Platinum Taq Polymerase (Qiagen). Two probes for PCDH11X/Y were designed to recognize all PCDH11X and PCDH11Y isoforms. The PCR primer sequences used for generating the two probes and the PCDH19 probe are provided in Supplementary Table 1. In situ hybridization and image analysis were performed as previously described22. Nonoverlapping probe sets for both genes confirmed the same expression pattern. Sections hybridized with sense probes showed absence of specific staining.
Mouse in situ hybridization analysis
Embryonic day 15.5 embryonic heads and dissected P2 brains were fixed in 4% paraformaldehyde at 4 °C, cryoprotected in 30% sucrose and frozen in OCT embedding medium. In situ hybridization of 16-μM sections was performed as described previously23. The Pcdh19 probe was a digoxigenin-labeled antisense RNA probe prepared as described24. A total of three neonates and two embryos were analyzed and representative sections are shown. No signal was detected using a sense control probe. Images were taken on a Zeiss Axiophot microscope, compiled and minimally processed (adjusted for color and light/dark) using Adobe Photoshop CS.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the families studied for their participation and members of the International Genetics of Learning Disability (IGOLD) study for their collaboration. This work was supported by grants from the Australian National Health and Medical Research Council Program Grant 400121 (I.E.S., S.F.B., J.C.M. and J.G.), Thyne-Reid Charitable Trusts (L.M.D.) and the Wellcome Trust. We also acknowledge support to J.F.G. from US National Institutes of Health grant GM061354 and D.H.G. from US National Institute of Mental Health U.S. grant R01 MH 64547. We are grateful for access to the tissues used in these studies from the Developmental Brain and Tissue Bank at University of Maryland funded by the US National Institutes of Health (National Institute of Child Health and Human Development contracts NO1-HD-4-3368 and NO1-HD-4-3383).
Footnotes
URLs. VEGA, http://vega.sanger.ac.uk/index.html.
Accession codes. GenBank: Incomplete human PCDH19 mRNA, NM_020766; incomplete human PCDH19 protein, NP_065817; complete human PCDH19 mRNA and protein, EF676096; human PCDH11X mRNA, NM_014522, NM_032967, NM_032968 and NM_032969; human PCDH11X protein, NP_055337, NP_116749, NP_116750 and NP_116751; human PCDH11Y mRNA, NM_032971, NM_032972 and NM_032973; human PCDH11Y protein, NP_116753, NP_116754 and NP_116755.
References
- 1.Ryan SG, et al. Epilepsy and mental retardation limited to females: an X-linked dominant disorder with male sparing. Nat. Genet. 1997;17:92–95. doi: 10.1038/ng0997-92. [DOI] [PubMed] [Google Scholar]
- 2.Juberg RC, Hellman CD. A new familial form of convulsive disorder and mental retardation limited to females. J. Pediatr. 1971;79:726–732. doi: 10.1016/s0022-3476(71)80382-7. [DOI] [PubMed] [Google Scholar]
- 3.Fabisiak K, Erickson RP. A familial form of convulsive disorder with or without mental retardation limited to females: extension of a pedigree limits possible genetic mechanisms. Clin. Genet. 1990;38:353–358. doi: 10.1111/j.1399-0004.1990.tb03594.x. [DOI] [PubMed] [Google Scholar]
- 4.Scheffer IE, et al. Epilepsy and mental retardation limited to females: an under-recognised disorder. Brain. 2008;131:918–927. doi: 10.1093/brain/awm338. [DOI] [PubMed] [Google Scholar]
- 5.Wolverton T, Lalande M. Identification and characterization of three members of a novel subclass of protocadherins. Genomics. 2001;76:66–72. doi: 10.1006/geno.2001.6592. [DOI] [PubMed] [Google Scholar]
- 6.Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 7.Vanhalst K, Kools P, Staes K, van Roy F, Redies C. Proto-cadherins: a gene family expressed differentially in the mouse brain. Cell. Mol. Life Sci. 2005;62:1247–1259. doi: 10.1007/s00018-005-5021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim SY, et al. Spatiotemporal expression pattern of non-clustered protocadherin family members in the developing rat brain. Neuroscience. 2007;147:996–1021. doi: 10.1016/j.neuroscience.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 9.Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97:779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- 10.Yagi T, Takeichi M. Cadherin superfamily genes: functions, genomic organization, and neurologic diversity. Genes Dev. 2000;14:1169–1180. [PubMed] [Google Scholar]
- 11.Uemura M, Nakao S, Suzuki ST, Takeichi M, Hirano S. OL-protocadherin is essential for growth of striatal axons and thalamocortical projections. Nat. Neurosci. 2007;10:1151–1159. doi: 10.1038/nn1960. [DOI] [PubMed] [Google Scholar]
- 12.Ahmed ZM, et al. Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am.J.Hum. Genet. 2001;69:25–34. doi: 10.1086/321277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel SD, et al. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell. 2006;124:1255–1268. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 14.Morishita H, et al. Structure of the cadherin-related neuronal receptor/protocadherin-alpha first extracellular cadherin domain reveals diversity across cadherin families. J. Biol. Chem. 2006;281:33650–33663. doi: 10.1074/jbc.M603298200. [DOI] [PubMed] [Google Scholar]
- 15.Bauer R, Bosserhoff AK. Functional implication of truncated P-cadherin expression in malignant melanoma. Exp. Mol. Pathol. 2006;81:224–230. doi: 10.1016/j.yexmp.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Arnold AP. Sex chromosomes and brain gender. Nat. Rev. Neurosci. 2004;5:701–708. doi: 10.1038/nrn1494. [DOI] [PubMed] [Google Scholar]
- 17.Yang X, et al. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wieland I, et al. Mutations of the ephrin-B1 gene cause craniofrontonasal syndrome. Am. J. Hum. Genet. 2004;74:1209–1215. doi: 10.1086/421532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanco P, Sargent CA, Boucher CA, Mitchell M, Affara NA. Conservation of PCDHX in mammals; expression of human X/Y genes predominantly in brain. Mamm. Genome. 2000;11:906–914. doi: 10.1007/s003350010177. [DOI] [PubMed] [Google Scholar]
- 20.Durand CM, et al. Expression and genetic variability of PCDH11Y, a gene specific to Homo sapiens and candidate for susceptibility to psychiatric disorders. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2006;141:67–70. doi: 10.1002/ajmg.b.30229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeichi M. The cadherin superfamily in neuronal connections and interactions. Nat. Rev. Neurosci. 2007;8:11–20. doi: 10.1038/nrn2043. [DOI] [PubMed] [Google Scholar]
- 22.Teramitsu I, Kudo LC, London SE, Geschwind DH, White SA. Parallel Foxp1 and Foxp2 expression in songbird and human brain predicts functional interaction. J. Neurosci. 2004;24:3152–3163. doi: 10.1523/JNEUROSCI.5589-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson LD, et al. Developmentally regulated expression of the regulator of G-protein signaling gene 2 (Rgs2) in the embryonic mouse pituitary. Gene Expr. Patterns. 2005;5:305–311. doi: 10.1016/j.modgep.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Gaitan Y, Bouchard M. Expression of the δ-protocadherin gene Pcdh19 in the developing mouse embryo. Gene Expr. Patterns. 2006;6:893–899. doi: 10.1016/j.modgep.2006.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.