Abstract
Background
Assessment of outcome using a single prognostic or predictive marker is the current basis of targeted therapy, but is inherently limited by its simplicity. Multiplexing has provided better classification but only been done quantitatively using RNA or DNA. Automated quantitative analysis (AQUA) is a new technology that allows quantitative in situ assessment of protein expression. We hypothesize that multiplexed quantitative measurement of ErbB receptor family proteins may allow better prediction of outcome.
Methods
We quantitatively assessed the expression of six proteins in four subcellular compartments in 676 patients using breast carcinoma tissue microarrays (TMA). Then using Cox proportional hazards modeling and unsupervised hierarchical clustering, we assessed the prognostic value of the expression singly and multiplexed.
Results
EGFR, HER-2 and HER-3 expression were associated with decreased survival. Multivariate analysis showed high HER-2 and HER-3 expression maintained independence as prognostic markers. Hierarchical clustering of expression data defined a small class enriched for HER-2 expression with 40% 10 year survival, compared to 55% using conventional methods. Clustering also revealed a similarly poor-prognostic subgroup co-expressing EGFR and HER-3 (but low for ER, PR and HER-2) with a 42% 10 year survival.
Conclusions
This work shows that the combined analysis of protein expression improved prognostic classification and that multiplexed models were superior to any single marker-based method for prediction of 10-year survival. These methods illustrate a protein-based, multiplexed approach that could more accurately identify patients for targeted therapies.
Keywords: AQUA, HER2, EGFR, HER3, HER4, Immunohistochemistry, survival
Introduction
HER-2 (also known as ErbB2 or neu) belongs to the ErbB family of four Type I tyrosine kinase receptors, including EGFR, HER-3, and HER-4, that homo- and heterodimerize to activate distinct programs of proliferation, survival, migration, and angiogenesis (1). In breast cancer, this family also demonstrates cross-talk with the hormone receptors for estrogen (ER) and progesterone (PR) as well as other pathways (2).
ErbB2 amplification is an important molecular alteration in breast cancer, and we hypothesized that interactions of HER-2 with other ErbB family members might improve our ability to classify HER-2+ breast cancer for the purposes of prognosis. While members of the HER family have been measured for prognostic value (3–12), they have never previously been rigorously multiplexed, since nearly all previous studies have been scored by traditional pathologist-based methods.
In order to measure tumor-specific content, most quantitative protein measurement techniques such as mass spectrometry require micro-dissection and are optimized with frozen specimens. Automated quantitative analysis (AQUA) measures protein expression levels in situ in formalin-fixed, paraffin-embedded tumor samples and allows discrimination of subcellular compartments (13, 14). The AQUA method has been validated in breast cancer and shown to be comparable to protein levels measured by ELISA assay (15, 16) with coefficients of variation <5%. To test the hypothesis that quantitative multiplexed analysis will improve prognostic value, we have assessed the expression of six targets (ER, PR, EGFR, HER-2, HER-3, HER-4) in four subcellular compartments, using AQUA in an archival tissue microarray (TMA) collection of invasive breast carcinoma.
Methods
Cell lines
A TMA containing cores from formalin-fixed, paraffin embedded cell pellets was used as a control for staining and AQUA analysis. JEG-3, SKOV3, and CHO cells were obtained from the Maihle laboratory at Yale University. A431, HL60, MDA-MB-453, MDA-MB-231, MDA-MB-468, SW-480, SK-BR-3, MCF-7, BT-549, T-47D, MDA-MB-435S, and BT-474 cell lines were purchased from the American Type Culture Collection (Manassas, VA). BAF3 cells were obtained from a laboratory in the Department of Genetics at Yale University, and culture conditions and cell line TMA construction have been published in detail elsewhere (15, 16). Our laboratory protocol for processing cell lines is also available on the web (http://www.tissuearray.org).
Patients
The Yale breast cancer cohort consists of 676 samples of invasive breast carcinoma collected serially from the Yale University Department of Pathology archives from 1961 to 1983 (Table 1). Slides were reviewed for tumor volume, and all samples were included that could be adequately sampled for the study. This cohort contains approximately half node-positive specimens and half node-negative specimens. Patient outcome was collected from the medical records and the Connecticut Tumor Registry. TNM classification was applied retrospectively according to guidelines from the AJCC Cancer Staging Manual, 6th Edition (17). Of 630 patients with outcome data, the mean follow-up time is 12.5 years and the mean age of diagnosis is 58.1 years. The median follow-up time is 8.8 years and the median age of diagnosis is 58.0 years. 334 patients were censored at 10 years and 228 were uncensored at 10 years. Of the 334 censored patients, their median follow-up was 18.9 years, with the minimum at 4.2 months. Complete treatment information was not available for the entire cohort; however, most patients were treated with post-surgical local irradiation. None of the node-negative patients were given adjuvant systemic therapy.
Table 1.
Population characteristics for Yale breast carcinoma tissue microarray
| Variable | Number | % | Variable | Number | % |
|---|---|---|---|---|---|
| 630 | 100.00% | 630 | 100.00% | ||
| Age at diagnosis | Laterality | ||||
| <=50 | 443 | 70.32% | Left | 303 | 48.10% |
| >50 | 181 | 28.73% | Right | 297 | 47.14% |
| N/K | 6 | 0.95% | Bilateral | 19 | 3.02% |
| N/K | 11 | 1.75% | |||
| Nuclear Grade | Histologic Grade | ||||
| 1 | 109 | 17.30% | 1 | 12 | 1.90% |
| 2 | 306 | 48.57% | 2 | 174 | 27.62% |
| 3 | 166 | 26.35% | 3 | 129 | 20.48% |
| N/K | 49 | 7.78% | N/K | 315 | 50.00% |
| Tumor Type | Tumor Stage | ||||
| Ductal | 519 | 82.38% | T1 | 265 | 42.06% |
| Lobular | 46 | 7.30% | T2 | 227 | 36.03% |
| IDC-ILC | 36 | 5.71% | T3 | 53 | 8.41% |
| IDC-LowRisk | 29 | 4.60% | T4 | 38 | 6.03% |
| N/K | 47 | 7.46% | |||
| Nodal Stage | Distant Metastasis | ||||
| pN0 | 316 | 50.16% | M0 | 592 | 93.97% |
| pN1 | 154 | 24.44% | M1 | 29 | 4.60% |
| pN2 | 95 | 15.08% | N/K | 9 | 1.43% |
| pN3 | 64 | 10.16% | |||
| N/K | 1 | 0.16% | |||
Specimen Characteristics
Formalin-fixed paraffin embedded tumor blocks from each patient were utilized in the construction of the tissue microarrays with one 0.6mm core transferred to a recipient paraffin block. Slides cut from two independent constructions were used in this study for each target. A sequential hematoxylin and eosinstained slide was histologically assessed by a pathologist to ensure adequate tumor sampling. TMA construction was performed with a tissue-arraying instrument (Beecher Instruments, Silver Springs, MD) using a method that was described previously (18). All pre-cut sections were coated in paraffin and stored at room temperature in a nitrogen chamber prior to staining to prevent loss of antigenicity (19).
Assay methods
Slides were stained by a modified indirect immunofluorescence method as described previously (13). Primary antibodies used to define the tumor compartment of each histospot included mouse monoclonal Cytokeratin AE1/AE3 (M3515, Dako Corporation, Carpinteria, CA) or wide-spectrum screening rabbit anticow cytokeratin antibody (Dako Z0622) each at 1:100. Estrogen receptor (Dako Clone 1D5) and progesterone receptor (Dako clone PgR636) were each used at 1:50 and incubated for 1 hour at room temperature. Other target antibodies were incubated overnight at 4ºC and included EGFR used neat (Dako pharmDx™ kit clone 2–18C9), HER-2 at 1:8000 (Dako A0485), HER-3 at 1:200 (clone RTJ1, Vector Laboratories, Burlingame, CA), and HER-4 at 1:400 (sc-283, Santa Cruz Biotechnology, Santa Cruz, CA). Secondary labeling of targets was performed by signal amplification using horseradish peroxidase-labeled secondary reagents (species-specific Dako Envision) followed by Cy-5 tyramide incubation. 4′,6-Diamidino-2-phenylindole (DAPI) in an anti-fading mounting medium was used to stain the nuclear compartment (Prolong Gold, Invitrogen, Eugene, OR).
Positive and negative controls were included in a specialized “boutique” array stained simultaneously containing 40 cases from a previously-described breast carcinoma tissue microarray(16) as well as 15 formalin-fixed, paraffin-embedded cancer cell lines exhibiting variable levels of expression for each marker analyzed. In addition, a breast cancer test slide was stained with each experiment without primary antibody.
Automated Quantitative Analysis (AQUA®) of Tissue microarrays
A complete and detailed discussion of the AQUA™ method has been published previously (13, 20). Briefly, monochromatic images of each histospot were acquired on an Olympus AX-51 epifluorescence microscope (Olympus, Melville, NY) using a motor-driven stage and automated custom software, and high-resolution (1024 × 1024 pixel; 0.5-μm) digital images were analyzed using AQUA™. A binary image (tumor mask) was created from the cytokeratin image of each histospot, representing areas of tumor epithelium. Histospots were excluded if the tumor mask represented less than 5% of the total histospot area. DAPI images were used to define the nuclear compartment within each histospot, and the membrane compartment was defined by perimembranous coalescence of cytokeratin immunoreactivity with specific exclusion of the nuclear compartment.
Application of RESA (Rapid Exponential Subtraction Algorithm) was used to improve subcellular localization; it is an image processing methodology which accounts for compartment overlap due to the thickness of tissue sectioning on glass slides by subtracting out-of-focus from in-focus image data according to a specialized algorithm. Target protein expression was quantified by calculating Cy5 fluorescent signal intensity on a scale of 0–255 within each image pixel. The Cy-5 wavelength is used for target labeling because it is outside the range of tissue autofluorescence. An AQUA score was generated by dividing the sum of target signals within the tumor mask by compartment area. After validation of images to ensure adequate tumor sampling and exclude any normal epithelium, the AQUA scores were normalized to a 100 point scale and averaged from two tumor samples. Although AQUA scores were calculated for each biomarker in four subcellular compartments, we restricted survival analysis to the dominant subcellular localization (nuclear: ER, PR; nonnuclear: HER-3, membranous: EGFR, HER-2). In this cohort, HER-4 expression was observed in all three compartments and so the total AQUA score in the Tumor Mask was considered for analysis.
A recent analysis of AQUA for HER-2 measurement showed a strong correlation between AQUA scores, quantitative ELISA protein measurements, and HER-2/neu gene amplification for a standard set of breast cancer cell line controls (15). We repeated both cell line and breast tumor samples used in this study as a reference for HER-2 positivity.
Statistical methods
The statistical calculations were performed using JMP Version 5.0 (SAS, Cary, NC). Disease specific survival (DSS) was chosen as the end point in the present study. Kaplan-Meier plots were used to illustrate the survival in groups of HER-2+ patients classified by the methods studied, and the log-rank test, to test for equality of survival curves. Hazard ratios were estimated using Cox regression. All P values corresponded to two-sided tests, and values less than .05 were considered significant.
Unsupervised hierarchical average-linkage clustering was performed using Cluster and Treeview (Eisen Laboratory, Stanford University, Palo Alto, CA). Tumors in the Yale cohort which had a value for at least five of six biomarkers (n = 550) were included in the clustering. AQUA scores were converted to z scores prior to clustering to normalize between markers(21). For cluster assignment, the distance from root node was chosen to maximize number of clusters as well as ensure each cluster contained at least 5% of the population. No formal statistical test was used to select the number of clusters other than the limitation imposed by the number of subjects in each cluster.
Results
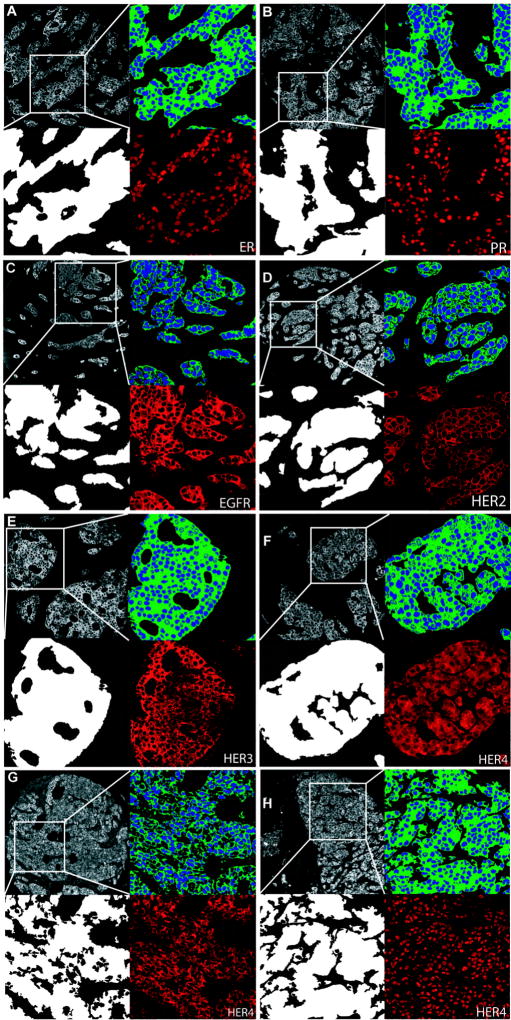
In the AQUA method, cellular compartments and targets are labeled in situ using antibodies conjugated to fluorochrome dyes. Figure 1 shows representative images from immunofluorescent labeling of six target biomarkers (ER, PR, EGFR, HER-2, HER-3, and HER-4) in the breast cancer samples studied, and each panel shows an enlarged view of the pixel area scored as tumor (Tumor Mask, lower left) and subcellular compartments (upper right panel), as well as Cy5 image for each target (lower right). Expression was predominantly nuclear for ER and PR, and membranous for EGFR and HER-2. HER-3 expression was both membranous and cytoplasmic, notably excluded from the nuclear compartment (non-nuclear). HER-4 however, showed three distinct patterns of expression: non-nuclear (Figure 1F), membranous (figure 1G), and nuclear (Figure 1H). Nuclear localization of HER-4 has been described previously, where it is thought to be involved in the transcription of target genes involved in mammary differentiation (22).
FIGURE 1. Immunofluorescent immunohistochemistry for automated quantitative analysis (AQUA).
In each panel, representative pseudo-colored images are shown of cytokeratin (upper left), tumor mask (lower left), nuclear (blue) and non-nuclear or membrane compartments (green) (upper right), and target expression (red) after RESA application (lower right). (A) Estrogen receptor, nuclear expression (B) Progesterone receptor, nuclear expression (C) EGFR, membranous expression (D) HER-2, membranous expression (E) HER-3, non-nuclear expression (F) HER-4, non-nuclear expression (G) HER-4, membranous expression (H) HER-4, nuclear expression
The YTMA49 cohort is composed of 676 breast cancer cases from the Yale Pathology archives with extensive annotation including long term follow-up, as described previously (Table 1)(16). After standardization by internal controls, AQUA measurements from two tumor samples were averaged. Expression data for at least five of six biomarkers and survival information were available from 550 patients while 126 patients were excluded due to insufficient data.
HER-2 and HER-3 are independent biomarkers of breast cancer survival
As demonstrated in Tables 2 and 3, Cox proportional hazards regression was used to assess the association of each marker with 10-year disease-specific survival (DSS) univariately and in multivariate models. As previously described, high AQUA-HER-2 (p=0.001) and low AQUA-ER (p=0.010) and AQUA-PR (p=0.002) scores were significantly associated with decreased survival (23). In contrast, using ordinal (0–3+) immunohistochemistry scores to stratify survival in the same way, low ER (p=0.007) and PR (p=0.010) scores are associated with decreased survival but HER-2 expression is not (p=.11). High AQUA-EGFR scores trended toward association with decreased survival but were only of borderline significance (p=0.065). In addition, AQUA-HER-3 scores were inversely associated with survival (p=0.003).
Table 2.
Association between continuous AQUA scores and breast cancer–specific survival by Cox univariate analysis with 10 year follow-up
| Univariate Cox Proportional Hazard Models | |||||
|---|---|---|---|---|---|
| 95% Confidence Intervals | |||||
| Variables | Risk Ratio | Lower | Upper | Effect (Chi-Square) | p-value |
| AQUA-ER | 0.989 | 0.980 | 0.997 | 6.686 | 0.010 |
| AQUA-PR | 0.986 | 0.975 | 0.995 | 9.178 | 0.002 |
| AQUA-EGFR | 1.009 | 0.999 | 1.018 | 3.408 | 0.065 |
| AQUA-HER-2 | 1.017 | 1.008 | 1.026 | 10.959 | 0.001 |
| AQUA-HER-3 | 1.016 | 1.006 | 1.026 | 9.076 | 0.003 |
| AQUA-HER-4 | 1.000 | 0.985 | 1.013 | 0.002 | 0.969 |
| Age at Diagnosis | 0.998 | 0.988 | 1.009 | 0.087 | 0.768 |
| Tumor Stage | 41.307 | 0.000 | |||
| [T2 v. T1] | 2.080 | 1.519 | 2.865 | ||
| [T3 v. T2] | 1.593 | 1.055 | 2.347 | ||
| [T4 v. T3] | 0.844 | 0.471 | 1.469 | ||
| Nodal Stage | 60.145 | 0.000 | |||
| [pN1 v. pN0] | 2.262 | 1.623 | 3.146 | ||
| [pN2 v. pN1] | 1.273 | 0.872 | 1.843 | ||
| [pN3 v. pN2] | 1.314 | 0.855 | 2.007 | ||
| Distant Metastasis | 28.735 | 0.000 | |||
| [M1 v. M0] | 4.646 | 2.837 | 7.185 | ||
| Nuclear Grade | 14.683 | 0.001 | |||
| [2 v. 1] | 1.214 | 0.834 | 1.813 | ||
| [3 v. 2] | 1.626 | 1.211 | 2.175 | ||
Table 3.
Association between multivariate Cox survival models and breast cancer–specific survival with 10 year follow-up (A) Models of each biomarker and including Tumor (pT) and Nodal (pN) Stage (B) All ErbB biomarkers and including Tumor (pT) and Nodal (pN) Stage (C) Cluster groups from multiplexed AQUA scores with TNM staging variables.
| A) Indivudual Models - Each AQUA biomarker + (pT, pN) | ||||||
|---|---|---|---|---|---|---|
| 95% Confidence Interval | ||||||
| Variables | Risk Ratio | Lower | Upper | Effect (Chi-Square) | p-value | |
| AQUA-ER | 0.987 | 0.978 | 0.996 | 8.132 | 0.004 | |
| AQUA-PR | 0.985 | 0.974 | 0.995 | 8.543 | 0.003 | |
| AQUA-EGFR | 1.011 | 1.001 | 1.021 | 4.387 | 0.036 | |
| AQUA-HER-2 | 1.017 | 1.007 | 1.026 | 10.540 | 0.001 | |
| AQUA-HER-3 | 1.012 | 1.001 | 1.022 | 4.520 | 0.034 | |
| AQUA-HER-4 | 1.006 | 0.988 | 1.021 | 0.478 | 0.489 | |
| B) All ErbB biomarkers + (pT, pN) | ||||||
| 95% Confidence Intervals | ||||||
| Variables | Risk Ratio | Lower | Upper | Effect (Chi-Square) | p-value | |
| AQUA-EGFR | 1.004 | 0.989 | 1.018 | 0.366 | 0.545 | |
| AQUA-HER-2 | 1.017 | 1.006 | 1.026 | 9.101 | 0.003 | |
| AQUA-HER-3 | 1.013 | 1.001 | 1.025 | 4.257 | 0.039 | |
| AQUA-HER-4 | 1.003 | 0.984 | 1.019 | 0.095 | 0.758 | |
| C) MultiplexAQUA + TNM | ||||||
| 95% Confidence Intervals | ||||||
| Variables | Risk Ratio | Lower | Upper | Effect (Chi-Square) | p-value | |
| Cluster Group (all v. ErbB-low VI) | 18.144 | 0.003 | ||||
| ER[I] | 0.629 | 0.429 | 0.892 | |||
| PR[II] | 0.754 | 0.554 | 1.009 | |||
| HER-2/HER-3[III] | 1.74 | 1.166 | 2.504 | |||
| EGFR/HER-3[IV] | 1.383 | 0.996 | 1.88 | |||
| HER-3/HER-4[V] | 0.993 | 0.605 | 1.533 | |||
| Tumor Stage | 26.686 | >0.001 | ||||
| [T2 v. T1] | 2.072 | 1.474 | 2.931 | |||
| [T3 v. T2] | 1.199 | 0.754 | 1.857 | |||
| [T4 v. T3] | 1.025 | 0.548 | 1.868 | |||
| Nodal Stage | 24.07 | >0.001 | ||||
| [pN1 v. pN0] | 1.743 | 1.199 | 2.529 | |||
| [pN2 v. pN1] | 1.264 | 0.819 | 1.926 | |||
| [pN3 v. pN2] | 1.249 | 0.763 | 2.036 | |||
| Distant Metastasis | 12.276 | >0.001 | ||||
| [M1 v. M0] | 3.447 | 1.801 | 6.102 | |||
The prognostic significance of HER-3 was further explored using the X-tile software program(19) to define optimal population cutpoints in a training set of half the patients in the cohort with both HER-3 expression data and outcome information (n=260) and validated by Kaplan Meier analysis in the remaining half (n=261). In the validation set half of the cohort, grouping by HER-3 expression with a cutpoint of AQUA>25, we find high levels of HER-3 show a 53% 10 year survival compared to 69% in the low HER-3 group (Log-rank p=0.0096).
Next, we constructed a multivariate Cox model including pathlogical tumor (pT) and nodal (pN) stage with AQUA scores from each of the biomarkers tested and observed that five of the six biomarkers (ER, PR, EGFR, HER-2, HER-3) were independently correlated with patient outcome when assayed using AQUA. While AQUA-ER and AQUA-PR were associated with more favorable prognosis, AQUA-EGFR, AQUA-HER-2, and AQUA-HER-3 were associated with decreased survival (Table 3A). When we included AQUA expression scores from all ErbB receptors in a multivariate model including pT and pN, both AQUA-HER-2 and AQUA-HER-3 remained independent prognostic factors ( Table 3B).
ErbB family co-expression is associated with prognosis
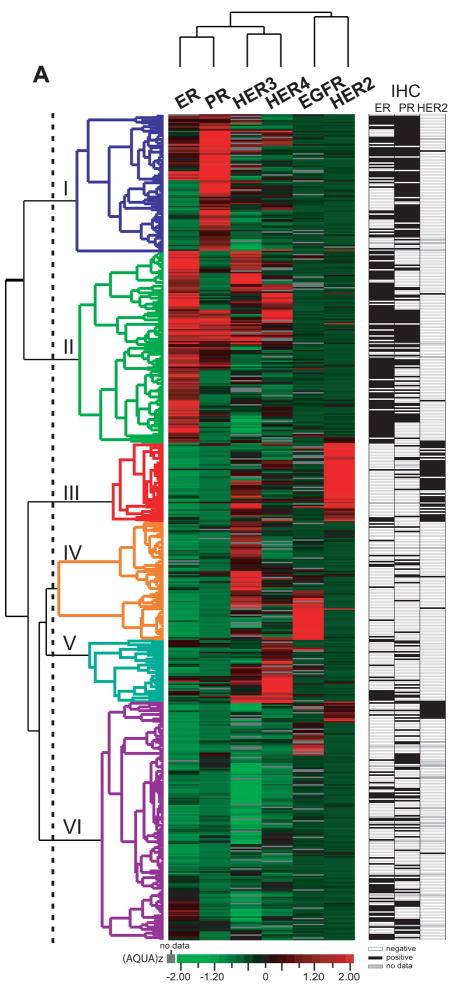
We used unsupervised average linkage hierarchical clustering to examine the relative co-expression of the AQUA targets measured (Figure 2). Prior to clustering, data were normalized for variance between experiments by z-score transformation (AQUAz). Six distinct clusters were observed, labeled Cluster I-VI in Figure 2 and colored red (high) to green (low) by the distance from the mean for each target. Clusters I and II were notable for high expression of ER and PR and separated by higher levels of HER-3 and HER-4 in Cluster II. Cluster III was enriched for HER-2 and HER-3 expression and had low levels of hormone receptor expression, while high HER-3 and EGFR expression were found in Cluster IV. HER-3 and HER-4 expression was enriched in Cluster V. The largest cluster (VI) included some cases with high expression of EGFR or HER-2, but it had relatively low levels of all targets. Of note, high expression of both EGFR and HER-2 was rarely observed.
FIGURE 2. Unsupervised hierarchical clustering of protein expression data measured in the Yale archival cohort.
Tumor samples with scores for at least five of six biomarkers (n = 550), and data from one subcellular compartment was included for each marker: ER (nuclear), PR (nuclear), EGFR (membranous), HER-2(membranous), HER-3 (non-nuclear), and HER-4(non-nuclear). AQUA scores were converted to z scores(21) (AQUAz) prior to clustering to normalize between markers, as described in Methods. Values for protein expression are shown as a heat map, and each data point is represented by a bar colored by its value’s distance from the target mean of the cohort in units of standard deviation. Black indicates protein expression level equal to the mean; green indicates protein expression level below the mean; red indicates protein expression level above the mean; and Gray indicates missing values. The branch lengths and pattern of the dendrogram demonstrate the relatedness of the tumors on the vertical axis and the antibody staining on the horizontal axis. Note that ER scores lower than the mean are not necessarily negative, but need to be displayed and assess in this manner for clustering analysis. Further analysis of the ER scores in this cohort may be found in previous work (13). To the right of the colored heat map is a second black and white heat map that shows a binary indication of the standard biomarkers as assessed by a pathologist for each case. White is negative, black is positive, and grey is unknown or missing data.
We calculated five and ten-year disease-specific survival (DSS) rates in the Cluster groups. Despite differential ErbB family expression, the hormone-receptor high groups had comparable survival rates (ER-I, 79.8% 5-yr and 58.3% 10-yr vs. PR-II, 77.7% 5-yr and 60.4% 10-yr). The HER-2/HER-3 Cluster III had the lowest survival rate (DSS 5-yr 44.9%, 10-yr 39.4%). Only 2/3 of HER-2 positive patients by conventional immunohistochemistry (HercepTest 3+) are included in this group, with the majority of remaining HER-2+ patients (20%, 12/60) found in Cluster VI. Low survival rates were also observed in the HER-3/EGFR Cluster IV (5-yr DSS 56.2%, 10-yr 42.0%). This group may be defining the so called “triple negative” class (9, 24, 25) of breast cancer since these cases are low for ER, PR and HER-2. The HER-3/HER-4 Cluster V is associated with relatively good outcome (DSS 5-yr 70.0%, 10-yr 51.7%). Survival in the HER family-low Cluster VI was similar to that in the hormone-receptor positive groups I and II (5-yr DSS 74.4%, 10-yr 61.7%). The association of clustering subgroups with outcome was independent of TNM staging parameters when assessed by a multivariate Cox proportional hazards model (p=0.003, Table 3C).
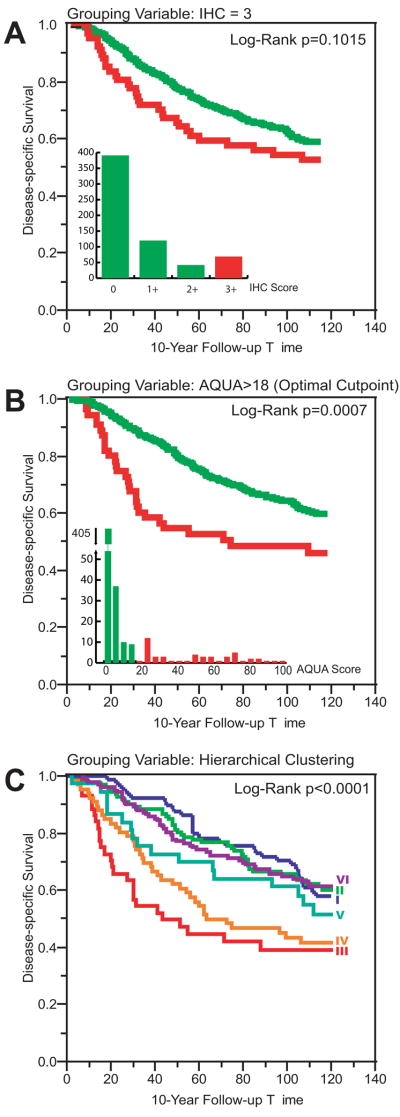
Multiplexing AQUA scores improves classification of HER-2+ breast cancer
To compare the multiplexed AQUA method to conventional methods we have done a Kaplan-Meier analysis (Figure 3). Traditional immunohistochemistry (IHC) on the TMA failed to reach significance in this cohort (IHC 3+, Figure 3A). The addition of quantitative analysis and the use of a previously determined optimized AQUA cutpoint score(16) resulted in an improvement in the prognostic value and achieves statistical significance (Figure 3B). However, selection of a class of HER-2+ patients defined by hierarchical clustering (Cluster III, Figure 3C) defines a smaller subset with substantially worse outcome. The median time from diagnosis to death from breast cancer was 98 months for the group defined by IHC, 55 months by AQUA, and only 43 months in the group defined by clustering. In contrast, HER-2 “low” patients had a median survival of almost 200 months by all three classification methods.
FIGURE 3. HER-2 status and patient outcome by conventional immunohistochemistry (IHC), AQUA-HER-2, or Multiplexed AQUA.
Kaplan-Meier survival curve analysis, using 10-year disease specific survival as clinical endpoint of interest (A) Patients grouped by immunohistochemical score using the HercepTest antibody and scoring guidelines. Green, HER-2 equivocal or negative patients by IHC score of 0, 1+, or 2+: 5-Year Survival 72.1%, Median Survival 198 months; Red, HER-2 positive patients by IHC score of 3+: 5-Year Survival 56.1%, Median Survival 98 months. Inset, frequency distribution of IHC scores. (B) Kaplan-Meier survival curve analysis of Yale patients, grouped by HER-2 AQUA score. The optimal cutpoint of 18 was chosen based on a previously published analysis (16). Green, HER-2 negative patients by AQUA: 5-Year Survival 73.5%, Median Survival 198 months; Red, HER-2 positive patients by AQUA: 5-Year Survival 48.6%, Median Survival 55 months. Inset, frequency distribution histogram of average HER-2 AQUA scores. AQUA HER-2+ group includes 71.67% of IHC 3+ tumors. (C) Kaplan-Meier survival curve analysis of Yale patients, grouped by AQUA multiplexed analysis with clustering as shown in Figure 1. Survival curves are colored according to the corresponding cluster on the heat map y-axis. The HER-2 enriched cluster (III) is depicted in red: 5-Year Survival 44.9%, Median Survival 43 months. This group includes 66.7% of the IHC 3+ tumors. The EGFR/HER-3 Cluster is depicted in orange: 5-Year Survival 44.9%, Median Survival 43 months. All other Cluster groups: 5-Year Survival 73.2%, Median Survival 199 months.
Discussion
In this study, we measured the protein expression of the ErbB family (EGFR, HER-2, HER-3, HER-4) and the hormone receptors ER and PR using the AQUA method in a large retrospective cohort of breast cancer patients and assessed target co-expression and association with breast cancer survival using proportional hazards modeling and hierarchical clustering. The implications of aberrant ErbB expression have been explored by many previous investigations, and overall, HER-2 has been consistently associated with a shorter time to progression and decreased survival time, while correlative findings of the other ErbB receptors have varied widely. (3–12) This is the first report of a quantitative protein detection method linking multiplexed ErbB expression to long-term patient outcome.
Most clinicians currently rely on clinicopathological parameters such as tumor size and nodal status, as well as ER, PR and Her2 tissue biomarkers to assess an individual’s prognosis after surgery for primary breast cancer. We find that among the biomarkers measured, AQUA-ER, AQUA-PR, AQUA-EGFR, AQUA-HER-2, and AQUA-HER-3 were significantly associated with long-term survival and independent of parameters of tumor size and nodal metastasis. We were unable to reproduce previous observations that HER-4 is a favorable prognostic biomarker.(7, 8) In addition, the poor prognostic association of AQUA-HER-2 and AQUA-HER-3 were independent of other ErbB expression patterns. Clustering of the protein expression data revealed groups of breast cancer patients that co-express sets of ErbB family members. Multiplexing of AQUA scores by hierarchical clustering classification was superior to conventional IHC or univariate AQUA classification of HER-2+ breast cancer for prognosis, and this effect is independent of current clinical staging variables.
This sort of clustering analysis has potential for use in classification of breast cancers in a manner similar to that done by cDNA array type studies (24). Although only six markers are used in this study, we included the three standard markers that are used in standard management of breast cancer (ER, PR, and HER-2) which allowed us to identify the “triple negative” subset of breast cancers. Examination of Figure 2 shows that the triple negative class self-assorts into Cluster IV. It is interesting to note that this study suggests that there are probably two biological classes within that group; the subset that is triple negative, but expressing high levels of EGFR and a second subset that is triple negative with high levels of HER-3. This observation confirms previous work reporting a high correlation between triple negative cases and EGFR over-expression (9, 25). Further studies are needed to assess the significance of the HER-3+ subdivision, but it could have implications for new ErbB targeted therapies such as pertuzumab and cannertinib (26).
Limitations of this study include its retrospective nature and the incompleteness of the treatment data. However, the collection and investigation of archival cohorts such as this allow valuable insights into the relationship of breast cancer outcome with the molecular features of primary tumors. These correlative studies suggest the investigation of multiplexed assessment of biomarkers as a method to predict response to therapy. In this study, assay conditions were carefully controlled using a specialized control cell and tissue microarray, which should ensure reproducibility in future studies now underway in cohorts treated with ErbB-targeted therapies. The results reported here show the power of quantitative protein-based multiplexed analysis. By collection of continuous scores proportional to protein expression of ErbB family members, we are able to define subsets of our cohort that show grouping that is analogous to cDNA-based classifications and is more specific and informative for prediction of outcome.
Acknowledgments
This work was supported by the an Avon-NCI Progress for Patients grant and NCI grants R33 CA 106709 and R33 CA 110511 to DLR and by an MSTP grant to JMG.
Abbreviations
- AQUA
automated quantitative analysis
- EGFR
epidermal growth factor receptor
- ErbB
originally from avian erythoblastosis viral oncogene (v-erbB)
- ELISA
enzyme-linked immunosorbent assay
- ER
estrogen receptor
- IHC
immunohistochemistry
- PR
progesterone receptor
- TMA
tissue microarray
References
- 1.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 2.Osborne CK, Shou J, Massarweh S, Schiff R. Crosstalk between estrogen receptor and growth factor receptor pathways as a cause for endocrine therapy resistance in breast cancer. Clinical cancer research. 2005;11:865s–870s. [PubMed] [Google Scholar]
- 3.Abd El-Rehim DM, Pinder SE, Paish CE, Bell JA, Rampaul RS, Blamey RW, Robertson JF, Nicholson RI, Ellis IO. Expression and co-expression of the members of the epidermal growth factor receptor (EGFR) family in invasive breast carcinoma. Br J Cancer. 2004;91:1532–1542. doi: 10.1038/sj.bjc.6602184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esteva FJ, Hortobagyi GN, Sahin AA, Smith TL, Chin DM, Liang SY, Pusztai L, Buzdar AU, Bacus SS. Expression of erbB/HER receptors, heregulin and P38 in primary breast cancer using quantitative immunohistochemistry. Pathol Oncol Res. 2001;7:171–177. doi: 10.1007/BF03032345. [DOI] [PubMed] [Google Scholar]
- 5.Hudelist G, Singer CF, Manavi M, Pischinger K, Kubista E, Czerwenka K. Co-expression of ErbB-family members in human breast cancer: Her-2/neu is the preferred dimerization candidate in nodal-positive tumors. Breast Cancer Res Treat. 2003;80:353–361. doi: 10.1023/A:1024929522376. [DOI] [PubMed] [Google Scholar]
- 6.Kew TY, Bell JA, Pinder SE, Denley H, Srinivasan R, Gullick WJ, Nicholson RI, Blamey RW, Ellis IO. c-erbB-4 protein expression in human breast cancer. Br J Cancer. 2000;82:1163–1170. doi: 10.1054/bjoc.1999.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suo Z, Risberg B, Kalsson MG, Willman K, Tierens A, Skovlund E, Nesland JM. EGFR family expression in breast carcinomas. c-erbB-2 and c-erbB-4 receptors have different effects on survival. Journal of Pathology. 2002;196:17–25. doi: 10.1002/path.1003. [DOI] [PubMed] [Google Scholar]
- 8.Suo Z, Yang H, Mei Q, Skovlund E, Cui J, Nesland JM. Type 1 protein tyrosine kinases in Chinese breast carcinomas: a clinicopathologic study. Int J Surg Pathol. 2001;9:177–187. doi: 10.1177/106689690100900303. [DOI] [PubMed] [Google Scholar]
- 9.Tischkowitz M, Brunet JS, Begin LR, Huntsman DG, Cheang MC, Akslen LA, Nielsen TO, Foulkes WD. Use of immunohistochemical markers can refine prognosis in triple negative breast cancer. BMC Cancer. 2007;7:134. doi: 10.1186/1471-2407-7-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Travis A, Pinder SE, Robertson JF, Bell JA, Wencyk P, Gullick WJ, Nicholson RI, Poller DN, Blamey RW, Elston CW, Ellis IO. C-erbB-3 in human breast carcinoma: expression and relation to prognosis and established prognostic indicators. Br J Cancer. 1996;74:229–233. doi: 10.1038/bjc.1996.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiseman SM, Makretsov N, Nielsen TO, Gilks B, Yorida E, Cheang M, Turbin D, Gelmon K, Huntsman DG. Coexpression of the type 1 growth factor receptor family members HER-1, HER-2, and HER-3 has a synergistic negative prognostic effect on breast carcinoma survival. Cancer. 2005;103:1770–1777. doi: 10.1002/cncr.20970. [DOI] [PubMed] [Google Scholar]
- 12.Witton CJ, Reeves JR, Going JJ, Cooke TG, Bartlett JM. Expression of the HER1-4 family of receptor tyrosine kinases in breast cancer. J Pathol. 2003;200:290–297. doi: 10.1002/path.1370. [DOI] [PubMed] [Google Scholar]
- 13.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 14.Camp RL, Dolled-Filhart M, King BL, Rimm DL. Quantitative analysis of breast cancer tissue microarrays shows that both high and normal levels of HER2 expression are associated with poor outcome. Cancer Res. 2003;63:1445–1448. [PubMed] [Google Scholar]
- 15.McCabe A, Dolled-Filhart M, Camp RL, Rimm DL. Automated quantitative analysis (AQUA) of in situ protein expression, antibody concentration, and prognosis. Journal of the National Cancer Institute. 2005;97:1808–1815. doi: 10.1093/jnci/dji427. [DOI] [PubMed] [Google Scholar]
- 16.Dolled-Filhart M, McCabe A, Giltnane J, Cregger M, Camp RL, Rimm DL. Quantitative in situ analysis of beta-catenin expression in breast cancer shows decreased expression is associated with poor outcome. Cancer Res. 2006;66:5487–5494. doi: 10.1158/0008-5472.CAN-06-0100. [DOI] [PubMed] [Google Scholar]
- 17.Singletary SE, Connolly JL. Breast cancer staging: working with the sixth edition of the AJCC Cancer Staging Manual. CA Cancer J Clin. 2006;56:37–47. doi: 10.3322/canjclin.56.1.37. quiz 50–31. [DOI] [PubMed] [Google Scholar]
- 18.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 19.Berger AJ, Camp RL, DiVito KA, Kluger HM, Halaban R, Rimm DL. Automated quantitative analysis of HDM2 expression in malignant melanoma shows association with early-stage disease and improved outcome. Cancer Res. 2004;64:8767–8772. doi: 10.1158/0008-5472.CAN-04-1384. [DOI] [PubMed] [Google Scholar]
- 20.Rubin MA, Zerkowski MP, Camp RL, Kuefer R, Hofer MD, Chinnaiyan AM, Rimm DL. Quantitative determination of expression of the prostate cancer protein alpha-methylacyl-CoA racemase using automated quantitative analysis (AQUA): a novel paradigm for automated and continuous biomarker measurements. Am J Pathol. 2004;164:831–840. doi: 10.1016/s0002-9440(10)63171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyd JC, Lacher DA. A multi-stage Gaussian transformation algorithm for clinical laboratory data. Clinical chemistry. 1982;28:1735–1741. [PubMed] [Google Scholar]
- 22.Linggi B, Carpenter G. ErbB receptors: new insights on mechanisms and biology. Trends Cell Biol. 2006;16:649–656. doi: 10.1016/j.tcb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 23.Pick E, Kluger Y, Giltnane JM, Moeder C, Camp RL, Rimm DL, Kluger HM. High HSP90 expression is associated with decreased survival in breast cancer. Cancer Res. 2007;67:2932–2937. doi: 10.1158/0008-5472.CAN-06-4511. [DOI] [PubMed] [Google Scholar]
- 24.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 25.Siziopikou KP, Ariga R, Proussaloglou KE, Gattuso P, Cobleigh M. The challenging estrogen receptor-negative/progesterone receptor-negative/HER-2-negative patient: a promising candidate for epidermal growth factor receptor-targeted therapy? Breast J. 2006;12:360–362. doi: 10.1111/j.1075-122X.2006.00276.x. [DOI] [PubMed] [Google Scholar]
- 26.Hobday TJ, Perez EA. Molecularly targeted therapies for breast cancer. Cancer Control. 2005;12:73–81. doi: 10.1177/107327480501200202. [DOI] [PubMed] [Google Scholar]