Abstract
Autism is a heterogeneous syndrome defined by impairments in three core domains: social interaction, language and range of interests. Recent work has led to the identification of several autism susceptibility genes and an increased appreciation of the contribution of de novo and inherited copy number variation. Promising strategies are also being applied to identify common genetic risk variants. Systems biology approaches, including array-based expression profiling, are poised to provide additional insights into this group of disorders, in which heterogeneity, both genetic and phenotypic, is emerging as a dominant theme.
Autistic disorder is the most severe end of a group of neurodevelopmental disorders referred to as autism spectrum disorders (ASDs), which share the common feature of dysfunctional reciprocal social interaction. A meta-analysis of ASD prevalence rates suggests that approximately 37 in 10,000 individuals are affected1. ASDs encompass several clinically defined conditions (see BOX 1 and the Diagnostic and Statistical Manual of Mental Disorders), pervasive developmental disorder - not otherwise specified and autistic disorder are the most common, whereas Asperger syndrome appears less frequently. Boys are at increased risk for the ASDs, an effect that becomes even more pronounced in so-called high-functioning cases.
Box 1. Classification and prevalence of ASDs.
Specific impairments in each of three core domains before age three are required for a diagnosis of autistic disorder (13 per 10,000). Within the social domain, impaired use of nonverbal communication (facial expressions or body language) or a reduction in spontaneous attempts to share interests with others are common. Features in the language domain manifest as delayed or absent speech or difficulties initiating or sustaining a conversation. Abnormalities in the restricted and/or repetitive domain can present as abnormal preoccupations, inflexible adherence to routines or rituals, or repetitive motor behaviours. Males are over-represented compared with females (approximately 4:1), an effect that does not seem to be driven by X chromosomal loci. Interestingly, this male-to-female ratio approaches 1:1 when only severe cases of autistic disorder are considered.
Additional terms are useful for describing affected children on the basis of phenotypic presentation, although one should note that diagnosis in the autism spectrum disorders (ASDs) is in many cases complicated by the presence of severe cognitive delay. Individuals with Asperger syndrome (2.6 per 10,000) show impairments in the social and restricted and/or repetitive domains, but most use language in an age-appropriate manner and are not mentally retarded. Males are also over-represented among these cases (approximately 8:1). Individuals with pervasive developmental disorder - not otherwise specified (PDD-NOS; 20.8 per 10,000) show marked impairment in each core domain but do not meet diagnostic criteria for autistic disorder proper. Rett disorder (see main text; TABLE 1; Supplementary information S1 (table)) and childhood disintegrative disorder (normal development until age two with subsequent regression) are less common but are also listed among the ASDs in the current version of the Diagnostic and Statistical Manual of Mental Disorders.
Other important terms worth noting here include the concept of ‘broad-spectrum cases’, a classification typically encompassing a range of presentations, including autistic disorder, Asperger syndrome and PDD-NOS. Likewise, the term idiopathic is used to describe the large number of cases with no known aetiology. Although methodological problems make it difficult to accurately assess how prevalence rates change over time, evidence exists for as much as a twofold increased prevalence in the ASDs in recent years. Most attribute this increase to heightened awareness and the use of broader diagnostic criteria but this does not exclude the involvement of environmental factors in the modulation of ASD risk. Prenatal and perinatal complications are elevated in cases85, and viral exposures — particularly rubella — are thought to elevate risk. It is also recognized that paternal age is increased among the fathers of affected children86, a finding that might be related to elevated rates of de novo copy number variation in the ASDs4. Unpublished estimates for concordance rates for autistic disorder among dizygotic twins may be as high as 25% (J. Hallmayer, personal communication). If this figure is confirmed, it would allow for the involvement of in utero factors while still supporting the high heritability value of 0.7. The contribution of epigenetic modifications have also been championed87,88 but, although they are probably important, the manner and extent of their involvement remains to be defined. As additional genetic ASD risk factors are identified, the way by which these molecules interact with the environment can be addressed formally.
A chronological overview of research in the ASDs underscores the short history of genetic work in this area as well as the diversity of the methods used. Before the 1970s, autism was not widely appreciated to have a strong biological basis. Instead, various psychodynamic interpretations, including the role of a cold or aloof style of mothering, were invoked as potential causes. The importance of genetic contributions became clear in the 1980s, when the co-occurrence of chromosomal disorders and rare syndromes with the ASDs were noted2. Subsequent twin and family studies provided additional support for a complex genetic aetiology, but these were limited by the lack of uniform diagnostic criteria. The development of validated diagnostic and assessment tools in the early 1990s, most notably the Autism Diagnostic Interview - Revised (ADI-R) and the Autism Diagnostic Observation Schedule (ADOS), addressed these concerns and these tools have proven crucial to the advancement of international research into the ASDs. This work, in concert with important technical advances, made it possible to carry out the first candidate gene association studies and resequencing efforts in the late 1990s. Whole-genome linkage studies followed, and were used to identify additional loci of potential interest. Although the application of genome-wide techniques to assess copy number variation (CNV) has only just begun3-5, these studies have already identified a large number of potentially important novel candidate loci.
Thus, in contrast to the complete absence of any biological understanding of the ASDs as recently as 30 years ago, we now know that defined mutations, genetic syndromes and de novo CNV account for about 10–20% of ASD cases (BOX 2; TABLE 1; Supplementary information S1 (table)). However, the striking finding that none of these known causes accounts for more than 1–2% of cases is reminiscent of mental retardation (MR), an overlapping but distinct neurodevelopmental syndrome for which there is no single major genetic cause, but rather many relatively rare mutations. Despite this heterogeneity in the ASDs, several biological themes, including defective synaptic function6 and abnormal brain connectivity7 (BOX 3), have been hypothesized to link rare and common variants at the level of biological function. Still, the relative proportion of ASDs that are explained by either rare or common genetic variation (or both) remains to be determined.
Box 2. Exploiting heterogeneity in the ASDs.
Available data suggest that autism spectrum disorder (ASD)-related syndromes individually account for no more than 1–2% of ASD cases (see TABLE 1). This is compared with unclassified cytogenetic lesions visible by G-banding (∼6–7% of cases) and unclassified de novo copy number variation (CNV) visible by molecular techniques (∼2–10% of cases). Taken together it is likely that known syndromes, observable cytogenetics lesions and rare de novo mutations account for between 10–20% of cases. It might be possible to leverage this heterogeneity to identify core features that are shared across autism types and are thus central to pathogenesis.
For instance, within etiologically defined subgroups (for example, fragile X syndrome), the phenotypes under investigation would be contrasted not only between mutation carriers and typically developing controls (mutation-specific features in panel a) but also between carriers with and without an ASD (ASD-specific features in panel a). Comparisons between fragile X mutation carriers and mutation-free controls will identify phenotypes that are related to mutation status but that do not inform us about ASDs per se. By contrast, comparisons between fragile X mutation carriers with and without an ASD should highlight factors that are more directly related to risk and presentation. Similarly, this same approach might be used to identify factors within fragile X carriers that modulate presentation or performance on any quantitative trait.
Given this experimental design (see panel a) — and sufficiently detailed characterization (see panel b) — one might expect such contrasts to identify phenotypic ‘signatures’ that define each of mutation-specific features and ASD-specific features. Mutation-specific signatures will be important to our understanding of gene function (not shown). ASD-specific signatures will clarify the relationship between ASD subtypes. Panel c illustrates hypothetical relationships between distinct syndromic ASDs, where coloured circles represent shared phenotypes. CACNA1C, calcium channel voltage-dependent L type alpha 1C subunit; Chr, chromosome; CNTNAP2, contactin associated protein-like 2; FMR1, fragile X mental retardation 1; SHANK3, SH3 and multiple ankyrin repeat domains 3. 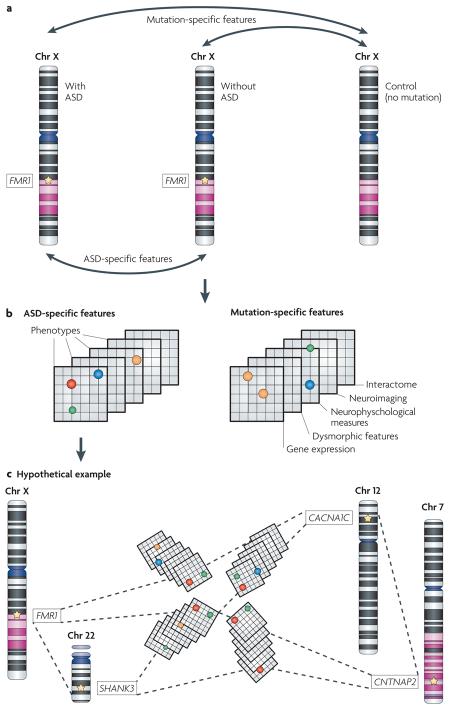
Table 1.
ASD-related syndromes
| Syndrome | Gene(s) associated with the syndrome |
Proportion of patients with the syndrome that have an ASD |
Proportion of patients with an ASD that have the syndrome |
Refs |
|---|---|---|---|---|
| 15q duplication — Angelman syndrome |
UBE3A (and others) | >40% | 1–2% | 101-103 |
| 16p11 deletion | Unknown | High | ∼1% | 20, 35, 44 |
| 22q deletion | SHANK3 | High | ∼1% | 21, 22, 104 |
| Cortical dysplasia-focal epilepsy syndrome |
CNTNAP2 | ∼70% | Rare | 37 |
| Fragile X syndrome | FMR1 | 25% of males; 6% of females | 1–2% | 105 |
| Joubert syndrome | Several loci | 25% | Rare | 106 |
| Potocki–Lupski syndrome | Chromosome position 17p11 |
∼90% | Unknown | 107 |
| Smith–Lemli–Optiz syndrome |
DHCR7 | 50% | Rare | 108 |
| Rett syndrome | MECP2 | All individuals have Rett syndrome |
∼0.5% | 109 |
| Timothy syndrome | CACNA1C | 60–80% | Unknown | 24 |
| Tuberous sclerosis | TSC1 and TSC2 | 20% | ∼1% | 110 |
The rates quoted in the table depend on the population that is being evaluated. For example, rates are higher in individuals from simplex families compared with multiplex families, and are higher in dysmorphic and mental retardation populations compared with idiopathic populations. ‘High’ is used for syndromes in which no good estimates exist (that is, only a handful of individuals with the syndrome in question have been identified). It should also be noted that none of the studies cited here indicates that assessment for the autism spectrum disorder (ASD) was performed blind to a patient's primary diagnosis. An expanded version of the table with additional variables can be found in Supplementary information S1 (table). CACNA1C, calcium channel voltage-dependent L type alpha 1C subunit; CNTNAP2, contactin associated protein-like 2; DHCR7, 7-dehydrocholesterol reductase; FMR1, fragile X mental retardation 1; MECP2, methyl CpG binding protein 2; SHANK3, SH3 and multiple ankyrin repeat domains 3; TSC1, tuberous sclerosis 1; TSC2, tuberous sclerosis 2; UBE3A, ubiquitin protein ligase E3A.
Box 3. Neurobiological hypotheses in the ASDs.
No single neurobiology currently dominates the autism spectrum disorders (ASDs), an observation that is presumably reflective of both the heterogeneity of the mechanisms at play and the short history of molecular work in the study of these disorders. Despite the general consensus regarding a developmental onset, little agreement exists around the primary nature of the insult(s). Although varied, dominant hypotheses can be attributed to cellular, regional or systemic dysfunction.
Among the cellular explanations, those involving the synapse currently dominate the field, although the manner by which generalized synaptic dysfunction might lead to specific behavioural impairments while largely sparing many aspects of cognition remains an important question. One hypothesis suggests a unifying role for glutamatergic neurotransmission89, a theory that is bolstered by the observation that reduction of GluR5 gene dosage can ameliorate ASD-like features in fragile X mice90. Others have posited that the defect lies at the inhibitory synapse31; this model could also account for the seizures that are observed in a subset of patients with ASDs. At the same time, strong arguments exist for the involvement of serotonin91, potentially accounting for abnormalities both in the brain and outside the central nervous system. Abnormal calcium signalling has likewise been suggested as a possible mechanism92. In support of this hypothesis several of the molecules underlying syndromic ASDs are known to act through intracellular signalling pathways. An important question that needs to be answered by any single unifying molecular hypothesis, synaptic or otherwise, is how cognitive and behavioural specificity is attained. The broad functional categories of genes identified thus far suggest that no single molecular explanation will suffice.
Histological abnormalities, particularly those involving the cerebellum, are among the earliest regional hypotheses, although such findings have not been observed consistently among cases. Reduced hemispheric asymmetry93, blunted mirror neuron activity94 and aberrant connectivity7 are alternative regional hypotheses that have emerged more recently. An area of convergence from pathological95 and imaging studies96 involves frontal and anterior temporal regions of the brain and their long-distance reciprocal and parietal connections7, presumably focusing on brain regions involved in joint attention, an early social behaviour and important precursor of pragmatic language97.
Layered over the top of these two paradigms are systemic abnormalities that are thought to have a role in the ASDs. The differential effects of maternal and paternal gains of 15q11, for example, are an important example of epigenetics at work. Although the role of epigenetic factors in the ASDs is still in its infancy, a handful of careful studies provides concrete molecular links between pervasive developmental disorders87,88. Hypocholesterolaemia98, exposure to prenatal testosterone99 and hyperactivation of the immune system100 have also been offered as explanations for the ASDs. The relative merits of these hypotheses will be tested as gene-finding efforts move forward and aspects of the ASDs are modelled in cells and in animal systems. Approaches that are rooted in network biology will be crucial for understanding how these diverse molecular pathways act together at the systems level. One potential way to reconcile the known molecular abnormalities (which include those that affect synaptic transmission, cell–cell interactions and intracellular signalling pathways (TABLE 2; TABLE 4)) with histological and anatomical systems findings is to suggest that they cause a developmental disconnection between higher-order association areas7.
In the face of this uncertainty, multiple parallel approaches are necessary to advance our understanding of the genetic factors underlying the ASDs. These approaches include whole-genome and pathway-based association studies, dense resequencing to identify mutations, and the continued collection of large well-characterized patient cohorts and their relatives for genotype–phenotype studies. Here we review this exciting and rapidly evolving field in which diverse genetic findings have begun to define potential biological mechanisms of disease.
- Gene association studies
A set of methods that is used to determine the correlation (positive or negative) between a defined genetic variant and a phenotype of interest.
- Whole-genome linkage study
A statistical evaluation of genetic variation throughout the genome that is used to identify polymorphic loci that segregate with a phenotype of interest.
- Copy number variation
(CNV). The insertion or deletion of a relatively large DNA fragment (>50 kb).
ASD as a complex genetic trait
Autism has a strong genetic basis
Several lines of evidence support genetic factors as a predominant cause of the ASDs. First is the growing body of literature demonstrating that mutations or structural variation in any of several genes can dramatically increase disease risk. Second, the relative risk of a child being diagnosed with autism is increased at least 25-fold over the population prevalence in families in which a sibling is affected8. Third, siblings and parents of an affected child are more likely than controls to show subtle cognitive or behavioural features that are qualitatively similar to those observed in probands9,10 (the broader autism phenotype); this is consistent with the segregation of quantitative sub-threshold traits within these families. Fourth, independent twin studies, although small, indicate that concordance rates for monozygotic twins (70–90%) are several-fold higher than the corresponding values for dizygotic twins (0–10%)11,12. An important question for future work will be to clarify how environmental and genetic factors interact to influence risk and presentation (BOX 1).
Genetic models of ASDs
Central to this question of how genetic variation comes to influence phenotypic presentation is whether distinct aspects of ASDs are subject to independent genetic modulation or, alternatively, they are sensitive to largely overlapping risk factors. Recent work in a community-based cohort revealed that, despite high heritability values (>0.64) for social, communication and repetitive and/or restrictive domains13, only modest co-variation was observed between them. Similarly, individuals with extreme scores in one domain did not necessarily have extreme scores in others. Additional support for this oligogenic model, in which disease results from the combined action of multiple interacting genes, comes from linkage studies that have identified distinct loci for endophenotypes that are related to different core domains14-16. Such results are also in keeping with the view that several risk alleles act together to modulate risk in families with multiple affected siblings8.
At the same time, it should be recognized that the relationship between core domains in the ASDs might not be properly reflected in community-based samples. For example, within samples ascertained for ASDs, data-mining techniques such as hierarchical clustering and principal components analysis identify a single continuously distributed factor that contributes to multiple aspects of disease17. Similarly, statistical analysis of ASD family data suggests that a significant proportion of ASD cases may be the result of dominantly acting de novo mutations that have a reduced penetrance in females18. Further support for the idea that autism might be a single continuum comes from a growing list of single genetic lesions, each of which seems to be largely sufficient to cause an ASD.
Together, these observations put into relief two contrasting but valid and potentially compatible paradigms that dominate current thinking regarding the role of genetic variation in ASD susceptibility. The independent heritability of distinct ASD core domains supports the importance of common variation in disease risk and phenotypic presentation. At the same time, the fact that functional disruption of single molecules seems to be sufficient to cause disease suggests that the identification of rare variants is also important. The primary technical approaches used in ASD research are largely rooted in one of these two models, as discussed in the following sections.
- Relative risk
The ratio of disease incidence in two groups of individuals that differ with regards to any associated factor (such as genetic polymorphism, environmental exposure or perinatal insult).
- Community-based cohort
A group of individuals that have been selected randomly from a population (as opposed to those showing a specific phenotype of interest).
Rare mutations
Cytogenetics
Chromosomal abnormalities offered the first glimpse at the potential role of rare variants in ASD susceptibility19. Mutation detection in candidate genes and identification of de novo CNV are more recent approaches, but are nevertheless rooted in a rare-variant framework. It is estimated that cytogenetically identified lesions are present in 6–7% of ASDs20, although this proportion is higher in dysmorphic populations with MR. Inherited duplications involving the chromosomal region 15q11–15q13 are among the most common cytogenetic abnormalities in the ASDs, accounting for 1–2% of cases, with maternal interstitial duplications and isodicentric marker chromosomes observed in most cases. As with all cytogenetics work, only large regions (typically containing fifty or more genes) are initially identified. Isolation of the molecules that are contributory requires the serendipitous recovery of overlapping rearrangements in unrelated individuals or the discovery of point mutations within individual genes.
- Heritability
The proportion of phenotypic variation that is attributable to inherited genetic factors (in contrast to environmental ones).
The clues obtained by these studies have proven important in our understanding of ASD aetiology. Within the 15q11–15q13 locus, ubiquitin protein ligase E3A (UBE3A) and gamma-aminobutyric acid A receptor beta 3 (GABRB3; an inhibitory neurotransmitter receptor) are currently thought to be central. Similarly, deletions involving 22q13 have been recognized for some time, but the important role of SH3 and multiple ankyrin repeat domains 3 (SHANK3; a synaptic adaptor protein) was appreciated only after resequencing and CNV analysis21,22. Deletions involving 2q37 are also common, having been observed in more than 70 cases. Although it is less clear which gene or genes within this region are contributory, patient-specific missense substitutions and positive linkage results highlight the potential involvement of centaurin gamma 2 (CENTG2; a GTPase-activating protein)23. Other regions that are implicated in the ASDs by chromosomal abnormalities in multiple patients include 5p15, 17p11 and Xp22 (Ref. 19). As a result of the large regions that are typically isolated by analysis of chromosomal anomalies, these methods cannot alone inform us about particular molecular functions that might be impaired in the ASDs.
ASD-related syndromes
Considerable insight into potential candidate genes was obtained from the study of molecularly defined syndromes in which ASD was observed at higher than expected frequencies. Several of these conditions, such as fragile X syndrome and Rett syndrome, suggest synaptic dysfunction as a unifying aetiology6, whereas others, such as tuberous sclerosis, highlight the diversity of signalling pathways that seem to be related to the ASDs. Note that not all ASD-related syndromes are limited to the brain. For example, in Timothy syndrome, mutations in the calcium channel voltage-dependent L type alpha 1C subunit gene (CACNA1C) cause a multisystem disorder presenting with cardiac arrhythmia, webbing of digits, MR and an ASD in ∼70% of patients24. Syndromes such as this — with prominent features outside the central nervous system — reinforce the notion of pleiotropy and argue for caution in the pursuit of candidates on the basis of tissue-restricted gene expression.
Although these ASD-associated syndromes involve genes with multiple molecular functions, it seems increasingly plausible that they converge on common biological pathways or brain circuits to give rise to ASDs7. In support of this notion, links between these syndromes are beginning to emerge. Levels of UBE3A and GABRB3, for example, are reduced in each of Angelman syndrome, Rett syndrome and idiopathic autism25. Further evidence for molecular overlap between different forms of syndromic ASDs comes from a subset of clinically identified ‘Angelman cases’ that are due to mutations in the Rett Syndrome gene, methyl CpG binding protein 2 (MECP2) (Ref. 26). Comparative study of these rare syndromes should be useful in identifying molecular features common to a variety of ASDs. Within disorders, contrasting cases with and without ASD-like features is also likely to prove informative (BOX 2; TABLE 1), although such studies have seldom been performed. As discussed below, support for such an approach comes from significant overlap in differentially expressed genes between each of fragile X syndrome and 15q duplication patients and controls27. Unlike common variants that may subtly modulate ASD-related phenotypes, the more pronounced effects that are typical of syndromic mutations allow for clearer assessment of causality, greatly facilitating downstream functional analysis. A potentially important observation is that a substantial proportion of these syndromic cases (TABLE 1; Supplementary information S1 (table)) are associated with seizures (8 out of 12) and/or congenital cardiovascular anomalies (7 out of 12). This observation is consistent with the idea that defects in electrical conduction and neural transmission might be important in ASD pathogenesis. These data also suggest that genes known to modulate such properties merit careful consideration in candidate gene studies. Furthermore, although motor delay is not recognized as a core feature among ASDs, it is noteworthy that such abnormalities are observed in 9 out of 12 of these ASD-linked syndromes.
- Endophenotype
A measurable trait that is both heritable and related to a specific aspect of a condition under investigation.
- Hierarchical clustering
A statistical method in which a collection of objects (observations, individuals or risk loci) are grouped into subsets, such that those within each cluster are more closely related to one another than objects that are assigned to different clusters.
- Principal components analysis
A statistical method used to simplify data sets by transforming a series of correlated variables into a smaller number of uncorrelated factors.
- Penetrance
The frequency with which individuals that carry an allele of a given gene will show the manifestations associated with the variant. If the penetrance of a disease allele is 100% then all individuals carrying that allele will express the associated disorder.
- Dysmorphic
Showing a structural abnormality of a body part or facial feature.
- Interstitial
A chromosomal segment that is located between the centromere and telomere.
- Isodicentric
A genetic abnormality characterized by the presence of two additional and identical DNA segment copies that are joined end to end to form a forty-seventh chromosome.
- Syndromic ASD
An ASD case that is observed in the context of a recognized syndrome (for example, fragile X syndrome).
Candidate resequencing
The introduction of cost-effective resequencing has made it possible to build on cytogenetic studies and obtain evidence for the involvement of specific candidate genes in the ASDs. The identification of rare ASD-linked mutations in neuroligin 3 (NLGN3) and neuroligin 4 X-linked (NLGN4X) was an important advance linking the ASDs to specific molecules that are involved in synaptic function28. Although coding variants in these genes do not seem to be a common explanation for ASDs29, the neuroligins provide a salient example of how the study of rare disease-linked variants can inform our understanding of disease mechanisms. For example, the presence of the disease-linked R451C mutation in NLGN3 interferes with cell-surface localization and appropriate protein–protein interactions30. Similarly, subsequent characterization of mice harbouring this same mutation in Nlgn3 highlights a possible role for excess inhibitory neurotransmission in disease31.
Mutations in SHANK3, the product of which interacts with neuroligins32, were identified by a similar route, and provide further evidence for the potential role of defective synaptogenesis in ASDs. This work also demonstrates the successful use of patients with structural chromosomal variation to guide gene choice in resequencing in large case–control cohorts. Three genetic lesions were identified in probands in three separate families from among the several hundred analysed21, and a follow-up screen for SHANK3 mutations in 400 non-overlapping ASD probands identified rare de novo variants in nearly 1% of cases, most of whom were female22.
- Mirror neuron
A neuron that is active when a subject is observing or imagining a motor movement; mirror neurons are thought to underlie imitation.
- Joint attention
Sharing interest or experience with another person by pointing or following gaze.
- Pragmatic language
Practical, social use of language.
- Neuroligin
A member of a family of postsynaptic cell-adhesion molecules that is important in regulating the balance of inhibitory and excitatory neurotransmission.
Beta-neurexins also interact with neuroligins33 and as such are compelling candidates for involvement in the ASDs. Identification of de novo deletions overlapping with neurexin 1 (NRXN1) in affected sisters5, along with rare missense mutations in other cases34, provides strong support for the involvement of this molecule in the ASDs. That NRXN1 deletions can be inherited from unaffected parents and are sometimes found in only a subset of affected siblings35 also suggests incomplete penetrance, consistent with the notion of substantial complexity. A recessive frameshift mutation was also identified in contactin associated protein-like 2 (CNTNAP2), a member of the neurexin family36, in Amish individuals presenting with cortical dysplasia-focal epilepsy syndrome, a congenital disorder characterized by seizures and language regression37. Notably, two-thirds of affected individuals also met the criteria for an ASD. Other recent work demonstrates that both rare single-base-pair mutations38 and common variation in CNTNAP2 (Refs 39,40) could also contribute to ASD risk. Although CNTNAP2 is best known for clustering potassium channels along myelinated axons, data from the human fetal brain indicate expression at high levels before myelination41. Along with evidence for abnormalities in neuronal migration in Amish patients who are homozygous for the frameshift mutation, these data suggest additional unappreciated functionality and an important role for this gene in the development of brain regions that are likely to be important for autism.
Copy number variation
The identification of CNV in addition to single-base-pair polymorphisms has opened a new window on human genetic variation42. De novo and inherited CNV are emerging as important causes of ASDs, either as rare variants that strongly modulate risk or as potentially new syndromes linked to the ASDs (see the autism chromosome rearrangement database for an extensive compilation)3-5. The concept of structural variation is not entirely new, as chromosomal disorders were among the earliest identified genetic abnormalities leading to ASDs. However, the increased resolution of array-based approaches suggests that the proportion of cases that might ultimately be attributable to rare structural variants is probably much higher than the 6–7% identified by standard cytogenetics. In addition to the identification of novel loci that are important for our understanding of the ASDs (FIG. 1; TABLE 2), observed differences in the extent of de novo variation between controls (1%), multiplex ASD families (2–3%) and simplex ASD families (7–10%)4,20 are consistent with other complex inherited diseases in which different genetic mechanisms are observed in sporadic compared with familial cases. These results are also relevant to the notion of the broader autism phenotype that is observed in the general population. The manyfold higher frequency of de novo CNV in simplex versus multiplex families would predict that ‘unaffected’ individuals from multiplex families would be more likely to harbour these lesser disease-related phenotypes than comparable individuals in simplex families (in whom the disease is more likely to arise de novo), as has been recently observed43.
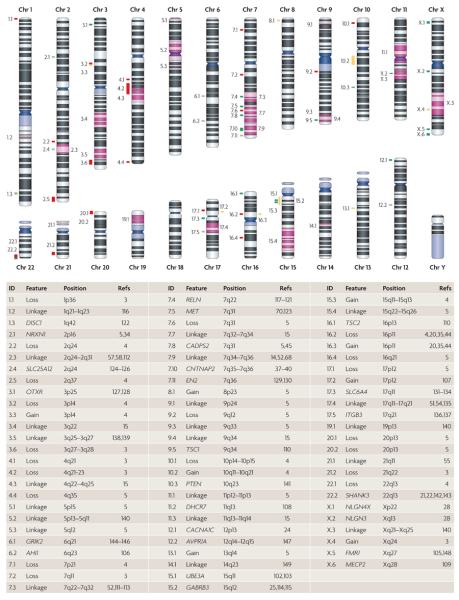
Figure 1. Loci implicated in ASD etiology.
Entries in the ID column of the table map link entries to the ideograms of individual chromosomes. Red and yellow bars correspond to de novo losses and gains, respectively, that are observed in cases but not in controls. Green bars correspond to genes that are observed to modulate autism spectrum disorder (ASD) risk (either through a rare syndrome or genetic association): light green and dark green bars represent promising or probable candidate genes, respectively, as defined in TABLE 2. Regions shaded in purple correspond to linkage peaks. Only human data were considered in the assembly of the table. AHI1, Abelson helper intergration site 1; AVPR1A, arginine vasopressin receptor 1A; CACNA1C, calcium channel voltage-dependent L type alpha 1C subunit; CADPS2, Ca2+-dependent activator protein for secretion 2; CNTNAP2, contactin associated protein-like 2; DHCR7, 7-dehydrocholesterol reductase; DISC1, disrupted in schizophrenia 1; EN2, engrailed homeobox 2; FMR1, fragile X mental retardation 1; GABRB3, gamma-aminobutyric acid (GABA) A receptor beta 3; GRIK2, glutamate receptor ionotropic kainate 2; ITGB3, integrin beta 3; MECP2, methyl CpG binding protein 2; MET, met proto-oncogene; NLGN3, neuroligin 3; NLGN4X, neuroligin 4 X-linked; NRXN1, neurexin 1; OXTR, oxytocin receptor; PTEN, phosphatase and tensin homologue; RELN, reelin; SHANK3, SH3 and multiple ankyrin repeat domains protein 3; SLC25A12, solute carrier family 25 (mitochondrial carrier, Aralar) member 12; SLC6A4, solute carrier family 6 (neurotransmitter transporter, serotonin) member 4; TSC1, tuberous sclerosis 1; TSC2, tuberous sclerosis 2; UBE3A, ubiquitin protein ligase E3A.
Table 2.
Evidence scores for ASD gene candidates
| Gene | Syndrome or mutation(s) |
Replicated association |
Analysis of variant |
Mouse model |
Other evidence | Total score |
|---|---|---|---|---|---|---|
| Promising | ||||||
| AVPR1A | 0 | 0 | 0 | 1 | 0 | 1 |
| DISC1 | 0 | 0 | 0 | 1 | 0 | 1 |
| ITGB3 | 0 | 1 | 0 | 0 | 0 | 1 |
| AHI1 | 2 | 0 | 0 | 0 | 0 | 2 |
| EN2 | 0 | 1 | 0 | 1 | 0 | 2 |
| GRIK2 | 0 | 1 | 0 | 0 | 1; homozygous mutation results in non-syndromic mental retardation | 2 |
| NRXN1 | 2 | 0 | 0 | 0 | 0 | 2 |
| SLC25A12 | 0 | 1 | 0 | 0 | 1; associated with neurite outgrowth, expression is upregulated in ASD brain | 2 |
| Probable | ||||||
| CACNA1C | 2 | 0 | 1 | 0 | 0 | 3 |
| CNTNAP2 | 2 | 1 | 0 | 0 | 0 | 3 |
| MET | 0 | 1 | 1 | 0 | 1; expression reduced in brains of cases versus controls | 3 |
| OXTR | 0 | 1 | 0 | 1 | 1; expression reduced in blood of cases versus controls | 3 |
| SHANK3 | 2 | 0 | 0 | 0 | 1; modulates glutamate-dependent reconfiguration of dendritic spines | 3 |
| SLC6A4 | 0 | 1 | 1 | 0 | 1; clinical benefit from inhibitors, variation linked to gray-matter volume | 3 |
| CADPS2 | 2 | 0 | 1 | 1 | 0 | 4 |
| DHCR7 | 2 | 0 | 1 | 0 | 1; hypocholesterolaemia in a proportion of probands | 4 |
| FMR1 | 2 | 0 | 1 | 1 | 0 | 4 |
| NLGN3 | 2 | 0 | 1 | 1 | 0 | 4 |
| NLGN4X | 2 | 0 | 1 | 1 | 0 | 4 |
| PTEN | 2 | 0 | 0 | 1 | 1; mutations result in abnormal structure and function of the synapse | 4 |
| TSC2 | 2 | 0 | 1 | 0 | 1; regulates dendrite morphology and function of glutamatergic synapses | 4 |
| GABRB3 | 2 | 1 | 0 | 1 | 1; expression is dysregulated in pervasive developmental disorders | 5 |
| MECP2 | 2 | 0 | 1 | 1 | 1; MECP2 deficiency causes reduced expression of UBE3A and GABRB3 | 5 |
| TSC1 | 2 | 0 | 1 | 1 | 1; regulates dendrite morphology and function of glutamatergic synapses | 5 |
| UBE3A | 2 | 0 | 1 | 1 | 1; expression is dysregulated in pervasive developmental disorders | 5 |
| RELN | 2 | 1 | 1 | 1 | 1; levels reduced in brains of cases versus controls | 6 |
Because the relationship to disease is most clear for rare variants, we biased our scoring accordingly. Genes associated with an autism spectrum disorder (ASD)-linked syndrome or mutation resulted in 2 points, whereas other lines of evidence resulted in 1 point. To qualify as a mouse model, 2 out of 3 core features were required to be present. The mean and standard deviation for evidence scores are 3.3 and 1.4, respectively. We assigned genes with scores greater than 3 as probable. Although we have done our best to systematically and comprehensively evaluate available data for each of the different molecules discussed, we recognize that these evidence scores are largely arbitrary. Together with unintended omissions of genes that are likely to be important in the ASDs, we hope that this table might serve as a useful starting point for discussion. AHI1, Abelson helper intergration site 1; AVPR1A, arginine vasopressin receptor 1A; CACNA1C, calcium channel voltage-dependent L type alpha 1C subunit; CADPS2, Ca2+-dependent activator protein for secretion 2; CNTNAP2, contactin associated protein-like 2; DHCR7, 7-dehydrocholesterol reductase; DISC1, disrupted in schizophrenia 1; EN2, engrailed homeobox 2; FMR1, fragile X mental retardation 1; GABRB3, gamma-aminobutyric acid (GABA) A receptor beta 3; GRIK2, glutamate receptor ionotropic kainate 2 precursor; ITGB3, integrin beta 3; MECP2, methyl CpG binding protein 2; MET, met proto-oncogene; NLGN3, neuroligin 3; NLGN4X, neuroligin 4 X-linked; NRXN1, neurexin 1; OXTR, oxytocin receptor; PTEN, phosphatase and tensin homologue; RELN, reelin; SHANK3, SH3 and multiple ankyrin repeat domains 3; SLC25A12, solute carrier family 25 (mitochondrial carrier, Aralar) member 12; SLC6A4, solute carrier family 6 (neurotransmitter transporter, serotonin) member 4; TSC1, tuberous sclerosis 1; TSC2, tuberous sclerosis 2; UBE3A, ubiquitin protein ligase E3A.
With regards to individual loci, the recent discovery of a recurrent de novo deletion involving an estimated 30 genes across 500 kb on chromosomal region 16p11 is of particular importance44 as it is present in about 1% of cases in several sizeable cohorts20,35,44. Importantly, a large population-based analysis35 indicates that, whereas the deletion is observed in controls, it is enriched 100-fold in ASD cases. Understanding the contribution of individual genes within this deletion, and the corresponding duplication that is also observed in cases, is an important question for future work. Another interesting CNV is a 750 kb de novo deletion involving the Ca2+-dependent activator protein for secretion 2 gene (CADPS2) and 7 other genes at 7q31, a replicated linkage region5 (see FIG. 1, item 7.7). This result, together with abnormalities in Cadps2 knockout mice and rare missense variants in patients45, supports a potential role for this positional candidate in disease. Similarly, a de novo loss at 20p13 (including the arginine vasopressin gene, the oxytocin gene and an additional 30 genes4) is intriguing given the associations between the variation in each of the corresponding receptors and ASDs (see later section on association studies) as well as the well-established role of both molecules in the modulation of social behaviour.
At the same time, it must be emphasized that the presence of a rare variant in a patient with a common disease need not always be meaningful46, a point underscored by the 1% frequency of de novo CNV detected in controls4. Empirical determination of how a CNV or any rare mutation affects gene function or expression — as was done in a patient with a deletion in the neuronal specific splicing factor ataxin 2-binding protein 1 (A2BP1)47 — will be important. Given that many of these rare variants span many genes and have only been observed in a single proband, much work is required to determine the subset causally related to disease. For some of the very rare, virtually unique, mutations even large sample sizes will not be sufficient to demonstrate statistical association, although the biological significance of the mutation may be clear.
- Multiplex
Multiplex families are those in which multiple individuals have a clinically diagnosed ASD.
- Simplex
Simplex families are those in which only a single individual has a clinically diagnosed ASD.
Specificity of rare variants
Overall, none of the molecules or syndromes currently linked to the ASDs have been shown to selectively cause autism. Instead, each seems to result in an array of abnormal neurobehavioural phenotypes, including autism, Asperger syndrome, non-syndromic MR and other neurodevelopmental abnormalities. For example, within a large French family harbouring a truncating mutation in NLGN4X, most male carriers presented with non-specific MR (10 out of 13), with only the remaining minority showing additional features consistent with an ASD48. Given that mutations in NLGN4X seem to cause MR more frequently than ASDs, a different consensus regarding involvement and specificity might have emerged had this paper been published earlier than the original autism paper. A similar range of phenotypes is observed for each of SHANK3, NRXN1 and CNTNAP2 mutations. Lastly, an extensive study of the 16p11 CNV in multiple clinical populations showed that it is at least as prevalent in a clinical population with global developmental delay or language delay as it is in autism35. Given this apparent lack of specificity for the ASDs, it will not be surprising if variation in some of these genes contribute to other neuropsychiatric disorders. As we discuss below, an understanding of the contribution of common variation — and the manner by which rare variants could modulate presentation even in cases with rare ‘major’ mutations — is an important future step.
Linkage studies
Substantial effort in autism genetics over the last 10 years has been focused on genetic linkage analysis using an affected sibling-pair design in multiplex families. Most studies have identified linkage regions reaching the threshold of suggestive linkage at best (see Ref. 49 for a recent review). Despite large increases in patient cohorts, linkage signals have not increased concomitantly with sample size. Loci on most chromosomes have been suggested to harbour ASD risk loci, but only a minority have been independently identified (FIG. 1; TABLE 3). To date, only loci on 17q11–17q21 and 7q have been replicated at levels that could be considered genome-wide significant50-52.
Table 3.
Linkage peaks with support from independent studies*
| Locus | Endpoint | Statistics | Cohort (subsetting) | Best marker |
Candidate genes | Refs |
|---|---|---|---|---|---|---|
| 1q21– 1q23 |
Asperger diagnosis | Zmax = 3.6 (p = 0.0002) | 30 extended families of Finnish origin | D1S484 | – | 150 |
| ASD diagnosis | MLS = 2.6 (p = 0.002) | 38 extended families (strict autism in 21) | D1S1653 | – | 138 | |
| 2q24– 2q31 |
ASD diagnosis | MLS = 4.8 (p = 2 × 10−5) | 152 IMGSAC families (strict autism in 127) | D2S2188 | CENTG2 | 112 |
| ASD diagnosis | NPL = 3.3 (p = 0.0005) | 100 families (speech delay in 49) | D2S364 | – | 57 | |
| 3q25– 3q27 |
ASD diagnosis | MLS = 4.8 (p = 2 × 10−5) | 38 extended families | D3S3037 | – | 138 |
| ASD diagnosis | NPL = 3.5 (p = 0.0003) | Large Utah pedigree with 7 affected children | rs1402229 | – | 139 | |
| 5p13– 5p14 |
ASD diagnosis | Z = 3.4 (p = 0.0003) | 1181 AGP families (194 FC families with ≥2 children with strictly defined autism) |
rs1968011 | – | 5 |
| ASD diagnosis | MLS = 2.5 (p = 0.003) | 110 AGRE families | D5S2494 | – | 140 | |
| ASD diagnosis | MLS = 2.5 (p = 0.003) | 345 AGRE families | D5S1473 | – | 135 | |
| 7q22– 7q31 |
ASD diagnosis | MLS = 3.2 (p = 0.0006) | 152 IMGSAC families | D7S477 |
RELN, MET, CADPS2 |
70,112,117- 121,123 |
| ASD diagnosis | MLS = 3.1 (p = 0.0008) | 170 IMGSAC families | D7S477 | – | 45,111 | |
| 7q34– 7q36 |
Age at first word | Zmax = 3.0 (p = 0.001) | 152 AGRE families | D7S1824 to D7S3058 |
CNTNAP2, EN2 |
14,39,40,129, 130,151,152 |
| Age at first word | Zmax = 2.1 (p = 0.02) | 291 AGRE families (including the 152 from above) |
D7S2426 | – | 68 | |
| 9q33– 9q34 |
Age at first word | Z = 3.5 (p = 0.0002) | 222 CPEA families | D9S164 | – | 15 |
| ASD diagnosis | Z = 3.3 (p = 0.0005) | 1181 AGP families (741 male only) | rs536861 | – | 5 | |
| 11p12– 11p13 |
ASD diagnosis | Z = 4.0 (p = 3 × 10−5) | 1181 AGP families (335 FC; batch CNV‡) | rs1358054 | – | 5 |
| Social responsiveness score |
Zmax = 3.2 (p = 0.0007) | 99 AGRE families | ATA34E08 | – | 69 | |
| 17q11– 17q21 |
ASD diagnosis | MLS = 4.3 (p = 5 × 10−5) | 257 AGRE families (148 male only) | D17S1294 |
ITGB3, SLC6A4 |
54,131-133, 136,137 |
| ASD diagnosis | MLS = 4.1 (p = 8 × 10−5) | 91 AGRE families (48 male only) | D17S2180 | – | 51 | |
| ASD diagnosis | MLS = 2.8 (p = 0.002) | 345 AGRE families | D17S1800 | – | 135 |
An attempt has been made to include all linkage peaks for which at least one study obtained a log10 of odds (LOD) score > 3.0 and a second obtained a LOD > 2.0. LOD is a measure of significance given by the logarithm of the odds under the null (no linkage) and alternative (linkage present) hypotheses. P values for Z scores were obtained in R using the formula pnorm(-abs(Z)); pnorm refers to normal distribution with a mean of 0 and a standard deviation of 1 and abs refers to absolute value. Values for LOD scores were obtained in R using the formula 1/10^(LOD).
Batch CNV refers to the analytic method that was used to identify and remove families with copy number variation (CNV). The candidate genes in the table are: CADPS2, Ca2+-dependent activator protein for secretion 2; CENTG2, centaurin gamma 2; CNTNAP2, contactin associated protein-like 2; EN2, engrailed homeobox 2; ITGB3, integrin beta 3; MET, met proto-oncogene; RELN, reelin; SLC6A4, solute carrier family 6 (neurotransmitter transporter, serotonin) member 4. Other abbreviations: AGP, Autism Genome Project; AGRE, Autism Genetic Resource Exchange; ASD, autism spectrum disorder; CPEA, Collaborative Programs of Excellence in Autism Network at the National Institutes of Health; FC, female containing; IMGSAC, International Molecular Genetic Study of Autism Consortium; MLS, multipoint LOD score; NPL, nonparametric LOD score.
The lack of genome-wide significant results in most published linkage studies is probably a consequence of the small effect size that is attributable to any particular gene. Early recognition of substantial genetic heterogeneity in the ASDs, and the need for large patient cohorts, led to the development of the Autism Genetic Resource Exchange (AGRE)53. AGRE is a publicly available resource of phenotypic data and bio-materials that has been widely used in these and other genetic studies of ASDs. Despite the replication of the chromosome 17q locus in the AGRE sample51, this signal has not been observed in every subsequent study15. Even the linkage scan published by the Autism Genome Project Consortium5, a large collaborative effort containing approximately 1,400 families, showed only minor overlap with previous genome scans. This might be because samples were contributed from multiple groups throughout the world, increasing the genetic heterogeneity in this cohort. Similarly, diagnostic differences used by participating groups might have increased phenotypic heterogeneity.
Subsetting cohorts to increase homogeneity
It is therefore not surprising that efforts to reduce heterogeneity by subsetting the sample according to specific phenotypes54,55 and refining phenotype definitions, have improved linkage signals. For example, subsetting by sex of proband51,54,56, language57 or other neurobehavioural features such as autistic regression or behavioural inflexibility55,58,59 has increased signals relative to non-stratified samples, including identification of genome-wide significant loci51,54. Given these successes, additional heritable phenotypes such as macrocephaly, structural features defined by magnetic resonance imaging, or the presence of seizures or electroencephalographic abnormalities might offer promise in defining subgroups of probands with autism.
Endophenotypes and mapping QTLs
In QTL mapping, autism endophenotypes are studied as opposed to affection status. The premise underlying a QTL approach complements models in which key aspects of the ASDs might be at one end of a continuum of normal behaviour and cognition60. Such work also recognize that the diagnostic categories used in clinical practice might not properly represent underlying genetic risk. That subtle ASD-like symptoms are elevated in frequency in siblings and parents of cases relative to controls9,10 makes the use of a QTL-based approach reasonable. Moreover, because many of these traits — including aspects of social behaviour, language and repetitive and/or restricted behaviours — vary within populations61,62 mapping efforts can be extended to large community-based cohorts. Important studies within diabetes63, psychiatric genetics64,65 and other human disorders66,67 highlight the power of this approach. As such, identification of refined quantitative end-points within the ASDs should be seen as a high priority.
Because a significant percentage of the non-autistic siblings of probands have a history of speech and language delay9, Alarcon et al. used a measure of language delay to identify a QTL on 7q34–7q36 (Ref. 14). Subsequent attempts at replication did not yield genome-wide significant results, but did provide suggestive support for linkage of language delay to this locus in ASDs as well as evidence for substantial genetic heterogeneity68. Schellenberg and colleagues15 identified a locus at 9q34 using the same age-at-first-word measure, taken from the ADI-R. Loci underlying quantitative assessment of non-verbal communication have also been identified16, and similar advances have been obtained through analysis of quantitative aspects of social behaviour, a core feature of the ASD diagnosis; this identified several regions already linked with ASDs, including a locus on chromosome 11 (Ref. 69). These preliminary QTL studies hold promise as a means of increasing power in linkage and association studies. Such methods will become more useful as our knowledge of ASD-related endophenotypes grows through twin, family and case–control studies.
The fact that linkage studies have not identified ‘the ASD gene’ is not a failure of these methods but rather an accurate reflection of the complexity of this group of disorders and the need for larger sample sizes. Although refined quantitative end points should improve the power of these approaches, insights that have already been obtained from these studies are important and should not be underestimated. Our identification of an association between common variation in CNTNAP2 and age at first word, for example, was the direct result of high-density SNP genotyping across the entire 7q34–7q36 linkage region. In a two-stage analysis, only variation in this gene survived correction for multiple comparisons. In this context, it is interesting to speculate about how many of the other ASD candidate genes in the more proximal 7q region (FIG. 1) might have been missed were it not for the well-replicated linkage to this region.
Association studies
Although several large whole-genome association studies are underway, including work in the AGRe sample, not one of these studies has yet been published. Numerous studies have, however, evaluated common variation in biological and positional candidates for association with ASDs or ASD-related phenotypes. A summary of findings for genes with the strongest support is provided in TABLE 4. The fact that most of these associated SNPs appear to be intronic — the met proto-oncogene (MET) promoter variant70 is an important exception — suggests that substantial work will be required to understand how disease-linked variants modulate clinical phenotypes.
Table 4.
Association studies in the ASDs*
| Gene (map position) | Relevant findings‡ | Replication of results | Refs |
|---|---|---|---|
|
AVPR1A (12q14–12q15) |
A positional candidate for ASD and important in rodent social behaviour. Interacts with OXTR. Association with affection status (p = 0.004) and a quantitative trait — Vineland Subscale Score (p = 0.007) |
No replication | 147 |
| CNTNAP2 (7q35) | A positional candidate gene with observed mutations. mRNA distribution markedly different in human versus rodent species. Association with affection status (p = 2 × 10−5) and a quantitative trait — age at first word (p = 0.05) |
Independent replication |
37-41 |
| DISC1 (1q42) | A key gene in psychiatric disorders and functionally linked to neuroligin signalling. Associated with ASD in a Finnish isolate by family-based association (p = 0.0007) and a case–control analysis (p = 9 × 10−5) |
No replication | 122 |
| EN2 (7q36) | A positional candidate that is linked to cerebellar abnormalities in mutant mice. SNP and haplotype-based associations to affection in multiple cohorts (p = 5 × 10−6, 0.001 and 0.04) |
Independent replication |
129,130 |
|
GABRB3 (15q11–15q12) |
A positional candidate; dysregulated in Rett syndrome, Angelman syndrome and autism brain25. Association with autism in multiple cohorts (p = 0.0014 and 0.0011) |
Independent replication |
25,114, 115 |
| GRIK2 (6q21) | A positional candidate that is linked to neuroligin signalling by PSD95. A rare homozygous mutation in an Iranian pedigree results in non-syndromic mental retardation. Association with autism in Caucasian (p = 0.0002) and Chinese populations (p = 0.01) |
Independent replication |
144,145 |
| ITGB3 (17q21) | Positional candidate involved in regulation of serotonin. Evidence for functional interaction with SLC6A4. Association with whole-blood serotonin levels in multiple populations (p = 0.01 and 0.0055) and autism (p = 0.00076) |
Serotonin QTL replicated |
136,137 |
| MET (7q31) | Positional candidate and reduced expression in brains of cases versus controls. Association between promoter variant and affection status in two large family-based cohorts (p = 0.0005 and 0.001) and a case–control analysis (p = 0.001). A function difference was observed between the two alleles of the associated variant |
Internal replication | 70,123 |
| OXTR (3p25) | Important in rodent social behaviour, reduced in blood of cases versus controls and interacts with AVPR1A. Association with affection status (p = 0.0094 and 5 × 10−5) and a quantitative trait — intelligence quotient (p = 0.0002) |
Association with affection status has been replicated |
127,128 |
| RELN (7q22) | Positional candidate and identification of rare variants among cases. Expression levels are reduced in brains of cases versus controls. Association with affection status by case–control analyses (p = 0.001) and family-based analyses (p = 0.001 and 0.002) |
Independent replication |
117- 121 |
| SLC25A12 (2q24) | A positional candidate that is related to neurite outgrowth and is upregulated in the prefrontal cortex of autistic subjects. Association between affection status and both single SNPs (p = 0.0094 and 0.02) and haplotypes (p = 5 × 10−5 and 0.03) |
Independent replication |
124,126 |
| SLC6A4 (17q11) | A positional candidate that is involved in the regulation of serotonin. Variation in SLC6A4 is linked to grey matter volume in the cortex. Evidence for functional interaction with ITGB3. Some clinical benefit is seen with SLC6A4 inhibitors. Association between promoter variant and both affection status (p = 0.007 and 0.007) and cortical grey matter volume (p = 0.004). Rare variants transmitted to cases at greater than expected frequency |
Replicated, although complex and controversial |
131- 134 |
No single approach was used to identify the genes we highlight here, but there were three factors important in determining which genes were highlighted. First, the extent of statistical support. To limit type II error, we have not required genome-wide significant results for inclusion in the table. Because these genes were tested in either a regional-based or candidate-based fashion, we instead required within-study significance corrected for multiple comparisons. Second, the availability of convergent biological evidence and third, the presence of independent replication. Given the extreme heterogeneity in autism spectrum disorders (ASDs), negative findings were not used as evidence against other positive results and are not included here.
Statistical significance of the findings is given by a p value; when multiple studies have been reported, the p value from each study is given. AVPR1A, arginine vasopressin receptor 1A; CNTNAP2, contactin associated protein-like 2; DISC1, disrupted in schizophrenia 1; EN2, engrailed homeobox 2; GABRB3, gamma-aminobutyric acid (GABA) A receptor beta 3; GRIK2, glutamate receptor ionotropic kainate 2 precursor; ITGB3, integrin beta 3; MET, met proto-oncogene; OXTR, oxytocin receptor; RELN, reelin; SLC25A12, solute carrier family 25 (mitochondrial carrier, Aralar) member 12; SLC6A4, solute carrier family 6 (neurotransmitter transporter, serotonin) member 4.
Genes for which rare ASD-linked mutations have been identified have not been shown to modulate risk through common variants. However, this is a potentially promising area and some interesting exceptions — including CNTNAP2 (Refs 39,40) and the Abelson helper integration site 1 gene (AHI1; A. Alvarez-Retuerto and D. G., unpublished results), in which mutations cause 6q23-linked Joubert syndrome71 — are emerging. At the same time, comprehensive evaluation of the genome-wide significant chromosome 17q locus by high-density SNP analysis72 did not reveal any single common variant that survived correction for multiple comparisons. Similarly, the CNTNAP2 variant, recently found to be associated with age at first word40, does not fully account for the linkage signal across the 7q34–7q36 interval14. Together, these data suggest that other variants, yet to be identified, might contribute to both peaks, necessitating much larger cohorts as has been necessary in other common diseases. These data also suggest that rare mutations are more important than previously anticipated.
At this point, it is perhaps reasonable to question whether any of the common variants identified thus far, each only subtly modulating risk, have added anything other than confusion to an otherwise productive field. In most cases, the true functional variant remains unidentified. Similarly, there is no case in which independent groups consistently replicated any of the existing associations. At the same time, such arguments belie the truth. None of the known mutations consistently results in autistic disorder, Asperger syndrome or any other defined spectrum disorders, necessitating the involvement of other factors. Moreover, given the heterogeneity in the ASDs it is not surprising that associations are not always replicated in each population evaluated. Holding associations to a standard of 100% replication would be similar to dismissing the role of a particular mutation after not seeing it in a small case series. Additional support for the common variant approach comes from data indicating that mice with mutations in genes for which association is well established (for example, reelin and Gabrb3) show ASD-like abnormalities73,74. Not only should rare and common variant approaches be pursued in parallel, but an integration of the results from both methods will also be necessary. Larger cohorts will also be needed to identify more definitive associations.
- Autistic regression
Normal development until the age of two with a subsequent loss of skills within core ASD domains.
- Macrocephaly
Head circumference greater than two standard deviations above the mean (the ninety fifth percentile).
- Multiple comparisons
Refers to the problem that arises when many null hypotheses are tested and ‘significant’ differences are observed when in reality there are no differences.
Array analyses — from genes to pathways
Microarray studies are beginning to provide important insights into molecules and pathways that might be dysregulated across the ASDs and within individual subtypes of pervasive developmental disorders. Early work using cDNA arrays and subtraction strategies contrasted cerebellar cortex from cases and controls and identified expression changes in genes relating to glutamatergic neurotransmission75. Interestingly, among the most differentially expressed genes identified by subtraction was erythrocyte membrane protein band 4.1-like 3 (EPB41L3; also known as 4.1B), a known interactor of CNTNAP2. Identification of molecules that might be central to the aetiology in fragile X syndrome — and, by extension, the ASDs — comes from other work identifying mRNAs that are associated with fragile X syndrome in vivo and a subset that seem to be dysregulated in patient-derived lymphoblastoid cells76.
Nishimura and colleagues27 showed that gene expression can distinguish among ASD cases caused by known genetic lesions and identify common pathways that were validated both in neural tissues and cases of idiopathic autism. This work defined 68 molecules that are differentially expressed in cells from fragile X syndrome and 15q duplication patients with an ASD relative to controls and identified cytoplasmic FMR1 interacting protein 1 (CYFIP1; an interactor of fragile X protein) and janus kinase and microtubule interacting protein 1 (JAKMIP1, also known as MARLIN1, which traffics gamma-aminobutyric acid receptors) as providing a direct molecular link between the two conditions. Several other studies have profiled gene expression in peripheral blood77-80 to identify biomarkers or pathophysiological pathways in ASDs and related neurodevelopmental disorders; these await validation using independent methods and samples.
It is notable that although less than 1% of the ∼900 unique genes collectively identified by these array studies were independently identified in separate experiments, among this set are several genes within the 15q interval (CYFIP1, non imprinted in Prader–Willi/Angelman syndrome 2 (NIPA2) and UBE3A). Additionally, several ontological categories are strongly over-represented across the entire data set, including ubiquitin conjugation (n = 18, p = 2.7 × 10−5), SH3 domain-containing proteins (n = 28, p = 8.7 × 10−9), GTPase regulator activity (n = 41, p = 4.0 × 10−11), α-protocadherin genes (n = 14, p = 1.2 × 10−15) and alternative splicing (n = 272, p = 4.0 × 10−34), raising these pathways as potential candidates for future mutation screening and association studies.
The use of gene expression data will increase as sample sizes grow, as additional molecularly defined syndromes are characterized by expression arrays37,44 and as expression information is better integrated with comprehensive phenotypic data81. It is also likely that the integration of expression studies with high-density SNP arrays will ultimately come to elucidate how disease-associated variants affect cellular function82. Furthermore, such expression QTL analysis might offer more power than traditional phenotypes, as the genotype relative risk associated with levels of specific transcripts might be higher than that associated with a complex neurobehavioural phenotype, such as social behaviour.
Looking forward
The identification of multiple rare mutations has implicated numerous genes of diverse function in the aetiology of the ASDs. Although causality remains to be demonstrated in most cases, a substantial subset seems to be important in modulating disease risk. These de novo mutations, together with those inherited in the context of a rare syndrome, each represent no more than 1–2% of cases individually, but account for at least 10–20% of the ASDs (TABLE 1; Supplementary information S1 (table)). The proportion of ASDs that might eventually come to be explained in terms of such major gene effects (as opposed to multigene interactions) should be clarified within the next year or two.
Given variable expressivity and incomplete penetrance among individuals carrying the same rare mutation, understanding the manner by which these rare variants interact with common alleles remains important. Similarly, the normal distribution of ASD-like features in populations62 as well as the elevated frequency of the broader autism phenotypes in close relatives of probands9,10 argues for an important role of common variation. That several common variants have now been independently shown to modulate risk and/or presentation is also evidence of important progress. A significant challenge here, however, is a continued focus on categorical measures (such as whether an individual is affected or not). Instead, quantitative endophenotypes will be necessary to properly identify risk alleles and understand the manner by which variation contributes to pathology. From a biological perspective, common variants are likely to have much greater salience when studied with regards to more specific phenotypes, including measures of gene expression, brain structure and quantitative aspects of social behaviour or communication. Use of quantitative end-points will also allow evaluation of whether genes that modulate ASD risk can contribute to aspects of normal phenotypic variation. For example, the association of variation in CNTNAP2 with language onset raises the possibility that variation in this gene might also modulate language-related cognitive phenotypes in both the general population and other clinically distinct, but related, disorders.
Also important, and immediately tractable, is the question of whether ASDs of different etiologies share common molecular mechanisms or pathways, and, if they do, whether the relationship between the underlying genes be understood83,84. Given the observed heterogeneity, an understanding of how risk factors interact functionally is an important step towards therapeutic interventions. Although this etiological heterogeneity will complicate substantially the translation of genetic findings from the laboratory to the clinic, it might prove useful in the identification of common targets for clinical intervention (BOX 2). Related to the heterogeneity in the ASDs, and discussed above, is the absence of clarity surrounding the specifics of the relationship between the ASDs, MR and other neuropsychiatric conditions. Although each can appear together, that they are also seen independently provides an opportunity to understand the overlap between MR and autism at the level of brain structure and function. Other co-morbid disorders observed in families with ASD probands also provide an important entry-point for exploring the genetic and biological boundaries of these conditions. For example, some of the language deficits observed in the ASDs are also seen in other disorders such as specific language impairment. Similarly, aspects of frontal executive and social dysfunction seem to overlap with other childhood neurodevelopmental conditions such as attention deficit hyperactivity disorder. These concepts reinforce the idea that current clinical notions of boundaries between neuropsychiatric disorders need not be representative of the underlying genetic or biological etiologies. At a practical level, these data support evaluation of putative ASD-linked variants in unaffected family members, typically developing controls and cohorts with other neurodevelopmental disorders (for example, nonspecific MR, schizophrenia and bipolar disorder). Also helpful here could be the use of rare disease subtypes to define the relationships between ASDs, as well as links to clinically distinct disorders with overlapping features.
Finally, we must aim to integrate existing and emerging genetic candidates into our understanding of human brain function. Such efforts are likely to provide important insights not only into ASD but also into related disorders in which behaviour is compromised through impaired function of overlapping regions and circuits. Because genes modulate behaviour through complex temporal and positional expression, analyses of candidate genes both through development41 and in patient material (for example, the Autism Tissue Program) will be important. Advances in systems biology should provide an important platform on which to integrate the modulatory effects of multiple interacting genes with functional data compiled from many levels of analysis83,84.
From the dominance of psychodynamic theories of autism as recently as 1970 to the awareness of upwards of 20 bona fide risk genes today, it must be concluded that substantial progress has been made in a relatively short time. That the identification of virtually every ASD-related gene and syndrome occurred within the last 5–10 years is particularly telling. Linkage studies have not found ‘the autism gene’ but have unequivocally demonstrated that more sophisticated solutions will be required to explain this group of disorders. The availability of new technology — particularly the ability to engage families in research through the internet — will permit the initiation of population-based strategies that are likely to provide more satisfying answers. Despite the immense amount of work implied in this agenda, the landscape for the future study of the ASDs is coming into view and has never looked as promising.
- Frontal executive
Frontal executive function describes broad aspects of higher cognition (for example, attention, working memory and relational reasoning) that are mediated by the frontal lobe and interconnected subcortical circuitry.
Supplementary Material
Acknowledgements
We gratefully acknowledge the families who have made these studies possible — along with the vision and leadership of AGRE and Autism Speaks. We are similarly indebted to the investigators whose work drives this field forward, many of whom we were unable to cite owing to space limitations. Thanks also to E. Herman, R. Mar-Heyming, B. Fogel and other members of the Geschwind laboratory for discussions. We also thank the anonymous reviewers. Work in the Geschwind laboratory is supported by funding from Autism Speaks, the Cure Autism Now Foundation, the National Institue of Mental Health (STAART - U54 MH68172; ACE - P50 HD055784; AGRE R01 MH64547; Asymmetry R37 MH60233) and the Tourette Syndrome Association.
Footnotes
DATABASES
Entrez Gene: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene CADPS2 | CNTNAP2 | GABRB3 | NLGN3 | NLGN4X | NRXN1 | SHANK3 | UBE3A
OMIM: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM Angelman syndrome | autistic disorder | cortical dysplasia-focal epilepsy syndrome | fragile X syndrome | Joubert syndrome | Rett syndrome | Timothy syndrome | tuberous sclerosis
FURTHER INFORMATION
Geschwind laboratory homepage: http://geschwindlab.neurology.ucla.edu
Autism chromosome rearrangement database: http://projects.tcag.ca/autism
Autism Genetic Resource Exchange (AGRE): http://www.agre.org
Autism tissue program: http://www.brainbank.org
Database of Genomic Variants: http://projects.tcag.ca/variation
Diagnostic and Statistical Manual of Mental Disorders: http://allpsych.com/disorders/dsm.html
SUPPLEMENTARY INFORMATION
See online article: S1 (table)
References
- 1.Fombonne E. Epidemiology of autistic disorder and other pervasive developmental disorders. J. Clin. Psychiatry. 2005;66(Suppl 10):3–8. [PubMed] [Google Scholar]
- 2.Blomquist HK, et al. Frequency of the fragile X syndrome in infantile autism. A Swedish multicenter study. Clin. Genet. 1985;27:113–117. doi: 10.1111/j.1399-0004.1985.tb00196.x. [DOI] [PubMed] [Google Scholar]
- 3.Jacquemont ML, et al. Array-based comparative genomic hybridisation identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J. Med. Genet. 2006;43:843–849. doi: 10.1136/jmg.2006.043166. The numerous de novo deletions that are reported in this work have received relatively little attention but are probably important in the ASDs. These data also show that rare de novo mutations are likely to appear at particularly high frequencies in syndromic populations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. Beyond identifying important CNV likely to prove important to our understanding of the ASDs, this study highlights a significant difference in the frequency of de novo variants between simplex and multiplex families, raising the possibility that distinct mechanisms are involved in each. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szatmari P, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nature Genet. 2007;39:319–328. doi: 10.1038/ng1985. This work — the largest ASD linkage study published to date — identifies a number of new loci that merit additional attention. Lack of overlap with previously published studies underscores the importance of genetic and phenotypic heterogeneity in ASDs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zoghbi HY. Postnatal neurodevelopmental disorders: meeting at the synapse? Science. 2003;302:826–830. doi: 10.1126/science.1089071. [DOI] [PubMed] [Google Scholar]
- 7.Geschwind DH, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr. Opin. Neurobiol. 2007;17:103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Jorde LB, et al. Complex segregation analysis of autism. Am. J. Hum. Genet. 1991;49:932–938. [PMC free article] [PubMed] [Google Scholar]
- 9.Bolton P, et al. A case–control family history study of autism. J. Child Psychol. Psychiatry. 1994;35:877–900. doi: 10.1111/j.1469-7610.1994.tb02300.x. [DOI] [PubMed] [Google Scholar]
- 10.Bishop DV, et al. Using self-report to identify the broad phenotype in parents of children with autistic spectrum disorders: a study using the Autism-Spectrum Quotient. J. Child Psychol. Psychiatry. 2004;45:1431–1436. doi: 10.1111/j.1469-7610.2004.00849.x. [DOI] [PubMed] [Google Scholar]
- 11.Bailey A, et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol. Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- 12.Steffenburg S, et al. A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J. Child Psychol. Psychiatry. 1989;30:405–416. doi: 10.1111/j.1469-7610.1989.tb00254.x. [DOI] [PubMed] [Google Scholar]
- 13.Ronald A, et al. Genetic heterogeneity between the three components of the autism spectrum: a twin study. J. Am. Acad. Child Adolesc. Psychiatry. 2006;45:691–699. doi: 10.1097/01.chi.0000215325.13058.9d. [DOI] [PubMed] [Google Scholar]
- 14.Alarcon M, Cantor RM, Liu J, Gilliam TC, Geschwind DH. Evidence for a language quantitative trait locus on chromosome 7q in multiplex autism families. Am. J. Hum. Genet. 2002;70:60–71. doi: 10.1086/338241. This first QTL study in autism is important because of its identification of a language QTL on chromosome 7q. Subsequent work from this group shows that this effect is attributable, at least in part, to variation in CNTNAP2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schellenberg GD, et al. Evidence for multiple loci from a genome scan of autism kindreds. Mol. Psychiatry. 2006;11:1049–1060. doi: 10.1038/sj.mp.4001874. [DOI] [PubMed] [Google Scholar]
- 16.Chen GK, Kono N, Geschwind DH, Cantor RM. Quantitative trait locus analysis of nonverbal communication in autism spectrum disorder. Mol. Psychiatry. 2006;11:214–220. doi: 10.1038/sj.mp.4001753. Variation in multiple quantitative phenotypes is likely to underlie ASD risk. These data suggest that key loci contribute to non-verbal aspects of communication in patients and highlight a heritable phenotypic measure that might prove important to characterizing the ASDs. [DOI] [PubMed] [Google Scholar]
- 17.Constantino JN, et al. The factor structure of autistic traits. J. Child Psychol. Psychiatry. 2004;45:719–726. doi: 10.1111/j.1469-7610.2004.00266.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhao X, et al. A unified genetic theory for sporadic and inherited autism. Proc. Natl Acad. Sci. USA. 2007;104:12831–12836. doi: 10.1073/pnas.0705803104. This work challenges the notion that the ASDs are the result of interactions between multiple risk alleles. Modelling data produces a compelling argument for the notion that a significant proportion of ASD cases are the result of dominantly acting de novo mutations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vorstman JA, et al. Identification of novel autism candidate regions through analysis of reported cytogenetic abnormalities associated with autism. Mol. Psychiatry. 2006;11:18–28. doi: 10.1038/sj.mp.4001781. [DOI] [PubMed] [Google Scholar]
- 20.Marshall CR, et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durand CM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nature Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moessner R, et al. Contribution of SHANK3 mutations to autism spectrum disorder. Am. J. Hum. Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wassink TH, et al. Evaluation of the chromosome 2q37.3 gene CENTG2 as an autism susceptibility gene. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2005;136:36–44. doi: 10.1002/ajmg.b.30180. [DOI] [PubMed] [Google Scholar]
- 24.Splawski I, et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 25.Samaco RC, Hogart A, LaSalle JM. Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3. Hum. Mol. Genet. 2005;14:483–492. doi: 10.1093/hmg/ddi045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watson P, et al. Angelman syndrome phenotype associated with mutations in MECP2, a gene encoding a methyl CpG binding protein. J. Med. Genet. 2001;38:224–228. doi: 10.1136/jmg.38.4.224. This paper highlights the fact that the molecular classification of patients, as is the case in other disorders, will be important in our understanding of the relationships between ASD subtypes and in identifying common links between them. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishimura Y, et al. Genome-wide expression profiling of lymphoblastoid cell lines distinguishes different forms of autism and reveals shared pathways. Hum. Mol. Genet. 2007;16:1682–1698. doi: 10.1093/hmg/ddm116. [DOI] [PubMed] [Google Scholar]
- 28.Jamain S, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nature Genet. 2003;34:27–29. doi: 10.1038/ng1136. This work provides the first evidence for an important causative role for the neuroligin genes in the ASDs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan J, et al. Analysis of the neuroligin 3 and 4 genes in autism and other neuropsychiatric patients. Mol. Psychiatry. 2005;10:329–332. doi: 10.1038/sj.mp.4001629. [DOI] [PubMed] [Google Scholar]
- 30.Comoletti D, et al. The Arg451 Cys-neuroligin-3 mutation associated with autism reveals a defect in protein processing. J. Neurosci. 2004;24:4889–4893. doi: 10.1523/JNEUROSCI.0468-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tabuchi K, et al. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer G, Varoqueaux F, Neeb A, Oschlies M, Brose N. The complexity of PDZ domain-mediated interactions at glutamatergic synapses: a case study on neuroligin. Neuropharmacology. 2004;47:724–733. doi: 10.1016/j.neuropharm.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 33.Boucard AA, Chubykin AA, Comoletti D, Taylor P, Sudhof TC. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron. 2005;48:229–236. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 34.Feng J, et al. High frequency of neurexin 1 beta signal peptide structural variants in patients with autism. Neurosci. Lett. 2006;409:10–13. doi: 10.1016/j.neulet.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 35.Weiss LA, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 36.Poliak S, et al. Caspr2, a new member of the neurexin superfamily, is localized at the juxtaparanodes of myelinated axons and associates with K+ channels. Neuron. 1999;24:1037–1047. doi: 10.1016/s0896-6273(00)81049-1. [DOI] [PubMed] [Google Scholar]
- 37.Strauss KA, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N. Engl. J. Med. 2006;354:1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 38.Bakkaloglu B, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am. J. Hum. Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arking DE, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am. J. Hum. Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alarcon M, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am. J. Hum. Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abrahams BS, et al. Genome-wide analyses of human perisylvian cerebral cortical patterning. Proc. Natl Acad. Sci. USA. 2007;104:17849–17854. doi: 10.1073/pnas.0706128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 43.Losh M, Childress D, Lam K, Piven J. Defining key features of the broad autism phenotype: A comparison across parents of multiple- and single-incidence autism families. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007 July 25; doi: 10.1002/ajmg.b.30612. doi: 10.1002/ajmg.b.30612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar RA, et al. Recurrent 16p11.2 microdeletions in autism. Hum. Mol. Genet. 2007;17:628–638. doi: 10.1093/hmg/ddm376. Together with references 20 and 35, this paper highlights an important role for genes at the 16p11 locus in a substantial proportion of ASD cases. [DOI] [PubMed] [Google Scholar]
- 45.Sadakata T, et al. Autistic-like phenotypes in Cadps2-knockout mice and aberrant CADPS2 splicing in autistic patients. J. Clin. Invest. 2007;117:931–943. doi: 10.1172/JCI29031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Belloso JM, et al. Disruption of the CNTNAP2 gene in a t(7;15) translocation family without symptoms of Gilles de la Tourette syndrome. Eur. J. Hum. Genet. 2007;15:711–713. doi: 10.1038/sj.ejhg.5201824. [DOI] [PubMed] [Google Scholar]
- 47.Martin CL, et al. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007;144:869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- 48.Laumonnier F, et al. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am. J. Hum. Genet. 2004;74:552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freitag CM. The genetics of autistic disorders and its clinical relevance: a review of the literature. Mol. Psychiatry. 2007;12:2–22. doi: 10.1038/sj.mp.4001896. [DOI] [PubMed] [Google Scholar]
- 50.Badner JA, Gershon ES. Regional meta-analysis of published data supports linkage of autism with markers on chromosome 7. Mol. Psychiatry. 2002;7:56–66. doi: 10.1038/sj.mp.4000922. [DOI] [PubMed] [Google Scholar]
- 51.Cantor RM, et al. Replication of autism linkage: fine-mapping peak at 17q21. Am. J. Hum. Genet. 2005;76:1050–1056. doi: 10.1086/430278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trikalinos TA, et al. A heterogeneity-based genome search meta-analysis for autism-spectrum disorders. Mol. Psychiatry. 2006;11:29–36. doi: 10.1038/sj.mp.4001750. [DOI] [PubMed] [Google Scholar]
- 53.Geschwind DH, et al. The autism genetic resource exchange: a resource for the study of autism and related neuropsychiatric conditions. Am. J. Hum. Genet. 2001;69:463–466. doi: 10.1086/321292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stone JL, et al. Evidence for sex-specific risk alleles in autism spectrum disorder. Am. J. Hum. Genet. 2004;75:1117–1123. doi: 10.1086/426034. Beyond the clear identification of the 17q risk locus, this work highlights the usefulness of considering families with only affected males separately from families with both affected males and females. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Molloy CA, Keddache M, Martin LJ. Evidence for linkage on 21q and 7q in a subset of autism characterized by developmental regression. Mol. Psychiatry. 2005;10:741–746. doi: 10.1038/sj.mp.4001691. [DOI] [PubMed] [Google Scholar]
- 56.Lamb JA, et al. Analysis of IMGSAC autism susceptibility loci: evidence for sex limited and parent of origin specific effects. J. Med. Genet. 2005;42:132–137. doi: 10.1136/jmg.2004.025668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buxbaum JD, et al. Evidence for a susceptibility gene for autism on chromosome 2 and for genetic heterogeneity. Am. J. Hum. Genet. 2001;68:1514–1520. doi: 10.1086/320588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shao Y, et al. Phenotypic homogeneity provides increased support for linkage on chromosome 2 in autistic disorder. Am. J. Hum. Genet. 2002;70:1058–1061. doi: 10.1086/339765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buxbaum JD, et al. Linkage analysis for autism in a subset families with obsessive-compulsive behaviors: evidence for an autism susceptibility gene on chromosome 1 and further support for susceptibility genes on chromosome 6 and 19. Mol. Psychiatry. 2004;9:144–150. doi: 10.1038/sj.mp.4001465. [DOI] [PubMed] [Google Scholar]
- 60.Abrahams BS, Geschwind DH. In: Encyclopedia of Genetics, Genomics, Proteomics and Bioinformatics. Dunn MJ, Jorde LB, Little PFR, Subramaniam S, editors. John Wiley & Sons; 2005. pp. p1–12. section 1.5. [Google Scholar]
- 61.Constantino JN, Todd RD. Autistic traits in the general population: a twin study. Arch. Gen. Psychiatry. 2003;60:524–530. doi: 10.1001/archpsyc.60.5.524. [DOI] [PubMed] [Google Scholar]
- 62.Hoekstra RA, Bartels M, Verweij CJ, Boomsma DI. Heritability of autistic traits in the general population. Arch. Pediatr. Adolesc. Med. 2007;161:372–377. doi: 10.1001/archpedi.161.4.372. [DOI] [PubMed] [Google Scholar]
- 63.Kissebah AH, et al. Quantitative trait loci on chromosomes 3 and 17 influence phenotypes of the metabolic syndrome. Proc. Natl Acad. Sci. USA. 2000;97:14478–14483. doi: 10.1073/pnas.97.26.14478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Francks C, et al. LRRTM1 on chromosome 2p12 is a maternally suppressed gene that is associated paternally with handedness and schizophrenia. Mol. Psychiatry. 2007 doi: 10.1038/sj.mp.4002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fisher SE, et al. Independent genome-wide scans identify a chromosome 18 quantitative-trait locus influencing dyslexia. Nature Genet. 2002;30:86–91. doi: 10.1038/ng792. [DOI] [PubMed] [Google Scholar]
- 66.Arya R, et al. Linkage of high-density lipoprotein-cholesterol concentrations to a locus on chromosome 9p in Mexican Americans. Nature Genet. 2002;30:102–105. doi: 10.1038/ng810. [DOI] [PubMed] [Google Scholar]
- 67.Menzel S, et al. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nature Genet. 2007;39:1197–1199. doi: 10.1038/ng2108. [DOI] [PubMed] [Google Scholar]
- 68.Alarcon M, Yonan AL, Gilliam TC, Cantor RM, Geschwind DH. Quantitative genome scan and ordered-subsets analysis of autism endophenotypes support language QTLs. Mol. Psychiatry. 2005;10:747–757. doi: 10.1038/sj.mp.4001666. [DOI] [PubMed] [Google Scholar]
- 69.Duvall JA, et al. A quantitative trait locus analysis of social responsiveness in multiplex autism families. Am. J. Psychiatry. 2007;164:656–662. doi: 10.1176/ajp.2007.164.4.656. [DOI] [PubMed] [Google Scholar]
- 70.Campbell DB, et al. A genetic variant that disrupts MET transcription is associated with autism. Proc. Natl Acad. Sci. USA. 2006;103:16834–16839. doi: 10.1073/pnas.0605296103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ferland RJ, et al. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nature Genet. 2004;36:1008–1013. doi: 10.1038/ng1419. [DOI] [PubMed] [Google Scholar]
- 72.Stone JL, Merriman B, Cantor RM, Geschwind DH, Nelson SF. High density SNP association study of a major autism linkage region on chromosome 17. Hum. Mol. Genet. 2007;16:704–715. doi: 10.1093/hmg/ddm015. [DOI] [PubMed] [Google Scholar]
- 73.Salinger WL, Ladrow P, Wheeler C. Behavioral phenotype of the reeler mutant mouse: effects of RELN gene dosage and social isolation. Behav. Neurosci. 2003;117:1257–1275. doi: 10.1037/0735-7044.117.6.1257. [DOI] [PubMed] [Google Scholar]
- 74.DeLorey TM, Sahbaie P, Hashemi E, Homanics GE, Clark JD. Gabrb3 gene deficient mice exhibit impaired social and exploratory behaviors, deficits in non-selective attention and hypoplasia of cerebellar vermal lobules: a potential model of autism spectrum disorder. Behav. Brain Res. 2008;187:207–220. doi: 10.1016/j.bbr.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Purcell AE, Jeon OH, Zimmerman AW, Blue ME, Pevsner J. Postmortem brain abnormalities of the glutamate neurotransmitter system in autism. Neurology. 2001;57:1618–1628. doi: 10.1212/wnl.57.9.1618. [DOI] [PubMed] [Google Scholar]
- 76.Brown V, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 77.Baron CA, et al. Genomic and functional profiling of duplicated chromosome 15 cell lines reveal regulatory alterations in UBE3A-associated ubiquitin-proteasome pathway processes. Hum. Mol. Genet. 2006;15:853–869. doi: 10.1093/hmg/ddl004. [DOI] [PubMed] [Google Scholar]
- 78.Hu VW, Frank BC, Heine S, Lee NH, Quackenbush J. Gene expression profiling of lymphoblastoid cell lines from monozygotic twins discordant in severity of autism reveals differential regulation of neurologically relevant genes. BMC Genomics. 2006;7:118. doi: 10.1186/1471-2164-7-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gregg JP, et al. Gene expression changes in children with autism. Genomics. 2007;91:22–29. doi: 10.1016/j.ygeno.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 80.Bittel DC, Kibiryeva N, Butler MG. Whole genome microarray analysis of gene expression in subjects with fragile X syndrome. Genet. Med. 2007;9:464–472. doi: 10.1097/gim.0b013e3180ca9a9a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bittel DC, Kibiryeva N, Butler MG. Expression of 4 genes between chromosome 15 breakpoints 1 and 2 and behavioral outcomes in Prader–Willi syndrome. Pediatrics. 2006;118:e1276–e1283. doi: 10.1542/peds.2006-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Myers AJ, et al. A survey of genetic human cortical gene expression. Nature Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 83.Krauthammer M, Kaufmann CA, Gilliam TC, Rzhetsky A. Molecular triangulation: bridging linkage and molecular-network information for identifying candidate genes in Alzheimer's disease. Proc. Natl Acad. Sci. USA. 2004;101:15148–15153. doi: 10.1073/pnas.0404315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aerts S, et al. Gene prioritization through genomic data fusion. Nature Biotechnol. 2006;24:537–544. doi: 10.1038/nbt1203. [DOI] [PubMed] [Google Scholar]
- 85.Glasson EJ, et al. Perinatal factors and the development of autism: a population study. Arch. Gen. Psychiatry. 2004;61:618–627. doi: 10.1001/archpsyc.61.6.618. [DOI] [PubMed] [Google Scholar]
- 86.Reichenberg A, et al. Advancing paternal age and autism. Arch. Gen. Psychiatry. 2006;63:1026–1032. doi: 10.1001/archpsyc.63.9.1026. [DOI] [PubMed] [Google Scholar]
- 87.Jiang YH, et al. A mixed epigenetic/genetic model for oligogenic inheritance of autism with a limited role for UBE3A. Am. J. Med. Genet. A. 2004;131:1–10. doi: 10.1002/ajmg.a.30297. [DOI] [PubMed] [Google Scholar]
- 88.Hogart A, Nagarajan RP, Patzel KA, Yasui DH, Lasalle JM. 15q11–13 GABAA receptor genes are normally biallelically expressed in brain yet are subject to epigenetic dysregulation in autism-spectrum disorders. Hum. Mol. Genet. 2007;16:691–703. doi: 10.1093/hmg/ddm014. These results underscore the idea that the mechanisms underlying the ASDs could be much more complex than typically appreciated; in addition to alterations in absolute levels of gene expression, the authors show that the relative expression of parental GABAA alleles is altered in the ASDs, demonstrating a role for imprinting. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 90.Dolen G, et al. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chugani DC. Serotonin in autism and pediatric epilepsies. Ment. Retard. Dev. Disabil. Res. Rev. 2004;10:112–116. doi: 10.1002/mrdd.20021. [DOI] [PubMed] [Google Scholar]
- 92.Krey JF, Dolmetsch RE. Molecular mechanisms of autism: a possible role for Ca2+ signaling. Curr. Opin. Neurobiol. 2007;17:112–119. doi: 10.1016/j.conb.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 93.Herbert MR, et al. Brain asymmetries in autism and developmental language disorder: a nested whole-brain analysis. Brain. 2005;128:213–226. doi: 10.1093/brain/awh330. [DOI] [PubMed] [Google Scholar]
- 94.Dapretto M, et al. Understanding emotions in others: mirror neuron dysfunction in children with autism spectrum disorders. Nature Neurosci. 2006;9:28–30. doi: 10.1038/nn1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bailey A, et al. A clinicopathological study of autism. Brain. 1998;121:889–905. doi: 10.1093/brain/121.5.889. [DOI] [PubMed] [Google Scholar]
- 96.Courchesne E, Carper R, Akshoomoff N. Evidence of brain overgrowth in the first year of life in autism. JAMA. 2003;290:337–344. doi: 10.1001/jama.290.3.337. [DOI] [PubMed] [Google Scholar]
- 97.Mundy P. Annotation: the neural basis of social impairments in autism: the role of the dorsal medial-frontal cortex and anterior cingulate system. J. Child Psychol. Psychiatry. 2003;44:793–809. doi: 10.1111/1469-7610.00165. [DOI] [PubMed] [Google Scholar]
- 98.Tierney E, et al. Abnormalities of cholesterol metabolism in autism spectrum disorders. Am. J. Med. Genet. B Neuropsychiatr Genet. 2006;141:666–668. doi: 10.1002/ajmg.b.30368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Knickmeyer RC, Baron-Cohen S. Fetal testosterone and sex differences in typical social development and in autism. J. Child Neurol. 2006;21:825–845. doi: 10.1177/08830738060210101601. [DOI] [PubMed] [Google Scholar]
- 100.Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, Pardo CA. Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann. Neurol. 2005;57:67–81. doi: 10.1002/ana.20315. [DOI] [PubMed] [Google Scholar]
- 101.Cook EH, Jr, et al. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am. J. Hum. Genet. 1997;60:928–934. [PMC free article] [PubMed] [Google Scholar]
- 102.Matsuura T, et al. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nature Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- 103.Peters SU, Beaudet AL, Madduri N, Bacino CA. Autism in Angelman syndrome: implications for autism research. Clin. Genet. 2004;66:530–536. doi: 10.1111/j.1399-0004.2004.00362.x. [DOI] [PubMed] [Google Scholar]
- 104.Manning MA, et al. Terminal 22q deletion syndrome: a newly recognized cause of speech and language disability in the autism spectrum. Pediatrics. 2004;114:451–457. doi: 10.1542/peds.114.2.451. [DOI] [PubMed] [Google Scholar]
- 105.Hatton DD, et al. Autistic behavior in children with fragile X syndrome: prevalence, stability, and the impact of FMRP. Am. J. Med. Genet. A. 2006;140:1804–1813. doi: 10.1002/ajmg.a.31286. [DOI] [PubMed] [Google Scholar]
- 106.Ozonoff S, Williams BJ, Gale S, Miller JN. Autism and autistic behavior in Joubert syndrome. J. Child Neurol. 1999;14:636–641. doi: 10.1177/088307389901401003. [DOI] [PubMed] [Google Scholar]
- 107.Potocki L, et al. Characterization of Potocki–Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am. J. Hum. Genet. 2007;80:633–649. doi: 10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tierney E, et al. Behavior phenotype in the RSH/Smith–Lemli–Opitz syndrome. Am. J. Med. Genet. 2001;98:191–200. doi: 10.1002/1096-8628(20010115)98:2<191::aid-ajmg1030>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 109.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 110.Baker P, Piven J, Sato Y. Autism and tuberous sclerosis complex: prevalence and clinical features. J. Autism Dev. Disord. 1998;28:279–285. doi: 10.1023/a:1026004501631. [DOI] [PubMed] [Google Scholar]
- 111.International Molecular Genetic Study of Autism Consortium A full genome screen for autism with evidence for linkage to a region on chromosome 7q. Hum. Mol. Genet. 1998;7:571–578. doi: 10.1093/hmg/7.3.571. [DOI] [PubMed] [Google Scholar]
- 112.International Molecular Genetic Study of Autism Consortium A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am. J. Hum. Genet. 2001;69:570–581. doi: 10.1086/323264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.International Molecular Genetic Study of Autism Consortium Further characterization of the autism susceptibility locus AUTS1 on chromosome 7q. Hum. Mol. Genet. 2001;10:973–982. doi: 10.1093/hmg/10.9.973. [DOI] [PubMed] [Google Scholar]
- 114.Cook EH, Jr., et al. Linkage-disequilibrium mapping of autistic disorder, with 15q11–13 markers. Am. J. Hum. Genet. 1998;62:1077–1083. doi: 10.1086/301832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Buxbaum JD, et al. Association between a GABRB3 polymorphism and autism. Mol. Psychiatry. 2002;7:311–316. doi: 10.1038/sj.mp.4001011. [DOI] [PubMed] [Google Scholar]
- 116.Ylisaukko-Oja T, et al. Search for autism loci by combined analysis of Autism Genetic Resource Exchange and Finnish families. Ann. Neurol. 2006;59:145–155. doi: 10.1002/ana.20722. [DOI] [PubMed] [Google Scholar]
- 117.Persico AM, et al. Reelin gene alleles and haplotypes as a factor predisposing to autistic disorder. Mol. Psychiatry. 2001;6:150–159. doi: 10.1038/sj.mp.4000850. [DOI] [PubMed] [Google Scholar]
- 118.Bonora E, et al. Analysis of reelin as a candidate gene for autism. Mol. Psychiatry. 2003;8:885–892. doi: 10.1038/sj.mp.4001310. [DOI] [PubMed] [Google Scholar]
- 119.Skaar DA, et al. Analysis of the RELN gene as a genetic risk factor for autism. Mol. Psychiatry. 2005;10:563–571. doi: 10.1038/sj.mp.4001614. [DOI] [PubMed] [Google Scholar]
- 120.Serajee FJ, Zhong H, Mahbubul Huq AH. Association of reelin gene polymorphisms with autism. Genomics. 2006;87:75–83. doi: 10.1016/j.ygeno.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 121.Li H, et al. The association analysis of RELN and GRM8 genes with autistic spectrum disorder in Chinese Han population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007;147:194–200. doi: 10.1002/ajmg.b.30584. [DOI] [PubMed] [Google Scholar]
- 122.Kilpinen H, et al. Association of DISC1 with autism and Asperger syndrome. Mol. Psychiatry. 2007;13:187–196. doi: 10.1038/sj.mp.4002031. [DOI] [PubMed] [Google Scholar]
- 123.Campbell DB, et al. Disruption of cerebral cortex MET signaling in autism spectrum disorder. Ann. Neurol. 2007;62:243–250. doi: 10.1002/ana.21180. [DOI] [PubMed] [Google Scholar]
- 124.Ramoz N, et al. Linkage and association of the mitochondrial aspartate/glutamate carrier SLC25A12 gene with autism. Am. J. Psychiatry. 2004;161:662–669. doi: 10.1176/appi.ajp.161.4.662. [DOI] [PubMed] [Google Scholar]
- 125.Silverman JM, et al. Autism-related routines and rituals associated with a mitochondrial aspartate/glutamate carrier SLC25A12 polymorphism. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007;147:408–410. doi: 10.1002/ajmg.b.30614. [DOI] [PubMed] [Google Scholar]
- 126.Segurado R, et al. Confirmation of association between autism and the mitochondrial aspartate/glutamate carrier SLC25A12 gene on chromosome 2q31. Am. J. Psychiatry. 2005;162:2182–2184. doi: 10.1176/appi.ajp.162.11.2182. [DOI] [PubMed] [Google Scholar]
- 127.Wu S, et al. Positive association of the oxytocin receptor gene (OXTR) with autism in the Chinese Han population. Biol. Psychiatry. 2005;58:74–77. doi: 10.1016/j.biopsych.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 128.Lerer E, et al. Association between the oxytocin receptor (OXTR) gene and autism: relationship to Vineland Adaptive Behavior Scales and cognition. Mol. Psychiatry. 2007 Sept 25; doi: 10.1038/sj.mp.4002087. doi: 10.1038/sj.mp.4002087. [DOI] [PubMed] [Google Scholar]
- 129.Gharani N, Benayed R, Mancuso V, Brzustowicz LM, Millonig JH. Association of the homeobox transcription factor, ENGRAILED 2, 3, with autism spectrum disorder. Mol. Psychiatry. 2004;9:474–484. doi: 10.1038/sj.mp.4001498. [DOI] [PubMed] [Google Scholar]
- 130.Benayed R, et al. Support for the homeobox transcription factor gene ENGRAILED 2 as an autism spectrum disorder susceptibility locus. Am. J. Hum. Genet. 2005;77:851–868. doi: 10.1086/497705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kim SJ, et al. Transmission disequilibrium mapping at the serotonin transporter gene (SLC6A4) region in autistic disorder. Mol. Psychiatry. 2002;7:278–288. doi: 10.1038/sj.mp.4001033. [DOI] [PubMed] [Google Scholar]
- 132.Devlin B, et al. Autism and the serotonin transporter: the long and short of it. Mol. Psychiatry. 2005;10:1110–1116. doi: 10.1038/sj.mp.4001724. [DOI] [PubMed] [Google Scholar]
- 133.Wassink TH, et al. Cerebral cortical gray matter overgrowth and functional variation of the serotonin transporter gene in autism. Arch. Gen. Psychiatry. 2007;64:709–717. doi: 10.1001/archpsyc.64.6.709. [DOI] [PubMed] [Google Scholar]
- 134.Sutcliffe JS, et al. Allelic heterogeneity at the serotonin transporter locus (SLC6A4) confers susceptibility to autism and rigid-compulsive behaviors. Am. J. Hum. Genet. 2005;77:265–279. doi: 10.1086/432648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yonan AL, et al. A genomewide screen of 345 families for autism-susceptibility loci. Am. J. Hum. Genet. 2003;73:886–897. doi: 10.1086/378778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Weiss LA, et al. Variation in ITGB3 is associated with whole-blood serotonin level and autism susceptibility. Eur. J. Hum. Genet. 2006;14:923–931. doi: 10.1038/sj.ejhg.5201644. [DOI] [PubMed] [Google Scholar]
- 137.Coutinho AM, et al. Evidence for epistasis between SLC6A4 and ITGB3 in autism etiology and in the determination of platelet serotonin levels. Hum. Genet. 2007;121:243–256. doi: 10.1007/s00439-006-0301-3. [DOI] [PubMed] [Google Scholar]
- 138.Auranen M, et al. A genomewide screen for autism-spectrum disorders: evidence for a major susceptibility locus on chromosome 3q25–27. Am. J. Hum. Genet. 2002;71:777–790. doi: 10.1086/342720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Coon H, et al. Evidence for linkage on chromosome 3q25–27 in a large autism extended pedigree. Hum. Hered. 2005;60:220–226. doi: 10.1159/000090546. [DOI] [PubMed] [Google Scholar]
- 140.Liu J, et al. A genomewide screen for autism susceptibility loci. Am. J. Hum. Genet. 2001;69:327–340. doi: 10.1086/321980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Butler MG, et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J. Med. Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bonaglia MC, et al. Disruption of the ProSAP2 gene in a t(12;22)(q24.1;q13.3) is associated with the 22q13.3 deletion syndrome. Am. J. Hum. Genet. 2001;69:261–268. doi: 10.1086/321293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wilson HL, et al. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J. Med. Genet. 2003;40:575–584. doi: 10.1136/jmg.40.8.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jamain S, et al. Linkage and association of the glutamate receptor 6 gene with autism. Mol. Psychiatry. 2002;7:302–310. doi: 10.1038/sj.mp.4000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shuang M, et al. Family-based association study between autism and glutamate receptor 6 gene in Chinese Han trios. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2004;131:48–50. doi: 10.1002/ajmg.b.30025. [DOI] [PubMed] [Google Scholar]
- 146.Kim SA, Kim JH, Park M, Cho IH, Yoo HJ. Family-based association study between GRIK2 polymorphisms and autism spectrum disorders in the Korean trios. Neurosci. Res. 2007;58:332–335. doi: 10.1016/j.neures.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 147.Yirmiya N, et al. Association between the arginine vasopressin 1a receptor (AVPR1a) gene and autism in a family-based study: mediation by socialization skills. Mol. Psychiatry. 2006;11:488–494. doi: 10.1038/sj.mp.4001812. [DOI] [PubMed] [Google Scholar]
- 148.Bailey DB, Jr, et al. Autistic behavior in young boys with fragile X syndrome. J. Autism Dev. Disord. 1998;28:499–508. doi: 10.1023/a:1026048027397. [DOI] [PubMed] [Google Scholar]
- 149.Ma DQ, et al. Dissecting the locus heterogeneity of autism: significant linkage to chromosome 12q14. Mol. Psychiatry. 2007;12:376–384. doi: 10.1038/sj.mp.4001927. [DOI] [PubMed] [Google Scholar]
- 150.Ylisaukko-oja T, et al. Genome-wide scan for loci of Asperger syndrome. Mol. Psychiatry. 2004;9:161–168. doi: 10.1038/sj.mp.4001385. [DOI] [PubMed] [Google Scholar]
- 151.Wang L, et al. Association of the ENGRAILED 2 (EN2) gene with autism in Chinese Han population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007 Oct 19; doi: 10.1002/ajmg.b.30623. doi: 10.1002/ajmg.b.30623. [DOI] [PubMed] [Google Scholar]
- 152.Brune CW, et al. Heterogeneous association between engrailed-2 and autism in the CPEA network. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007 Oct 19; doi: 10.1002/ajmg.b.30585. doi: 10.1002/ajmg.b.30585. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.