Abstract
Hypoxia-inducible factor-1 is a known cancer progression factor, promoting growth, spread, and metastasis. However, in selected contexts HIF-1 is a tumor suppressor coordinating hypoxic cell cycle suppression and apoptosis. Prior studies focused on HIF-1 function in established malignancy, however little is known about its role during the entire process of carcinogenesis from neoplasia induction to malignancy. Here we tested HIF-1 gain of function during multistage murine skin chemical carcinogenesis in K14-HIF-1αPro402A564G (K14-HIF-1αDPM) transgenic mice. Transgenic papillomas appeared earlier and were more numerous, 6±3 transgenic versus 2±1.5 nontransgenic papillomas per mouse, yet they were more differentiated, their proliferation was lower, and their malignant conversion was profoundly inhibited, 7% in transgenic versus 40% in non transgenic mice. Moreover, transgenic cancers maintained squamous differentiation whereas epithelial mesenchymal transformation was frequent in nontransgenic malignancies. Transgenic basal keratinocytes upregulated the HIF-1 target N-myc downstream regulated gene-1, a known tumor suppressor gene in human malignancy, and its expression was maintained in transgenic papillomas and cancer. We also discovered a novel HIF-1 target gene, selenium binding protein-1 (selenbp1), a gene of unknown function whose expression is lost in human cancer. Thus, HIF-1 can function as a tumor suppressor through transactivation of genes that are themselves targets for negative selection in human cancers.
Keywords: HIF-1alpha, carcinogenesis, EMT, selenium, NDRG1
INTRODUCTION
The transcription factor HIF-1 is a fundamental mediator of cellular adaptation to microenvironmental stresses such as hypoxia, free radical exposure, activation of oncogenes, loss of tumor suppressor genes, or enhanced survival signaling (1, 2). HIF-1α is induced in multiple tissue biological contexts including embryonic development, hematopoiesis, inflammation, cerebral and myocardial ischemia, and carcinogenesis (3).
HIF-1 can contribute to cancer progression in multiple, nonoverlapping mechanisms by transactivating suites of genes regulating the microvasculature, glycolysis, oxidative phosphorylation, cell motility, migration, tissue invasion, and metastasis (1, 4). Detectable and elevated levels of HIF-1α protein are present in premalignant lesions, and in epithelial, soft tissue, CNS, and hematological malignancies (1, 5). HIF-1α protein stabilization is also associated with poor prognosis in most of these malignancies (1).
However, the concept that HIF-1α invariably facilitates growth and spread of cancer is counterbalanced by HIF-1α’s putative role as a tumor suppressor. Thus HIF-1α upregulation has been associated with increased patient survival in patients with squamous carcinomas of the head and neck, or the lung (6, 7). Hypoxic cell cycle arrest mediated by p21 and p27 induction appears to require HIF-1α (8-10). Moreover, loss of HIF-1α function is also associated with enhanced outgrowth of ES cell carcinomas (10). HIF-1α has also been shown to bind and sequester p53 as a mechanism for enhanced genomic instability associated with chronic cellular hypoxia (11).
Additionally, in certain cellular contexts such as renal cell or lung cancer, HIF-1α directly and indirectly inhibits c-Myc function, resulting in either p21-mediated cell cycle arrest in the former, or apoptosis in each cancer type (12, 13). Induction of the proapoptotic target genes BNIP3 or NIX may also explain the tumor suppressor capability of HIF-1α (14). However, each of these facets of HIF-1α-mediated tumor suppression is controversial. The precise roles of cyclin-dependent kinases in hypoxic cell cycle arrest and the functional significance of either HIF-1α-p53, or HIF-1α-c-myc interactions remain complex, and are dependent on cell-type, and levels of oncogene expression (15-17). The contribution of HIF-1α or hypoxia-induced BNIP3 to cancer cell apoptosis has also been questioned (18).
We designed our experiments to test HIF-1 gain of function throughout the entire process of epithelial carcinogenesis from cancer stem cell initiation and subsequent promotion to malignant conversion of high-risk premalignant precursors using a combination of two-stage chemical carcinogenesis and K14-HIF-1αPro402A/P564G (HIF-1αDPM) transgenic mice (19). Use of the keratin-14 promoter targeted transgene expression to basal keratinocytes (20), whereas mutation of prolines 402 and 564 rendered the HIF-1α protein resistant to binding by the ubiquitin ligase pVHL (see references within (19)). HIF-1 gain of function facilitated the outgrowth of initiated foci into papillomas, however differentiation of these premalignant neoplasias was maintained and malignant conversion was potently inhibited. Expression of N-Myc downstream-regulated gene-1 (NDRG1), a known HIF-1 target (21), and a gene with tumor suppressor functions (22, 23), was upregulated and redistributed to basal keratinocytes of transgenic skin and papillomas. We also discovered a novel HIF-1 target gene, selenium binding protein-1 (SELENBP1), whose loss of function in several common epithelial cancers (24-26) suggested that it too had tumor suppressor function.
METHODS
Animal Studies
K14-HIF-1αDPM transgenic mice, created and used in the FVB/n in bred strain, were described previously (19). Mice were housed in pathogen-free conditions, and the Washington University Animal Studies Committee approved all experiments described in this study. For mouse skin carcinogenesis, the back skin of each mouse was shaved 2 days before topical treatment, with DMBA, 25 μg in 200 μl of acetone. One week later, mice were topically treated with 12.5 μg of 12-O-tetradecanoylphorbol-13-actate (TPA) (Sigma) in 200 μl of acetone, once a week for 20 weeks. Mice were sacrificed either when lesions with an appearance consistent with cancers reached 1 cm in diameter, or arbitrarily at 25 weeks post-initiation.
Tissue harvest and histology
Tissues for routine histopathology and selected immunohistochemical analysis were obtained from formalin perfused mice, fixed using a microwave processing technique (19), and following a PBS wash, processing through graded alcohols and xylenes, were embedded in paraffin. Five micron tissue sections were stained with hematoxylin/eosin, or underwent immunohistochemical analysis.
Histopathology and Immunohistochemistry
All sections were deparaffinized, rehydrated, washed in PBS, and blocked with Dako protein block (Carpinteria, CA) for 30 minutes at room temperature (RT). Antigen retrieval was performed in a pressure cooker (Decloaking chamber, Biocare Medical, Concord, CA) in citrate buffer, pH 6.0, and used for anti-cytokeratin 10, biotinylated anti-cytokeratin 14, anti-desmin and anti-E-cadherin immunostaining. Sections immunostained for bromodeoxyuridine (BrdU) were pretreated with 2N HCl for one hour RT and 0.01% protease (Type XXIV, Sigma) for 15 seconds. Antibodies/dilutions for the following markers were used in Dako antibody diluent and applied overnight at 4°C: rabbit anti-cytokeratin 10 and 14 (1:50, 1:2,000, Covance, Princeton, NJ), biotinylated anti-cytokeratin 14 (1:1250, Lab Vision, Fremont, CA), goat anti-N-myc downstream-regulated gene 1 (NDRG1) (1:200, Santa Cruz, Santa Cruz, CA), rabbit anti-desmin (1:250), rabbit E-cadherin (1:200, Cell Signaling, Danvers, MA), biotinylated anti-BrdU (1:200, Caltag, Carlsbad, CA), and rat anti-Panendothelial Cell Antigen (MECA-32) (1:20 BD Pharmingen, San Diego, CA) cytokeratin 8 antibody (TROMA-1) (1:1000, Developmental Studies Hybridoma Bank, Iowa City, IA). Secondary antibodies labeled with Alexa Fluor 488, Alexa Fluor 594, or Alexa Fluor 594 strepavidin conjugate were placed on tissue sections for one hour at RT (1:400, Molecular Probes, Eugene, OR). Nuclei were counterstained using SlowFade Gold Anti-fade reagent with DAPI (Vector, Burlingame, CA).
BrdU incorporation
DMBA/TPA treated mice were injected i.p. with 100 mg/kg 5-bromodeoxyuridine (BrdU) (Sigma, St. Louis, MO). Tissue was collected after 3 hours after BrdU injection for measurement of cell replication. Incorporated BrdU was detected as described above.
Cell lines
PDV cells originated from a DMBA-treated C57BL/6 neonatal keratinocyte culture (27). HEK 293, and PDV cells were cultured in DMEM (Life Technologies, Grand Island, NY), supplemented with 10% fetal bovine serum (Sigma, St. Louis, MO), 1000 U/ml penicillin-streptomycin (Sigma, St. Louis, MO) at 37°C in 21% O2/5% CO2. Where indicated, PDV cell were treated with 100 μM CoCl2 (Sigma, St. Louis, MO) for 24 hours.
Keratinocyte culture
Primary mouse keratinocytes were isolated from newborn transgenic and wild-type littermate epidermis as described previously (19), and seeded at 5 × 106 cells per 60 mm dish (or equivalent concentrations) in Ca2+ - and Mg2+ -free Minimal Essential Media (Invitrogen Life Technologies, Carlsbad, CA) supplemented with 8% chelexed (Bio-Rad Laboratories, Hercules, CA) fetal bovine serum (Gemini Bioproducts, Woodland, CA) and 0.2 mM Ca2+. After 24 hr, cultures were switched to the same medium with 0.05 mM Ca2+ to select for basal cells.
Microarray analysis
Primary keratinocytes ((n=3) for nontransgenic and transgenic) were homogenized in TRIzol (Invitrogen, Carlsbad, CA); total RNA isolated using the manufacturer’s guidelines, and treated with RQ1 DNase (Promega, Madison, WI) for 30 minutes at 37°C. Microarray probes were synthesized using RNA samples and hybridized with Affymetrix MU430Av2 GeneChips in the Siteman Cancer Center Multiplexed Gene Analysis Core, and differentially expressed transcripts identified using unpaired t-tests.
Plasmid construction, transfections and luciferase reporter assays
A 479bp (-635 to –156) fragment from the selenbp1 (NC_000069) promoter was PCR amplified from mouse tail DNA and cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, CA). The insert was sequenced, and subcloned into pRL-null (Promega, Madison, WI) (pSELENBP1-Luc). The full length HIF-1αP402A/P564A/N803A cDNA, a kind gift from R. Bruick, was cloned into pIRES-hrGFP2a (Stratagene, La Jolla, CA). Human embryonic kidney 293 cells (HEK293), passaged in 48–well plates, were co-transfected with DNA-liposome complexes Lipofectamine 2000 (Invitrogen Life technology, Carlsbad, CA), containing 15 ng of either pRL-null reporter alone, or pSELENBP1-Luc; with either 200 ng of pIRES-hrGFP2a, or increasing amounts (50ng, 100ng, 200ng) of CMV-HIF-1α-PPN overnight. We determined luciferase activity in quadruplicate transfections using a Synergy HT luminometer (Bio-Tek, Winooski, VT).
Quantitative RT-PCR
Back skin, papillomas, and cancers were snap frozen in liquid nitrogen, homogenized in TRIzol (Invitrogen, Carlsbad, CA), and total RNA was treated in RQ1 DNase (Promega, Madison, WI) for 30 minutes at 37°C. RT-PCR was performed as described previously (19) using a Stratagene MX3000P (La Jolla, CA). Primer Express software (version 2.0 Applied Biosystems) was used to design primer/probe sets (Supplementary Table 1) to amplify the genes described below. All target cDNA’s were normalized to histone 3.3A (28).
Western blotting
Cells or tissues were lysed in radioimmunoprecipitation (RIPA) buffer (10 mM Tris (pH 7.4), 150 mM NaCl, 1 mM EDTA, 0.1% sodium dodecyl sulfate, 1% Triton X-100, 1% sodium deoxycholate, 0.25 mM phenylmethylsulfonylfluoride), 20 μg of total lysate loaded on polyacrylamide gels, transferred to polyvinylidene difluoride (PVDF) membranes (Amersham Biosciences, Buckinghamshire, United Kingdom), blocked in Blotto-Tween solution (5% nonfat dry milk, 0.1% Tween in PBS) for 1 hr, and incubated overnight in PBS with a rabbit polyclonal anti-SELENBP1 (1:20,000, a gift from D. Medina, Baylor College of Medicine), goat polyclonal anti-NDRG1 (1:1,000, Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-cytokeratin 10 and 14 (1:500, Covance, Princeton, NJ), rabbit polyclonal anti-Snail and E-cadherin (1:1,000, Cell Signaling, Danvers, MA), or beta-tubulin (1:5,000, Sigma, Saint Louis, MO). Peroxidase-coupled AffiniPure goat anti–rabbit or donkey anti-goat immunoglobulin G (1:200, Jackson Immunoresearch Laboratories, West Grove, PA) was used in secondary incubations for 1 hr followed by chemiluminescence detection (ECL Plus, Amersham Biosciences, Piscataway, NJ).
Microscopy
All microscopy images were obtained with a BX61 microscope, (Olympus America, Central Valley, PA) using the following objectives, UPlan Apochromatic 4X/NA 0.16, UPlan Apochromatic 10X/NA 0.40, UPlan Apochromatic 20X/NA 0.70, and UPlan Apochromatic 40X/NA 0.85. Tissue sections stained with hematoxylin/eosin or diaminobenzadine immunoperoxidase were mounted with Permount (SP15-500, Fisher Scientific, Pittsburgh, PA) and coverslipped Microscopy images were obtained with a DP70 color bayer mosaic digital camera, Peltier device cooled to -10°C, (Olympus). Tissue sections for fluorescence microscopy images were mounted with SlowFade Gold antifade reagent with4’,6-diamidino-2-phenylindole (DAPI) (Invitrogen, Molecular Probes, Eugene OR), coverslipped, and images were obtained with a Soft Imaging Solutions FVII cooled monochrome digital camera, Peltier cooled to -10°C (Olympus). All images were captured with MicroSuite Biological Suite version 5 software, (Olympus Soft Imaging Solutions, Lakewood, CO) and resized and formatted with Adobe Photoshop CS3 software (Adobe Systems Incorporated, San Jose, CA).
Statistical analysis
Data were analyzed using GraphPad PRISM (San Diego, CA), results expressed as the mean +/-SEM, and statistical significance determined using either the Student’s t-or Mann-Whitney U test, or contingency table analysis.
RESULTS
K14-HIF1-αDPM mice exhibit propensity for benign neoplasms with resistance to cancer formation
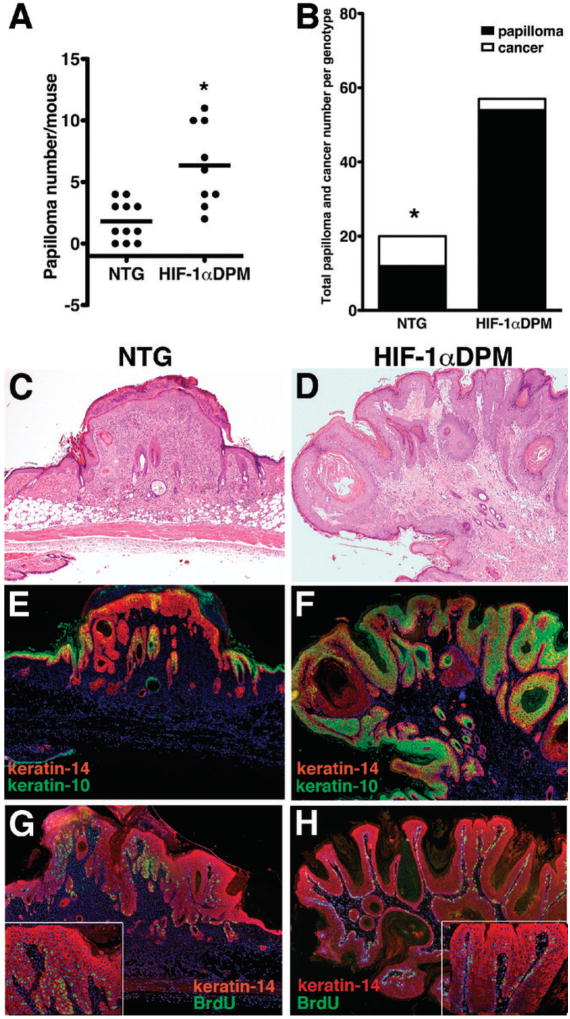
To study the effects of HIF-1 gain of function at initiation and then progression of epithelial carcinogenesis, we performed the classical two-stage carcinogenesis protocol on K14-HIF1-αDPM transgenic and nontransgenic mice. However, twice a week TPA application caused an exacerbated back skin inflammation in transgenic mice, consistent with our recent work (19). Therefore, we began a new study with an unchanged DMBA dose, but half the dose and frequency of TPA application that was well tolerated by transgenic mice for 20 weeks of promotion. The nontransgenic papilloma frequency, 2 ± 1.5 per mouse (Figure 1, Panel A), was consistent with previous work with a similar chemical carcinogenesis protocol in FVN/n mice (29). Moreover 40% of all papillomas, n=20 for the entire group of nontransgenic mice, converted to cancer (Figure 1, Panel B). Our low nontransgenic papilloma frequency with enhanced malignant conversion rate was consistent with induction of “high-risk” papillomas (30). Transgenic mice developed three-fold more papillomas, 6 ± 3 per mouse, than nontransgenic counterparts (Figure 1, Panel A), and the total number of papillomas in the entire transgenic cohort, 57, was also nearly three-fold higher than the 20 papillomas in the nontransgenic study group (Figure 1, Panel B). These data were consistent with an increase in initiation and growth facilitation of transgenic initiated foci, by HIF-1 gain of function. In contrast to our previous work (19) there was no differential increase in CD45 positive cells in transgenic papillomas, likely due to dose and interval reduction of TPA application (data not shown). Surprisingly, the malignant conversion rate of each individual transgenic papilloma, 7%, was seven-fold lower than nontransgenic counterparts (Figure 1, Panel B). Previously we discovered that HIF-1α overexpression induced angiogenesis (28) and this was present in both transgenic papillomas and cancers (Supplementary Figure 1, Panels A and C). Increased blood vessel density was previously documented to be an early event required for papilloma development (31, 32), and it could explain the enhanced papillomagenesis in transgenic mice.
Figure 1. K14-HIF-1αPro402A/564G (DPM) transgenic mice exhibit propensity for benign neoplasm with resistance to cancer formation.
Papilloma frequency per mouse is presented in Panel A, and malignant conversion rate for the collective number of papillomas for each entire experimental group, nontransgenic (NTG) and transgenic K14-HIF-1αPro402A/564G (HIF-1αDPM) depicted in Panel B. Histology of NTG, Panel C, and HIF-1αDPM papillomas, Panel D. Expression of epidermal keratins specific for proliferating keratinocytes (keratin-14 red fluorescence) and differentiated suprabasal keratinocytes (keratin-10, green fluorescence) is depicted in Panels E, NTG, and F, HIF-1αDPM. Note the loss of keratin-10 in the NTG, and its persistent expression in HIF-1αDPM transgenic papillomas. Bromodeoxyuridine (BrdU) positive neoplastic keratinocytes (green fluorescence) were frequent and detectable in both basal and suprabasal layers in nontransgenic papillomas, Panel G, whereas BrdU positive keratinocytes were less frequent and restricted to the basal layer in transgenic papillomas, Panel H, and inserts). Magnification is 40X in Panels B-D, 200X in inserts of Panels D).
The resistance of transgenic papillomas to malignant conversion resistance led us to further determine differences in their tissue biology compared to nontransgenic counterparts (Figure 1, Panels C and D). Nontransgenic papillomas (Figure 1 Panel E), evidenced loss of the differentiation-associated keratin-10 (Figure 1, Panel E), while transgenic papillomas retained keratin-10 expression (Figure 1, Panel F). Moreover, epidermal proliferation was enhanced in nontransgenic papillomas; BrdU-positive keratinocytes were detectable in both the basal and suprabasal layers (Figure 1, Panel G and insert, see also Supplementary Figure 2, Panel A), whereas BrdU-positive transgenic keratinocytes were confined to the papilloma basal layer Figure 1, Panel H, insert, and Supplementary Figure 2, Panel B). Diminution of mRNA levels of Ki67 and cyclin D1, (Supplementary Figure 3, Panels A and B) further supported the conclusion that transgenic papilloma proliferation was inhibited.
Suppression of epithelial-mesenchymal transition in transgenic squamous cancers
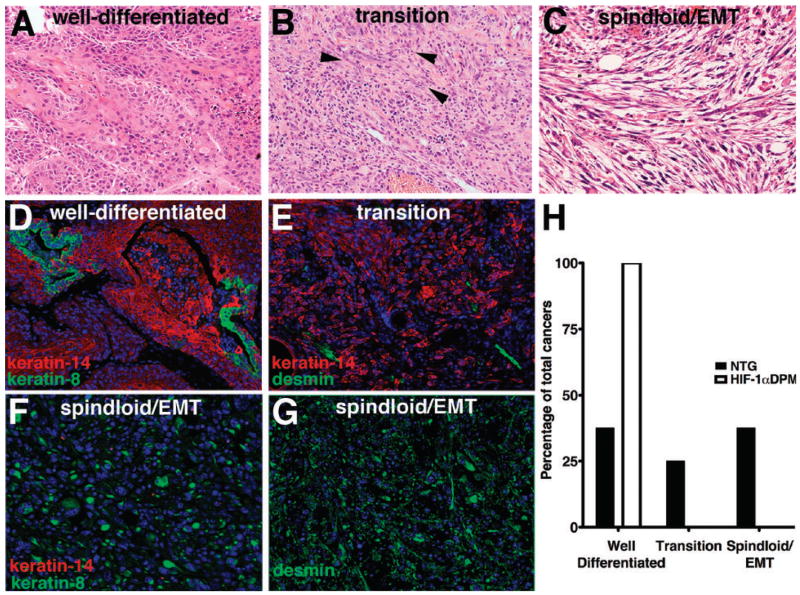
In order to determine whether resistance of transgenic papillomas to malignant conversion also produced a different cancer phenotype compared to transgenic counterparts, we histopathologically classified all squamous cancers in the study (31), into three histotype groups: well differentiated SCC-most of the tumor mass composed of epithelial cells, with eosinophilic cytoplasmic staining consistent with keratin expression (Figure 2, Panel A); transition lesions-consisting of epitheliod cells with a paucity of keratinization and apparent gaps between adjacent malignant cells consistent with decreased homotypic cellular adhesion (Figure 2, Panel B); and poorly differentiated spindloid tumors with no keratinization (Figure 2, Panel C). This histopathological differentiation classification was bolstered by immunohistochemical analysis of patterns of K14 and K8 expression (20) wherein well differentiated cancers displayed strong immunofluorescence for K14, with only sporadic K8 detection (Figure 2, Panel D). Transition cancers displayed an overall diminished signal for K14 with a sporadic desmin signal localized in linear-tube-like structures consistent microvessels (Figure 2, Panel E). Spindloid cancers evidenced strong, but scattered K8 immunofluorescence, undetectable K14 (Figure 2, Panel F), and a ubiquitous desmin signal (Figure 2, Panel G), a hallmark of epithelial-mesenchymal transition (EMT). Using these criteria, all three transgenic cancers were well differentiated (Figure 2, Panel H). In contrast, only 37.5% of nontransgenic cancers were well differentiated, another 25% were transition lesions, and 37.5% were spindloid/EMT cancers (Figure 2, Panel H). Thus HIF-1 gain of function not only inhibited conversion of premalignant lesions to cancer, it also suppressed development of poorly differentiated malignancies.
Figure 2. HIF-1 gain of function suppresses development of poorly differentiated malignancies in epithelial squamous cancers.
Histological classification of malignant differentiation (Panels A-C) produced in this model of two-stage DMBA/TPA squamous carcinogenesis. Well-differentiated cancers were composed of cells containing large amounts of eosinophilic cytoplasm (Panel A). Transition cancers were contained epithelioid cells (Panel B) with a tendency towards a fibroblastic phenotype (arrowheads). Poorly differentiated spindloid/epithelial mesenchymal transition (EMT) cancers evidenced a frank spindloid histopathology (Panel C). Well differentiated squamous cancers retained strong keratin-14 expression with small clusters of cells expressing the “simple epithelial, keratin-8 (Panel D), transition lesions expressed less prominent keratin-14 expression (Panel E), whereas spindloid/EMT lesions solely expressed keratin-8 and desmin (Panels F and G). The incidence of each of these histotypes is displayed in Panel H. Magnification 200X, Panels A-G.
Induction of the tumor suppressor genes NDRG1 and SELENBP1 by basal keratinocyte HIF-1α gain of function
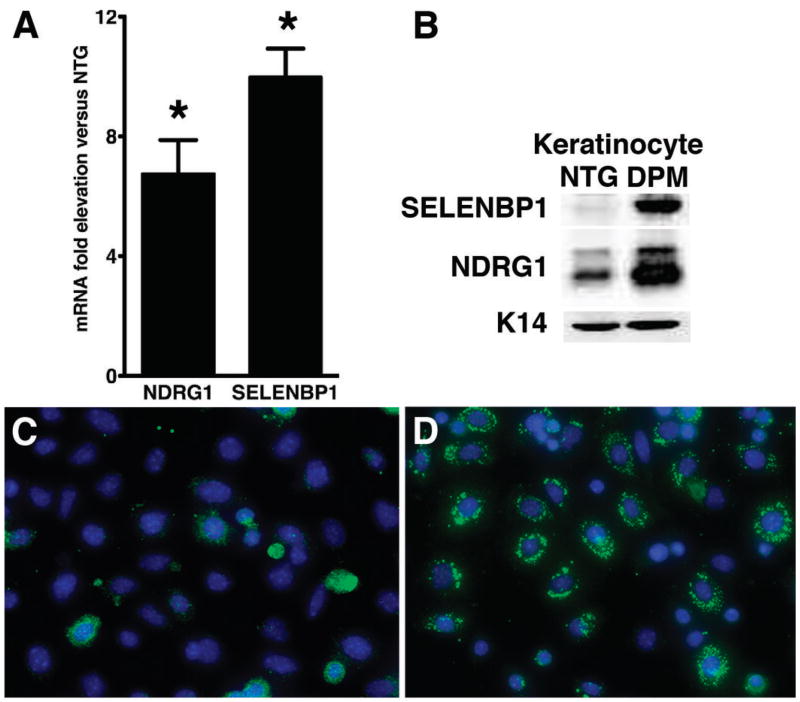
As our tissue-derived data suggested that HIF-1 was mediating repression of squamous carcinogenesis via cell autonomous mechanisms, we established primary keratinocyte cell cultures from 2-3 day old neonates and performed microarray analysis (Supplementary Table 2) focusing on differential expression of genes known to be involved in regulation of differentiation or proliferation. Surprisingly, and again consistent with the carcinogenesis data, we did not detect a significant differential downregulation of E-cadherin mRNA in transgenic keratinocytes (data not shown). However, we discovered a seven-fold increase of N-Myc downstream regulated gene-1 (NDRG1) mRNA expression, a previously characterized HIF-1 target gene (21). We also found a 80-fold induction in selenium binding protein-1 and -2 (SELENBP1, -2) mRNAs (33). Mouse SELENBP1 and SELENBP2 proteins are highly related, with only 20 nucleotides and 14 amino residues differentiating the two genes in the coding sequence (34), making it impossible to separate their expression by real time RT-PCR or even protein expression. Despite their similarity, the two genes are regulated differently, and likely have different functions in that selenbp2 appears to be involved in acetaminophen metabolism (34). Our subsequent analysis determined that alterations in protein expression during carcinogenesis were likely due to SELENBP1 expression (see below), so we used this terminology collectively when unable to determine the genes individually. As antibodies for immunohistochemical detection of SELENBP1 in mouse tissues were not available, in contrast to NDRG1, we restricted our analysis of expression of SELENBP1 protein or its mRNA to tissue or cell extracts. We validated the transgenic induction of NDRG1 and SELENBP1 in independent sets of keratinocyte cultures and found a ten-fold elevation of SELENBP1 mRNA and protein, and a seven-fold increase in NDRG1 mRNA and protein in transgenic compared to nontransgenic cultures (Figure 3, Panels A and B). Transgenic NDRG1 overexpression was further validated by immunofluorescence revealing markedly enhanced punctate cytoplasmic NDRG1 protein expression (Figure 3, Panels C and D) consistent with previously reported localization to recycling/sorting vesicles (35).
Figure 3. Differential increase NDRG1 and SELENBP1 expression is cell autonomous in transgenic keratinocytes.
Fold elevation of NDRG1 and SELENBP1 mRNA in primary transgenic keratinocyte cultures (Panel A) compared to nontransgenic counterparts. Western blotting of keratinocyte culture extracts revealed a similar marked induction of SELENBP1 and NDRG1 protein in transgenic versus nontransgenic cultures, keratin-14 is a loading control (Panel B). Immunofluorescence analysis of NDRG1 expression revealed enhanced punctate cytoplasmic NDRG1 protein expression in transgenic primary keratinocytes (Panel D, green fluorescence) compared to nontransgenic keratinocytes (Panel C). Error bars represent mean ± SEM.
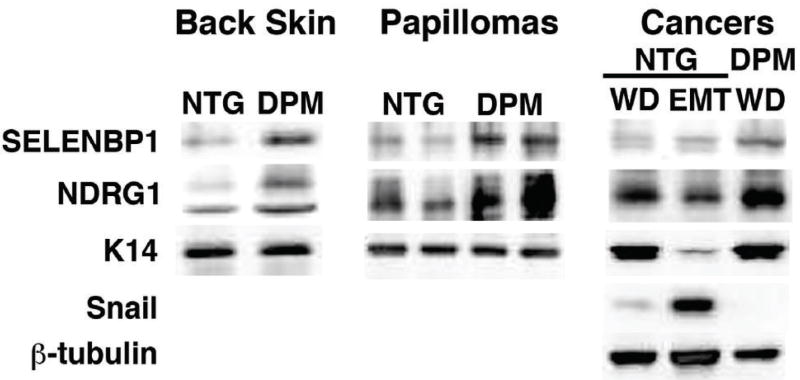
Enhanced NDRG1 and SELENBP1 mRNA (data not shown) and protein expression was also present in untreated transgenic back skin that contained a two fold increase in the former and a four-fold elevation of the latter protein (Figure 4) respectively. HIF-1 gain of function also redirected NDRG1 expression from the terminally differentiated suprabasal layer and inner root sheath of nontransgenic skin (Figure 5, Panel A and Supplementary Figure 4, Panels A and C) (36), to the proliferative basal cell compartment of transgenic epidermis (Figure 5, Panel B, and Supplementary Figure 4, Panels B and D). The paranuclear localization of NDRG1 expression in transgenic skin (arrowheads, Supplementary Figure 4, Panel B), was consistent with localization in Trans Golgi recycling/sorting vesicles (35).
Figure 4. Upregulation of ndrg1 and selenbp1 protein in transgenic back skin, papillomas and carcinomas.
NDRG1 and SELENBP1 protein (Panel A) were increased in transgenic (DPM, K14-HIF-1αDPM) back skin, left panel, compared to nontransgenic controls (NTG), and differentially elevated in papillomas, middle panel, keratin-14 was a loading control, in both blots. In cancers, right panel, NDRG1 protein was 3-fold lower in Snail-high (EMT) compared to Snail-low (WD) nontransgenic protein extracts, whereas SELENBP1 protein was expressed at a low level in nontransgenic cancers compared to SELENBP1 expression in transgenic cancers independent of Snail expression.
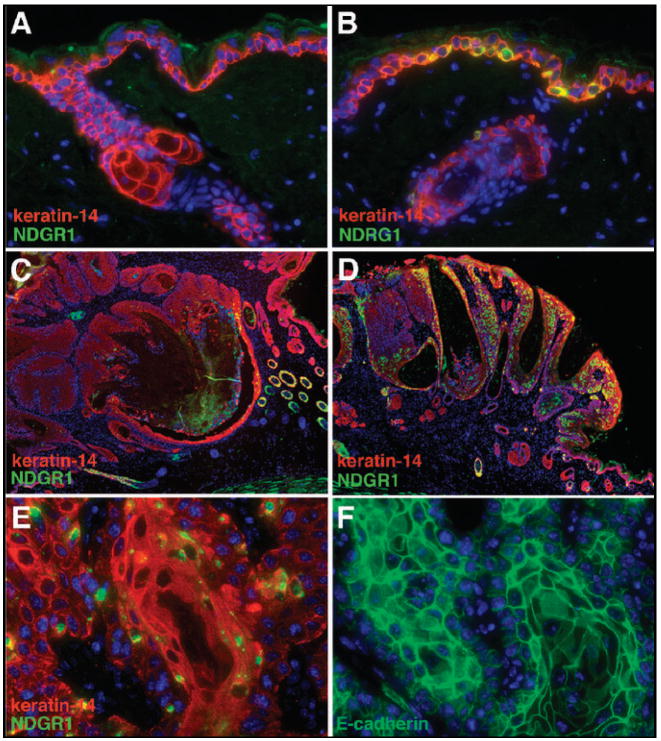
Figure 5. immunofluorescent localization of NDRG1 protein expression in NTG versus transgenic mice during each stage of carcinogenic progression.
Low-level NDRG1 expression (green fluorescence) in the differentiated suprabasal layer of NTG skin (Panel A) in contrast to strong paranuclear expression in proliferative basal keratinocytes in transgenic back skin (Panel B). Sporadic NDRG1 expression restricted to sporadic neoplastic epidermal cells in nontransgenic papillomas and adjacent, nonpapillomatous skin (Panel C) contrasted to prominent and diffuse expression in transgenic papillomas (Panel D). Enhanced NDRG1 expression in well-differentiated regions of squamous transgenic cancers (Panel E) with adjacent sections demonstrating retention of membrane bound E-cadherin (Panel F). Keratin-14 was used to mark basal cells in Panels A-E. Magnification 400X in Panels A, B, and E, and F; 40X in Panels C and D.
To further investigate potential NDRG1 and SELENBP1 tumor suppressor functions we determined the levels and patterns of expression of these molecules in papillomas, the premalignant precursor lesion of chemical carcinogenesis. NDRG1 protein expression was markedly downregulated in nontransgenic papillomas (Figure 5, Panel C), in contrast to its strong expression in the basal and suprabasal neoplastic epidermal layers of transgenic papillomas (Figure 5, Panel D). Western blotting revealed a two-fold increase of NDRG1 protein in transgenic compared to nontransgenic papillomas (Figure 4). SELENBP1 protein was also elevated 3-fold in transgenic papilloma extracts (Figure 4). Elevated NDRG1 and SELENBP1 protein expression also correlated with a three-fold elevation of expression of keratin-10, a differentiation marker, in transgenic, compared to nontransgenic papilloma extracts (data not shown). NDRG1 was known to induce differentiation in colon cancer cell lines and was also elevated by several different differentiation inducing agents (22). As commitment to terminal differentiation is initiated in the epidermal basal layer (20), expression of a molecule such as NDRG1 in stem-like/transient amplifying cells could have been responsible for differentiation maintenance and lower proliferation of transgenic papillomas.
As both NDRG1 and SELENBP1 expression were lost and correlated with poor prognosis in human cancers (22, 24-26, 37), we determined expression of these molecules in nontransgenic and transgenic cancers (Figure 4). Nontransgenic protein extracts were correlated to the histological presence or absence of EMT. Snail, an EMT marker and initiator (38) was not detectably expressed in transgenic cancers despite gain of HIF-1α function (Figure 4) (39, 40), whereas it was consistently expressed at either a low or high level in either histologically well differentiated or spindloid nontransgenic malignancies respectively (Figure 4). NDRG1 protein was 3-fold lower in Snail high compared to Snail low nontransgenic protein extracts (Figure 4). SELENBP1 protein was expressed at a low level in nontransgenic cancers, independent of Snail expression, compared to elevated SELENBP1 levels in transgenic cancers (Figure 4). These data were bolstered by immunofluorescence analysis of malignant tissue sections from transgenic (Figure 5, Panels E and F) and nontransgenic mice (data not shown). Strong NDRG1 expression was detectable in well-differentiated regions of transgenic squamous cancers (Figure 5, Panel E). Adjacent sections revealed that these malignant cells retained membrane bound E-cadherin (Figure 5, Panel F). NDRG1 expression was lost in malignant squamous cells of both transition and EMT cancers along with E-cadherin in the latter lesions (data not shown). The association of NDRG1 and E-cadherin expression patterns mirror NDRG1 siRNA knockout studies in prostate cancer cell lines wherein NDRG1 was shown to control E-cadherin recycling and E-cadherin plasma membrane localization (35).
Mouse selenbp1 is a HIF-1α target gene
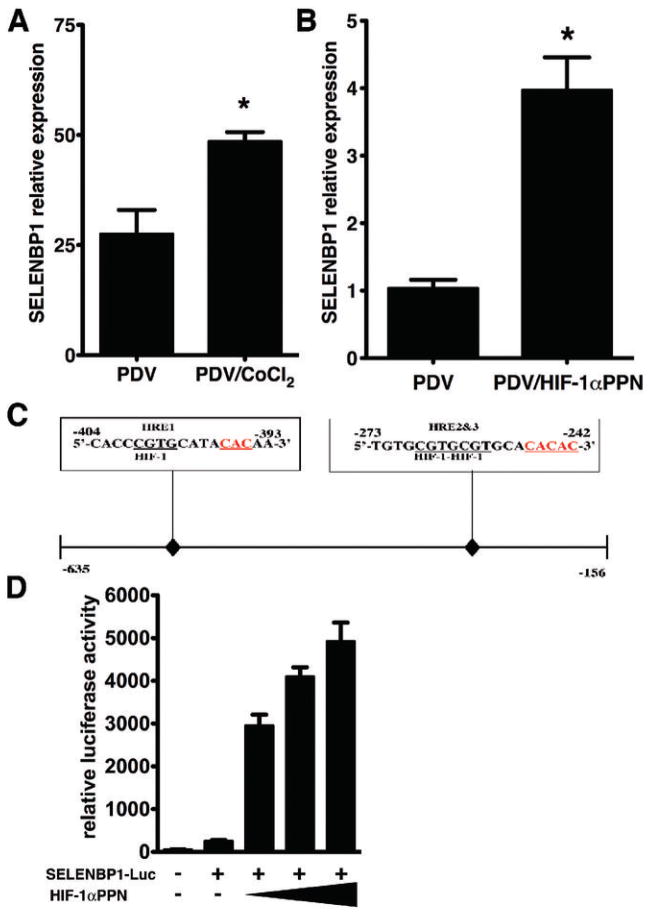
We focused further investigation on the regulation and function of SELENBP1 in keartinocyte and epithelial cell lines. Direct regulation of selenbp1 by HIF-1α was determined by the hypoxia mimetic CoCl2, which produced a two-fold elevation of SELENBP1 mRNA (Figure 6, Panel A) PDV cells (27). Transient overexpression of the triple HIF-1α point mutant, HIF-1αP402A/P564A/N802A (HIF-1αPPN), increased SELENBP1 mRNA four-fold (Figure 6, Panel B).
Figure 6. Mouse SELENBP1 is a bona fide HIF-1α target gene.
CoCl2, a hypoxia-mimetic, differentially elevated SELENBP1 mRNA expression in PDV cells (Panel A). Real-time RT-PCR demonstrated a four–fold elevation of SELENBP1 mRNA in PDV cells transiently transfected with a mutant constitutively active form of HIF-1α (HIF-1αPro402/564A/Asn803A, HIF-1αPPN) in PDV cells (Panel B). A 0.479 kb DNA fragment encompassing three hypoxia response elements, HRE’s, (boxes above sequences) in 5’-promoter region of selenbp1, inserted into a luciferase reporter plasmid (Panel C) demonstrated a titratable four-fold induction of SELENBP1 activity (Panel D) when transfected with 50, 100, and 200 ng of HIF-1αPPN. Error bars represent mean ± SEM. Results are representative of three independent experiments. (*P < 0.05, t-test).
We searched the promoter and first intron regions of the selenbp1 and selenbp2 genes for “hypoxia response elements” (HRE’s); in particular, matches to a consensus sequence containing the core HIF-1-binding site 5’-CGTG-3’, and a 1-8 nucleotide spacer followed by a CAC sequence (41). A total of eight HRE’s were detected in the first 5000bp of the mouse selenbp1 gene, four in the promoter and four in the first intron (data not shown). A 0.479 kb DNA fragment containing the three upstream selenbp1 HRE’s (Figure 6, Panel C) produced a titratable four-fold induction of selenbp1 promoter activity when co-transfected with CMV-HIF-1αP402A/P564A/N802A in HEK293 cells (Figure 6, Panel D). In marked contrast, expression from fragment spanning the homologous promoter region in selenbp2 (data not shown) was unaffected by HIF-1 overexpression. Collectively the data demonstrated that selenbp1 was a bona fide new HIF-1 target gene, and further emphasized the distinction and differential regulation of the murine selenbp1 versus selenbp-2 genes. Notably, the homologous region in the human SELENBP1 promoter contained four HRE’s within 1400 base pairs of the transcriptional start site (data not shown).
DISCUSSION
In this study K14-HIF-1αDPM mice subjected to two-stage carcinogenesis were sensitive to the outgrowth of initiated foci to form benign neoplasms with an inherent resistance to malignant progression. Rapid emergence and frequency of transgenic papillomas was likely facilitated by angiogenesis, a prominent component of this model of HIF-1 gain of function (19, 28). A robust angiogenesis was previously demonstrated in squamous cell hyperplastic foci, the immediate antecedents of nascent papillomas (31). Moreover epidermal VEGF overexpression itself accelerated papilloma development and enhancement of papilloma number, further supporting a functional role for angiogenesis in papillomagenesis (32).
A striking finding in our study was that despite enhanced papillomagenesis, proliferation was diminished and differentiation maintained in transgenic versus nontransgenic papilloma precursor lesions. Moreover, the resistance to EMT of transgenic malignancies was also surprising, as HIFα proteins were known to decrease E-cadherin expression via induction of the transcriptional repressors Snail, TCF3, ZFHX1A, and SIP1 (39, 40). However, one explanation for our findings could be the redirection and induction of NDRG1 in the basal keratinocytes of transgenic epidermis, and maintenance of expression of this molecule in transgenic papillomas and cancers compared to the loss of its expression in nontransgenic counterpart lesions. NDRG1 was a known HIF-1 target with two HRE’s identified upstream of its promoter (21). Several studies confirmed NDRG1’s role in epithelial cell differentiation. NDRG1 increased during epidermal keratinocyte differentiation in culture and localized to the differentiated layers of intact skin (36). However, in contrast to our work, NDRG1 was overexpressed in squamous cancers resulting from two-stage chemical carcinogenesis. The discrepancies between that study and our work was likely due to differences in promotion protocols with our once-weekly TPA administration favoring emergence of EMT and poorly differentiated malignancies, in which we demonstrated loss of NDGR1 expression. NDRG1 was also functionally linked to differentiation of colorectal cancer cells (22), and inhibited tumor cell matrix invasion in colon and prostate cancers (22, 23). Regulation of differentiation and invasion could be linked to the requirement of intact NDRG1 function for E-cadherin recycling and stabilization (35). One comprehensive explanation of cancer resistance in our work could be that NDRG1 upregulation in basal keratinocytes enhanced their fate choice from continual proliferation to commitment to terminal differentiation (20). Maintenance of NDRG1 expression mediated by HIF-1 gain of function could have facilitated continual E-cadherin expression and EMT inhibition in transgenic squamous cancers. The fact that NDRG1 expression is lost in many epithelial malignancies in which HIF-1α is overexpressed (22, 23, 42) could be due in part to negative selection similar to that seen for p53 in hypoxic cell cultures (43). However repression of NDRG1 expression by c-, or N-myc, PTEN, and p53, and DNA methylation (44, 45) could also contribute to loss of function of this gene in human carcinomas.
The resistance for malignant conversion in this experiment was also prima facie surprising given our previous work demonstrating enhanced inflammation in the same K14-HIF-1αDPM transgenic model. Inflammation is known to facilitate cancer development, however the nature and intensity of this process, govern recruitment of distinctive inflammatory cell subsets, helper, Th1, versus suppressor, Th2, T-cells, myeloid suppressor cells, or tumor associated macrophages, that either inhibit or stimulate carcinogenesis (46). In our study biweekly TPA application produced an extremely robust inflammatory reaction and transgenic mice were actually resistant to both papilloma formation and cancers, which mirrored our previous work (data not shown). In contrast, reduction of TPA dosage to once a week, while sufficient for induction of “high-risk” papillomas, did not produce a differential inflammatory reaction in transgenic versus nontransgenic mice as determined by density of CD45 cells. As such, we are confident that even though K14-HIF-1αDPM mice can possess an inflammatory hyperresponsiveness to TPA, we found a dose of this tumor promoter that rendered this variable irrelevant, and supported the potential primacy of the NDRG1 and SELENBP1 HIF-1 targets in carcinogenic inhibition in this model.
We also identified selenbp1 as a novel HIF-1 target gene, the homologue of SELENBP1, in humans (47). As the name suggests, SELENBP1 belongs to the selenium containing protein family. Selenium is an important micronutrient with novel anticancer, “nutraceutical” activities. There is a statistically significant inverse relationship between selenium levels and cancer risk (48) and mechanisms proposed for its nutraceutical activity include antioxidant protection, altered carcinogen metabolism, inhibition of proliferation and tumor cell invasion, and induction of apoptosis (49). SELENBP1 is the only human selenium associated protein in which selenium is covalently bound, in contrast to the more common incorporation of selenocysteine into the amino acid sequence via the TGA codon (33). Most importantly, the cellular levels of SELENBP1 protein are directly regulated by selenium, rising with elevated media selenium concentration (50).
Inferences on SELENBP1 function in normal epithelium and carcinomas were derived from several different types of studies. SELENBP1 expression was widespread in epithelial tissues including kidney, liver, lung, colon, prostate, pancreas, kidney, and ovary (47). SELENBP1 was commonly lost in many types of human epithelial cancers and its loss correlated with poor prognosis (25, 26, 51). The molecular mechanisms of SELENBP1 downregulation in cancer cells have remained a mystery; neither promoter hypermethylation nor gene deletion was responsible (26). Previous work suggested that SELENBP1 exerted tumor suppressor function by inhibition of proliferation. Androgen stimulated LNCaP cells were shown to downregulate SELENBP1 expression (51), and SELENBP1 protein levels correlated with DNA synthesis inhibition (50). However, we were unable to delineate an affect on proliferation or apoptosis in transfection rescue of malignant cells with absent SELENBP1 expression (data not shown). As such, the precise function of this protein remains to be determined.
In summary, we have demonstrated that HIF-1 could definitely function as a tumor suppressor gene potentially by upregulation of a known, NDRG1, and novel gene, SELENBP1, both of which are provocatively lost, and associated with poor prognosis in human epithelial malignancies. One inference of our data is that coordinate negative selection for both of these HIF-1 targets may be present in regions of human cancer with stabilization of HIF-1α protein expression. An outstanding corollary inference is that HIF-1 functions to protect cells against malignant conversion by direct or indirect HIF-1 regulation of cell cycle and apoptosis regulators (8, 9, 14-17), and the more recent demonstration of HIF-1a upregulation in response to keratinocyte UVB irradiation (52). As such, profligate application of HIF inhibitors as novel chemopreventive agents should be carefully evaluated.
Supplementary Material
Acknowledgments
The authors wish to thank Dan Medina for the gift of the rabbit anti-mouse SELENBP1 antibody, Greg Longmore for provision of the PDV cells, Stuart Yuspa, Loren Michel, and Greg Longmore for critical input on the manuscript, and Karline Bringe for technical assistance. The work was supported by NCI R01 CA90722.
References
- 1.Semenza GL. Targeting HIF-1 for cancer therapy. Nature Reviews Cancer. 2003;3(10):721–32. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 2.Zhong H, Chiles K, Feldser D, et al. Modulation of hypoxia-inducible factor 1alpha expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer Research. 2000;60(6):1541–5. [PubMed] [Google Scholar]
- 3.Semenza GL. HIF-1 and tumor progression: pathophysiology and therapeutics. Trends in Molecular Medicine. 2002;8(4 Suppl):S62–7. doi: 10.1016/s1471-4914(02)02317-1. [DOI] [PubMed] [Google Scholar]
- 4.Erler JT, Bennewith KL, Nicolau M, et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440(7088):1222–6. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 5.Shaw RJ. Glucose metabolism and cancer. Current Opinion in Cell Biology. 2006;18(6):598–608. doi: 10.1016/j.ceb.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Nakanishi K, Hiroi S, Tominaga S, et al. Expression of hypoxia-inducible factor-1alpha protein predicts survival in patients with transitional cell carcinoma of the upper urinary tract. Clinical Cancer Research. 2005;11(7):2583–90. doi: 10.1158/1078-0432.CCR-04-1685. [DOI] [PubMed] [Google Scholar]
- 7.Beasley NJ, Leek R, Alam M, et al. Hypoxia-inducible factors HIF-1alpha and HIF-2alpha in head and neck cancer: relationship to tumor biology and treatment outcome in surgically resected patients. Cancer Research. 2002;62(9):2493–7. [PubMed] [Google Scholar]
- 8.Koshiji M, Huang LE. Dynamic balancing of the dual nature of HIF-1alpha for cell survival. Cell Cycle (Georgetown, Tex) 2004;3(7):853–4. doi: 10.4161/cc.3.7.990. [DOI] [PubMed] [Google Scholar]
- 9.Goda N, Ryan HE, Khadivi B, McNulty W, Rickert RC, Johnson RS. Hypoxia-inducible factor 1alpha is essential for cell cycle arrest during hypoxia. Molecular and Cellular Biology. 2003;23(1):359–69. doi: 10.1128/MCB.23.1.359-369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carmeliet P, Dor Y, Herbert JM, et al. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394(6692):485–90. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 11.Koshiji M, To KK, Hammer S, et al. HIF-1alpha induces genetic instability by transcriptionally downregulating MutSalpha expression. Molecular Cell. 2005;17(6):793–803. doi: 10.1016/j.molcel.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 12.Savai R, Schermuly RT, Voswinckel R, et al. HIF-1alpha attenuates tumor growth in spite of augmented vascularization in an A549 adenocarcinoma mouse model. International Journal of Oncology. 2005;27(2):393–400. [PubMed] [Google Scholar]
- 13.Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007;11(4):335–47. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sowter HM, Ratcliffe PJ, Watson P, Greenberg AH, Harris AL. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Research. 2001;61(18):6669–73. [PubMed] [Google Scholar]
- 15.Hammond EM, Giaccia AJ. Hypoxia-inducible factor-1 and p53: friends, acquaintances, or strangers? Clinical Cancer Research. 2006;12(17):5007–9. doi: 10.1158/1078-0432.CCR-06-0613. [DOI] [PubMed] [Google Scholar]
- 16.Dang CV, Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nature Reviews Cancer. 2008;8(1):51–6. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- 17.Green SL, Freiberg RA, Giaccia AJ. p21(Cip1) and p27(Kip1) regulate cell cycle reentry after hypoxic stress but are not necessary for hypoxia-induced arrest. Molecular and Cellular Biology. 2001;21(4):1196–206. doi: 10.1128/MCB.21.4.1196-1206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papandreou I, Krishna C, Kaper F, Cai D, Giaccia AJ, Denko NC. Anoxia is necessary for tumor cell toxicity caused by a low-oxygen environment. Cancer Research. 2005;65(8):3171–8. doi: 10.1158/0008-5472.CAN-04-3395. [DOI] [PubMed] [Google Scholar]
- 19.Scortegagna M, Cataisson C, Martin RJ, et al. HIF-1alpha regulates epithelial inflammation by cell autonomous NFkappaB activation and paracrine stromal remodeling. Blood. 2008;111(7):3343–54. doi: 10.1182/blood-2007-10-115758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arbeit JM. Transgenic models of epidermal neoplasia and multistage carcinogenesis. Cancer Surveys. 1996;26:7–34. [PubMed] [Google Scholar]
- 21.Cangul H. Hypoxia upregulates the expression of the NDRG1 gene leading to its overexpression in various human cancers. BMC Genetics. 2004;5:27. doi: 10.1186/1471-2156-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan RJ, Ford HL, Fu Y, Li Y, Shaw LM, Pardee AB. Drg-1 as a differentiation-related, putative metastatic suppressor gene in human colon cancer. Cancer Research. 2000;60(3):749–55. [PubMed] [Google Scholar]
- 23.Bandyopadhyay S, Pai SK, Gross SC, et al. The Drg-1 gene suppresses tumor metastasis in prostate cancer. Cancer Research. 2003;63(8):1731–6. [PubMed] [Google Scholar]
- 24.Li T, Yang W, Li M, et al. Expression of selenium-binding protein 1 characterizes intestinal cell maturation and predicts survival for patients with colorectal cancer. Molecular Nutrition & Food Research. 2008 doi: 10.1002/mnfr.200700331. [DOI] [PubMed] [Google Scholar]
- 25.Huang KC, Park DC, Ng SK, et al. Selenium binding protein 1 in ovarian cancer. International Journal of Cancer. 2006;118(10):2433–40. doi: 10.1002/ijc.21671. [DOI] [PubMed] [Google Scholar]
- 26.Chen G, Wang H, Miller CT, et al. Reduced selenium-binding protein 1 expression is associated with poor outcome in lung adenocarcinomas. The Journal of Pathology. 2004;202(3):321–9. doi: 10.1002/path.1524. [DOI] [PubMed] [Google Scholar]
- 27.Fusenig NE, Amer SM, Boukamp P, Worst PK. Characteristics of chemically transformed mouse epidermal cells in vitro and in vivo. Bulletin du Cancer. 1978;65(3):271–9. [PubMed] [Google Scholar]
- 28.Elson DA, Thurston G, Huang LE, et al. Induction of hypervascularity without leakage or inflammation in transgenic mice overexpressing hypoxia-inducible factor-1alpha. Genes & Development. 2001;15(19):2520–32. doi: 10.1101/gad.914801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hennings H, Glick AB, Lowry DT, Krsmanovic LS, Sly LM, Yuspa SH. FVB/N mice: an inbred strain sensitive to the chemical induction of squamous cell carcinomas in the skin. Carcinogenesis. 1993;14(11):2353–8. doi: 10.1093/carcin/14.11.2353. [DOI] [PubMed] [Google Scholar]
- 30.Darwiche N, Ryscavage A, Perez-Lorenzo R, et al. Expression profile of skin papillomas with high cancer risk displays a unique genetic signature that clusters with squamous cell carcinomas and predicts risk for malignant conversion. Oncogene. 2007;26(48):6885–95. doi: 10.1038/sj.onc.1210491. [DOI] [PubMed] [Google Scholar]
- 31.Bolontrade MF, Stern MC, Binder RL, Zenklusen JC, Gimenez-Conti IB, Conti CJ. Angiogenesis is an early event in the development of chemically induced skin tumors. Carcinogenesis. 1998;19(12):2107–13. doi: 10.1093/carcin/19.12.2107. [DOI] [PubMed] [Google Scholar]
- 32.Larcher F, Murillas R, Bolontrade M, Conti CJ, Jorcano JL. VEGF/VPF overexpression in skin of transgenic mice induces angiogenesis, vascular hyperpermeability and accelerated tumor development. Oncogene. 1998;17(3):303–11. doi: 10.1038/sj.onc.1201928. [DOI] [PubMed] [Google Scholar]
- 33.Bansal MP, Mukhopadhyay T, Scott J, Cook RG, Mukhopadhyay R, Medina D. DNA sequencing of a mouse liver protein that binds selenium: implications for selenium’s mechanism of action in cancer prevention. Carcinogenesis. 1990;11(11):2071–3. doi: 10.1093/carcin/11.11.2071. [DOI] [PubMed] [Google Scholar]
- 34.Lanfear J, Fleming J, Walker M, Harrison P. Different patterns of regulation of the genes encoding the closely related 56 kDa selenium-and acetaminophen-binding proteins in normal tissues and during carcinogenesis. Carcinogenesis. 1993;14(3):335–40. doi: 10.1093/carcin/14.3.335. [DOI] [PubMed] [Google Scholar]
- 35.Kachhap SK, Faith D, Qian DZ, et al. The N-Myc down regulated Gene1 (NDRG1) Is a Rab4a effector involved in vesicular recycling of E-cadherin. PLoS ONE. 2007;2(9):e844. doi: 10.1371/journal.pone.0000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomez-Casero E, Navarro M, Rodriguez-Puebla ML, et al. Regulation of the differentiation-related gene Drg-1 during mouse skin carcinogenesis. Molecular Carcinogenesis. 2001;32(2):100–9. doi: 10.1002/mc.1069. [DOI] [PubMed] [Google Scholar]
- 37.Bandyopadhyay S, Pai SK, Hirota S, et al. Role of the putative tumor metastasis suppressor gene Drg-1 in breast cancer progression. Oncogene. 2004;23(33):5675–81. doi: 10.1038/sj.onc.1207734. [DOI] [PubMed] [Google Scholar]
- 38.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nature Reviews Cancer. 2007;7(6):415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 39.Evans AJ, Russell RC, Roche O, et al. VHL promotes E2 box-dependent E-cadherin transcription by HIF-mediated regulation of SIP1 and snail. Molecular and Cellular Biology. 2007;27(1):157–69. doi: 10.1128/MCB.00892-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krishnamachary B, Zagzag D, Nagasawa H, et al. Hypoxia-inducible factor-1-dependent repression of E-cadherin in von Hippel-Lindau tumor suppressor-null renal cell carcinoma mediated by TCF3, ZFHX1A, and ZFHX1B. Cancer Research. 2006;66(5):2725–31. doi: 10.1158/0008-5472.CAN-05-3719. [DOI] [PubMed] [Google Scholar]
- 41.Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129(1):111–22. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 42.Li J, Kretzner L. The growth-inhibitory Ndrg1 gene is a Myc negative target in human neuroblastomas and other cell types with overexpressed N-or c-myc. Molecular and Cellular Biochemistry. 2003;250(1-2):91–105. doi: 10.1023/a:1024918328162. [DOI] [PubMed] [Google Scholar]
- 43.Graeber TG, Osmanian C, Jacks T, et al. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature. 1996;379(6560):88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- 44.Stein S, Thomas EK, Herzog B, et al. NDRG1 is necessary for p53-dependent apoptosis. The Journal of Biological Chemistry. 2004;279(47):48930–40. doi: 10.1074/jbc.M400386200. [DOI] [PubMed] [Google Scholar]
- 45.Kurdistani SK, Arizti P, Reimer CL, Sugrue MM, Aaronson SA, Lee SW. Inhibition of tumor cell growth by RTP/rit42 and its responsiveness to p53 and DNA damage. Cancer Research. 1998;58(19):4439–44. [PubMed] [Google Scholar]
- 46.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454(7203):436–44. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 47.Chang PW, Tsui SK, Liew C, Lee CC, Waye MM, Fung KP. Isolation, characterization, and chromosomal mapping of a novel cDNA clone encoding human selenium binding protein. Journal of Cellular Biochemistry. 1997;64(2):217–24. doi: 10.1002/(sici)1097-4644(199702)64:2<217::aid-jcb5>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 48.Duffield-Lillico AJ, Reid ME, Turnbull BW, et al. Baseline characteristics and the effect of selenium supplementation on cancer incidence in a randomized clinical trial: a summary report of the Nutritional Prevention of Cancer Trial. Cancer Epidemiology Biomarkers and Prevention. 2002;11(7):630–9. [PubMed] [Google Scholar]
- 49.Zeng H, Combs GF., Jr Selenium as an anticancer nutrient: roles in cell proliferation and tumor cell invasion. The Journal of Nutritional Biochemistry. 2008;19(1):1–7. doi: 10.1016/j.jnutbio.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 50.Morrison DG, Dishart MK, Medina D. Intracellular 58-kd selenoprotein levels correlate with inhibition of DNA synthesis in mammary epithelial cells. Carcinogenesis. 1988;9(10):1801–10. doi: 10.1093/carcin/9.10.1801. [DOI] [PubMed] [Google Scholar]
- 51.Yang M, Sytkowski AJ. Differential expression and androgen regulation of the human selenium-binding protein gene hSP56 in prostate cancer cells. Cancer Research. 1998;58(14):3150–3. [PubMed] [Google Scholar]
- 52.Wunderlich L, Paragh G, Wikonkal NM, Banhegyi G, Karpati S, Mandl J. UVB induces a biphasic response of HIF-1alpha in cultured human keratinocytes. Experimental Dermatology. 2008;17(4):335–42. doi: 10.1111/j.1600-0625.2007.00640.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.