Abstract
Background
TTN-encoded titin, CSRP3-encoded muscle LIM protein, and TCAP-encoded telethonin are Z-disc proteins essential for the structural organization of the cardiac sarcomere and the cardiomyocyte’s stretch sensor. All three genes have been established as cardiomyopathy-associated genes for both dilated cardiomyopathy (DCM) and hypertrophic cardiomyopathy (HCM). Here, we sought to characterize the frequency, spectrum, and phenotype associated with HCM-associated mutations in these three genes in a large cohort of unrelated patients evaluated at a single tertiary outpatient center.
Methods
DNA was obtained from 389 patients with HCM (215 male, left ventricular wall thickness of 21.6 ± 6 mm) and analyzed for mutations involving all translated exons of CSRP3 and TCAP and targeted HCM-associated exons (2, 3, 4, and 14) of TTN using polymerase chain reaction (PCR), denaturing high performance liquid chromatography (DHPLC), and direct DNA sequencing. Clinical data were extracted from patient records and maintained independent of the genotype.
Results
Overall, 16 patients (4.1%) harbored a Z-disc mutation: 12 had a MLP mutation and 4 patients a TCAP mutation. No TTN mutations were detected. Seven patients were also found to have a concomitant myofilament mutation. Seven patients with a MLP-mutation were found to harbor the DCM-associated, functionally characterized W4R mutation. W4R-MLP was also noted in a single white control subject. Patients with MLP/TCAP-associated HCM clinically mimicked myofilament-HCM.
Conclusions
Approximately 4.1% of unrelated patients had HCM-associated MLP or TCAP mutations. MLP/TCAP-HCM phenotypically mirrors myofilament-HCM and is more severe than the subset of patients who still remain without a disease-causing mutation. The precise role of W4R-MLP in the pathogenesis of either DCM or HCM warrants further investigation.
Keywords: Genetics, Genes, Hypertrophy, Cardiomyopathy, Z-disc, Muscle LIM protein, Telethonin, TCAP, Titin
Introduction
Affecting one in 500 persons, hypertrophic cardiomyopathy (HCM) is a disease associated with remarkable genotypic and phenotypic heterogeneity [1,2]. Clinical outcomes range from an entirely asymptomatic course with normal longevity to chronic progressive heart failure or sudden cardiac death (SCD). Indeed, HCM is one of the leading causes of SCD in young persons [1].
The most common genetically mediated form of HCM is myofilament-HCM with hundreds of disease-associated mutations in eight genes encoding proteins critical to the sarcomere’s thick—[β-myosin heavy chain (MYH7) [3], regulatory myosin light chain (MYL2), and essential myosin light chain (MYL3)] [4], intermediate—[myosin binding protein C (MYBPC3)] [5], and thin myofilament [cardiac troponin T (TNNT2), α-tropomyosin (TPM1) [6], cardiac troponin I (TNNI3) [7], and actin (ACTC) [8,9]]. Myofilament-HCM accounts for approximately 40–65% of HCM among cohorts of unrelated patients [10]. In general, patients with myofilament-HCM have greater hypertrophy and present at a younger age than those who remain without an established disease-causing mutation [11]. The two most common genotypes of myofilament-HCM, MYBPC3- and MYH7-HCM, are phenotypically indistinguishable from each other [12–20].
Besides perturbations involving the sarcomere’s myofilaments, the Z-disc, which comprises a cadre of proteins involved in cardiomyocyte cytoarchitecture and mechano-sensor-signaling, has emerged recently as host to several HCM-associated mutations extending the spectrum of “sarcomeric”-HCM. To date, three genes encoding critical Z-disc proteins: TTN-encoded titin, CSRP3-encoded muscle LIM protein (MLP), and the TCAP-encoded telethonin, have been implicated in the pathogenesis of both dilated cardiomyopathy (DCM) and HCM [21–24].
As part of the cardiomyocyte stretch response machinery, TTN-encoded titin, which extends throughout half of the sarcomere from the M-line to the Z-disc is the largest of the three proteins; mapped on chromosome 2q31, TTN encodes for a giant 26,926 amino-acid protein with a molecular weight of 2993 kD [25]. CSRP3-encoded MLP and TCAP-encoded telethonin are mapped to chromosome 11p15.1 and 17q12, respectively, and contain 194 and 167 amino acids, respectively [26,27]. Prior to this study, one HCM-associated mutation in TTN (R740L-TTN) [28], three HCM-associated mutations in MLP (L44P-MLP, C58G-MLP, and S54R/E55G-MLP) [22], and two HCM-associated mutations in TCAP (T137I-TCAP and R153H-TCAP) have been reported [21].
Having completed a comprehensive mutational analysis involving all translated exons of the eight genes responsible for myofilament-HCM [14,15,29,30], we sought to determine the frequency, spectrum, and phenotype associated with these three genes that encode essential Z-disc proteins among a large cohort of unrelated patients diagnosed clinically with HCM.
Methods
Study population
Following a written informed consent for this IRB-approved research protocol, blood samples were obtained from 389 unrelated patients with HCM (215 male, left ventricular wall thickness of 21.6 ± 6 mm) evaluated at the Mayo Clinic’s HCM clinic between April 1997 and December 2001. Subsequently DNA was extracted from the blood samples using Purgene DNA extraction kits (Gentra, Minneapolis, Minnesota).
HCM-associated mutational analysis of TTN, CSRP3, and TCAP
Using polymerase chain reaction (PCR) and denaturing high performance liquid chromatography (DHPLC) (WAVE, Transgenomic, Omaha, Nebraska), the three genes implicated in Z-disc-HCM: TTN-encoded titin, CSRP3-encoded muscle LIM protein, and TCAP-encoded telethonin, were analyzed. Abnormal elution profiles were further characterized by direct DNA sequencing (ABI Prism 377; Applied Biosystem, Foster City, California).
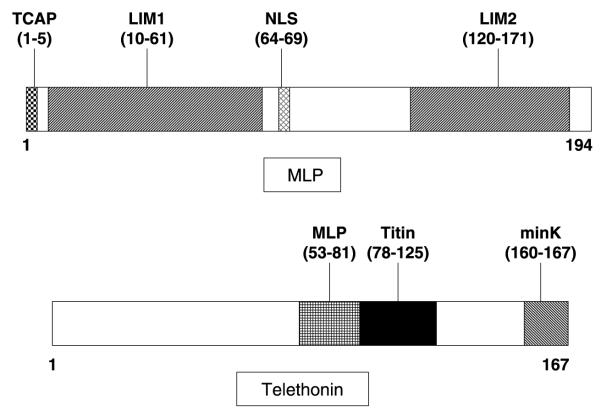
For TTN, only a targeted analysis of the exons (2, 3, 4, and 14) hosting cardiomyopathy-associated mutations was performed while a comprehensive open reading frame/splice-site analysis was conducted for all translated exons of CSRP3 (5 exons) and TCAP (2 exons). A topological schematic of both MLP and telethonin including key functional domains is depicted in Fig. 1. Primers, annealing temperatures and optimized WAVE conditions are available upon request. Four hundred reference alleles, derived from 100 white and 100 black healthy controls (Coriell Cell Repositories), were also examined to determine whether an identified amino acid variant was a common polymorphism. The non-synonymous mutations were annotated using the single letter convention as in L44P whereby the wild type leucine (L) at residue 44 has been replaced by proline (P).
Fig. 1.
Topological schematic of muscle LIM protein and telethonin. Shown are the important domains of the protein. For MLP, the TCAP-binding domain, both LIM-domains and its nuclear localization signal (NLS) are shown. For telethonin, the MLP -, titin-, and minK-binding domains are shown. Amino-acid localization of the specific domains between parentheses.
Statistical analysis
Analysis of variance tests were used to assess differences between continuous variables; contingency tables or z tests were used as appropriate to analyze nominal variables independency of the different variables. Student’s t tests were performed to elucidate differences between the different subgroups. A p value less than 0.05 was considered statistically significant.
Results
Table 1 summarizes the phenotype of the entire HCM cohort including those with perturbations involving either MLP or telethonin. The mean age at diagnosis for our total cohort was approximately 41 ± 19 years with 216 patients (56%) having cardiac symptoms at presentation and 60 (15%) having received an implantable cardioverter-defibrillator (ICD). The mean maximum left ventricular wall thickness (LVWT) was 21.6±6 mm. Of the 389, 161 (41%) were treated in part by a surgical myectomy, reflecting the surgical referral bias and subsequent over-representation of obstructive HCM in this cohort. Approximately one-third had a family history of HCM whereas one-seventh was found to have a family history of sudden cardiac death. Myofilament-HCM was demonstrated previously for 147 of the 389 subjects (38%) [14,15,30].
Table 1.
Clinical characteristics of HCM cohort
| HCM-cohort | Genotype negative | Single myofilament mutation |
MLP | TCAP | |
|---|---|---|---|---|---|
| No. of individuals | 389 | 233 | 140 | 12 | 4 |
| Sex, male/female | 215/174 | 127/106 | 79/61 | 7/5 | 2/2 |
| Age at Dx | 41.2 ± 19 | 45.1 ± 19 | 34.5 ± 17 | 48.5 ± 17 | 38.8 ± 9 |
| Cardiac symptoms | 216 (56%) | 128 (55%) | 74 (53%) | 9 (75%) | 4 (100%) |
| Max LVWT (mm) | 21.6 ± 6 | 20.6 ± 6 | 23.0 ± 7 | 20.1 ± 3 | 29.5 ± 12 |
| LVWT ≥ 25mm | 78 (20%) | 38 (16%) | 38 (28%) | 0 | 2 (50%) |
| Resting LVOTO (mmHg) | 46.6 ± 42 | 46.6 ± 42 | 42.8 ± 42 | 80 ± 43 | 75 ± 38 |
| Pos. FH for HCM | 121 (31%) | 54 (23%) | 61 (44%) | 3 (25%) | 1 (25%) |
| Pos. FH for SCD | 54 (14%) | 26 (11%) | 27 (19%) | 2 (17%) | 1 (25%) |
| Myectomy | 160 (41%) | 92 (39%) | 62 (44%) | 5 (42%) | 3 (75%) |
| Pacemaker | 67 (17%) | 35 (15%) | 26 (19%) | 5 (42%) | 2 (50%) |
| ICD | 60 (15%) | 23 (10%) | 36 (26%) | 1 (8%) | 0 |
| Multiple or concomitant myofilament mutation | 147 | — | 10/140 | 6/12 | 1/4 |
Values are means ± SD or % (n).
Dx indicates diagnosis; FH, family history; HCM, hypertrophic cardiomyopathy; ICD, implantable cardioverter-defibrillator; LVOTO, left ventricular outflow tract obstruction; LVWT, left ventricular wall thickness; SCD, sudden cardiac death.
Overall, 16 (4.1%) individuals with HCM hosted possible mutations in the genes underlying Z-disc-HCM: TTN (0), CSRP3 (12), and TCAP (4). The clinical phenotypes of these patients are described in Table 2. The average at diagnosis for MLP (CSRP3)- and TCAP-associated HCM was 48.5 ± 17 and 38.8 ± 9 years, respectively, while the mean maximal left ventricular wall thickness (MLVWT) was 20.1 ± 3 mm and 29.5 ± 12 mm, respectively. Three patients (25%) with a MLP-mutation and one patient (25%) with a TCAP-mutation reported a family history of HCM, while 2 and 1 patient (17 and 25%), respectively, had a family history of SCD. A total of eight patients underwent a surgical myectomy due to refractory symptoms despite optimal medical treatment.
Table 2.
Clinical profiles of patients with a HCM-associated CSPR3 (MLP) or TCAP mutation
| Case | Gene | Mutation (exon) |
Myofilament mutation |
Age (y) Sex |
Age at Dx |
Racea | Symptoms at presentation |
Subsequent symptoms | AF | Max. LVWT (mm) |
Resting LVOTO (mmHg) |
FH of HCM |
FH of SCD (age at SCD)b |
Treatment |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CSRP3 | W4R (1) | 80/F | 69 | 1 | Angina, dyspnea | Angina, dyspnea | Y | 15 | 21 | Yes | No | PM | |
| 2 | CSRP3 | W4R (1) | 29/M | 16 | 1 | Asymptomatic | Dyspnea | N | 25 | 0 | No | No | — | |
| 3 | CSRP3 | W4R (1) | 56/M | 41 | 1 | Asymptomatic | Angina, dyspnea, (pre)syncope | N | 17 | 32 | No | No | PM | |
| 4 | CSRP3 | W4R (1) | 78/F | 68 | 1 | n/a | Dyspnea | N | 20 | 0 | No | No | Myectomy | |
| 5 | CSRP3 | W4R (1) | F1113I-MYBPC3 | 59/M | 50 | 1 | Asymptomatic | Dyspnea, (pre)syncope | N | 23 | 117 | No | No | Myectomy, PM, ICD |
| 6 | CSRP3 | W4R (1) | T1377M-MYH7 | 50/F | 43 | 1 | n/a | Angina, dyspnea, (pre)syncope | N | 18 | 86 | Yes | No | Myectomy |
| 7 | CSRP3 | W4R (1) | I511T-MYH7 | 60/F | 53 | 1 | Dyspnea | Dyspnea, (pre)syncope | N | 16 | 0 | Yes | No | — |
| 8 | CSRP3 | K42 fs/165 (2) | 53/M | 46 | 2 | Angina, dyspnea | Angina, dyspnea, (pre)syncope | N | 18 | 112 | No | No | — | |
| 9 | CSRP3 | L44P (2) | G1041 fs/5-MYBPC3 | 71/F | 62 | n/a | Presyncope | Angina, dyspnea, (pre)syncope | N | 25 | 100 | Yes | Yes (40,32,39) | Myectomy, PM |
| 10 | CSRP3 | R64C (2) | I1131T-MYBPC3 | 72/M | 65 | n/a | Dyspnea, (pre)syncope | Angina, dyspnea | Y | 23 | 58 | No | No | — |
| 11 | CSRP3 | Y66C (2) | R162Q-TNNI3 | 36/M | 28 | 1 | n/a | Asymptomatic | N | 19 | 100 | No | Yes (42) | Myectomy |
| 12 | CSRP3 | Q91L (2) | 57/M | 44 | 1 | Angina | Angina, dyspnea, (pre)syncope | Y | 22 | 18 | No | No | PM | |
| 13 | TCAP | E13del (1) | 53/M | 47 | 1 | Dyspnea, (pre)syncope | Dyspnea, (pre)syncope | N | 22 | 100 | No | No | — | |
| 14 | TCAP | E13del (1) | 42/M | 37 | 1 | Angina, dyspnea | Angina, dyspnea | N | 30 | 81 | Yes | Yes (54) | Myectomy, ICD | |
| 15 | TCAP | R70W (2) | 65/F | 44 | 1 | Asymptomatic | Dyspnea | Y | 46 | 19 | Yes | No | Myectomy, PM | |
| 16 | TCAP | P90L (2) | Q998R-MYBPC3 | 45/F | 26 | 1 | Dyspnea | Angina, dyspnea, presyncope | Y | 20 | 100 | No | No | Myectomy, PM |
AF, atrial fibrillation; Dx, diagnosis; FH, family history; HCM, Hypertrophic cardiomyopathy; ICD, implantable cardioverter defibrillator; LVOTO, left ventricular outflow tract obstruction; LVWT, left ventricular wall thickness; n/a, not available; PM, pacemaker; SCD, sudden cardiac death.
1, Caucasian; 2, Hispanic.
In a first degree relative.
HCM-associated MLP mutations
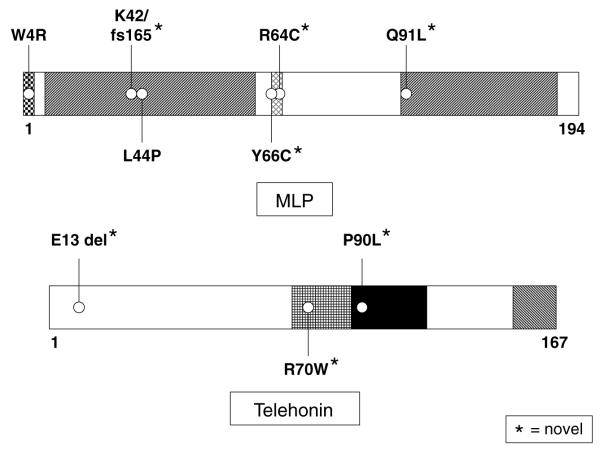
Fig. 2 depicts the mutations found in the CSRP3-encoded MLP; novel mutations are indicated by an asterisk. Five CSRP3 variants were identified in 12 patients, including 4 missense mutations and 1 frameshift mutation, involving residues highly conserved across species (data not shown) and not seen in 400 reference alleles. Clinical phenotypes are described in Table 2. K42fs/165 and Q91L were detected in patients having no HCM-associated myofilament mutations (cases 8 and 12). The previously published HCM-causing mutation (L44P, case 9) localized to the LIM1 α-actinin binding domain, while the R64C and Y66C mutations (cases 10 and 11) localized to the six amino acid nuclear localization signal (NLS). These three mutations (cases 9–11) were detected in patients also hosting HCM-associated myofilament mutations.
Fig. 2.
Schematic representation of mutations in muscle LIM protein and telethonin. Representation of mutations found in our cohort of 389 patients with HCM. The L44P-MLP has been previously published as a HCM-associated mutation. The W4R-MLP mutation has been previously published and functionally characterized in patients with DCM. Novel mutations are indicated with an asterisk.
The missense mutation, W4R-MLP, which localizes to telethonin’s binding domain, was noted in seven patients (cases 1–7). Three of these patients (cases 5–7) also had a mutation involving either the beta myosin heavy chain or myosin binding protein C. W4R was also observed in one of the 400 reference alleles examined (a healthy Caucasian control).
HCM-associated TCAP mutations
Three different, novel TCAP mutations were identified in 4 patients with HCM (Table 2, cases 13–16). Two patients (cases 13 and 14) had an in-frame deletion involving glutamic acid at position 13 (E13del). The R70W mutation was located in the reciprocal MLP-binding domain of telethonin in a patient (case 15) with a MLVWT of 46 mm and a positive family history for HCM. The titin-binding domain of telethonin was host to a missense mutation, P90L, for one patient (case 16) who also had a missense mutation involving myosin binding protein C.
Genotype—phenotype relationships in MLP/TCAP-HCM
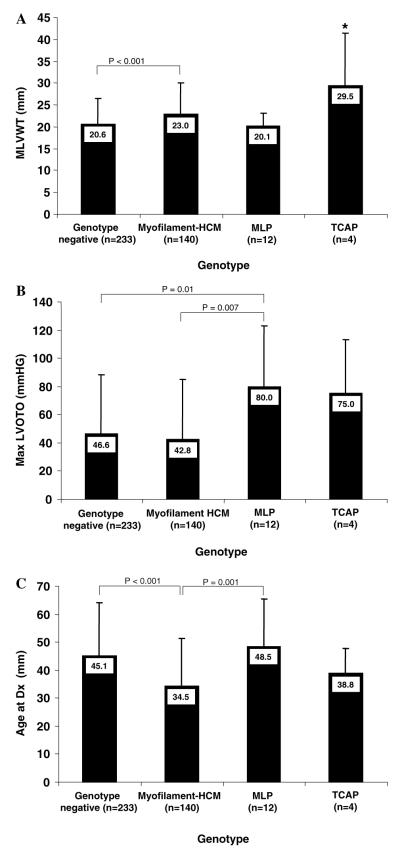
Compared to patients still lacking a mutation (genotype negative) and patients with myofilament-HCM, patients with mutations involving either MLP or TCAP more closely resembled the subset with myofilament-HCM (Fig. 3). The subset with MLP-HCM were, however, more obstructive (80 ± 43 mmHg) than both myofilament-HCM (42.8±42mmHg; p=0.01) and genotype negative-HCM (46.6±42mmHg; p=0.007). Despite the small sample size, patients with TCAP-HCM had significantly greater MLVWT (29.5±12mm) compared with either genotype negative-(20.6±6mm; p=0.006), myofilament-(23.0± 7.0mm; p=0.04), or MLP-HCM (20.1±3mm; p=0.01) and a similar age at diagnosis as myofilament positive-HCM (38.8±9 vs. 34.5±17 years old; p=0.6). When a subset analysis of patients with either Z-disc only (n=9) mutations or Z-disc mutation plus a concomitant myofilament (n=7) mutation was performed, the phenotypes of these two subgroups did not differ from each other on MLVWT (23.9±9mm vs. 20.6±3mm; p=0.3), MLVOTO (67.4±49mmHg vs. 93.5±20mmHg; p=0.2) or age at diagnosis (46.3±6 yrs vs. 45.8±6.8 yrs; p=0.9), supporting the role of MLP/TCAP mutations in pathogenesis of HCM.
Fig. 3.
Degree of hypertrophy (A), degree of left ventricular outflow tract obstruction (B), and age at diagnosis (C) for genotyped subjects. Genotyped patients with hypertrophic cardiomyopathy are grouped on the X-axis as hosting as hosting no putative mutation (genotype negative), hosting a myofilament mutation (myofilament-HCM), a MLP-mutation or a TCAP-mutation. Unless otherwise noted, all pair wise comparisons are not statistically significant. *p < 0.05 compared to all other groups.
Discussion
As critical components of the dynamic protein scaffolding between the sarcomere and cytoskeleton at the Z-line, the titin-muscle LIM protein—telethonin complex is involved in both cyto-architecture and mechano-signaling, thus serving as a potential link between myofilament-HCM and Z-disc-HCM. Prior to this study, one HCM-associated mutation in titin [28], four HCM-associated mutations in MLP [22,31] and two HCM-associated mutations in telethonin have been reported [21]. In addition, consistent with the notion that HCM and DCM are often allelic disorders, several DCM-associated mutation in these three Z-disc proteins have been discovered as well [23,24,32–34]. Based upon our observations in this study, the genes encoding Z-disc proteins currently implicated so far as only DCM-susceptibility genes constitute rational candidate genes to explore in HCM.
This study represents the largest series of patients examined for the three known subtypes of Z-disc-HCM whereby approximately 4% of unrelated patients harbored a mutation in either MLP (CSRP3) or TCAP. We did not observe any mutations in the giant protein, titin, which extends across half of the entire sarcomere. However, only those regions implicated previously in either HCM or DCM were examined. Among the 12 patients with a non-synonymous, amino-acid altering variant in the CSRP3-encoded MLP, a compelling case for disease-association exists at the present time for five patients (cases 8–12). Besides the L44P-MLP, R64C-MLP, and Y66C-MLP missense mutations, three patients (cases 9–11) also possessed a concomitant myofilament mutation: G1041fs/5-MYBPC3, I1131T-MYBPC3, and R162Q-TNNI3 respectively. The L44P-MLP variant along with the K42fs/165-MLP frameshift mutation localize to the LIM1-domain which is responsible for binding to α-actinin. In a yeast 2-hybrid assay, Geier [22] recently showed a significantly impaired binding affinity for α-actinin due to C58G-MLP.
The pathogenic mechanism for HCM in these patients hosting both MLP variants and myofilament mutations may be due to synergistic heterozygosity (two-hit hypothesis) as we have previously demonstrated in a patient hosting a known myosin binding protein C missense mutation and a functionally compromised frataxin mutation [35]. Previously, we demonstrated that among the 140 patients in our cohort previously established to have solely myofilament-HCM, 10 patients (7%) hosted two myofilament mutations with one of the variants usually involving myosin binding protein C [14]. Supporting the notion that both variants contributed to the expressed phenotype, these patients with multiple myofilament-HCM were younger at diagnosis and had greater hypertrophy than those having a single myofilament mutation. Herein, proportionately more patients with putative Z-disc-HCM also had a myofilament mutation raising the possibility that some of these variants may represent false positives. Future studies of the families represented by these HCM cases may shed light on the relative contributions of both the myofilament and the Z-disc mutation in the expressed phenotype.
The precise contribution of W4R-MLP (seen in seven patients, cases 1–7) in the pathogenesis of HCM remains an enigma. Four of the seven patients with W4R-MLP in the present study also have a published HCM-associated myofilament mutation. Initially, W4R was discovered as a DCM-associated mutation and was reportedly absent in 640 normal reference alleles [34]. Localizing to the telethonin-binding domain of MLP, it was not surprising to see in vitro assays demonstrating markedly reduced interaction/localization with telethonin [34]. Transgenic mouse models of W4R-MLP yield mice with a rather pronounced cardiomyopathy characterized by significant ventricular dilation and systolic dysfunction [36].
Recently, W4R-MLP was observed in 1 of 137 unrelated patients with HCM [31]. This variant was found in a patient with predominant apical HCM in which no myofilament mutations were identified. However, these investigators also observed W4R in 3 of 500 reference alleles (0.6% allelic frequency). We have now observed W4R in 1/400 reference alleles. While clearly a phenotype producing mutation in an overexpression transgenic mouse model, further studies are necessary to elucidate the precise role of W4R-MLP in the pathogenesis of cardiomyopathies in humans.
Finally, four patients (cases 13–16) hosted mutations in telethonin with one patient also having a myofilament Q998R-MYBPC3 genotype (case 16). These patients had severe hypertrophy (mean MLVWT=29.5 mm) whereas previously published TCAP probands had a mean MLVWT of 20 mm. In particular, the patient in our study with R70W-TCAP (case 15) had massive hypertrophy with a septal wall thickness of 46 mm. No other mutations in known HCM genes have been found in this individual. R70W-TCAP localizes to the functional domain essential for binding MLP.
Most of the HCM- and DCM-associated mutations reported in these three Z-disc proteins have not been characterized functionally. It remains to be determined whether or not the various mutations selectively perturb force generating (HCM-predisposing) or force transmitting (DCM-predisposing) functions.
Conclusions
In this study, HCM-susceptibility mutations in CSRP3 and TCAP represent uncommon causes of HCM, with a prevalence similar to troponin I- and actin-HCM. The combined clinical phenotype of MLP/TCAP-HCM resembles that of myofilament-HCM. Co-segregation and functional studies are now needed to dissect the relative contributions of the various Z-disc mutations to the pathogenesis and phenotypic expression of HCM.
Acknowledgments
We are grateful to the patients seen at the HCM clinic for their participation in this study and to Mr. Doug Kocer, the nurse coordinator of the HCM clinic.
Footnotes
M.J.A.’s research program is supported by the Dr. Scholl Foundation, the CJ Foundation for SIDS, the Doris Duke Charitable Foundation, the American Heart Association, and the National Institutes of Health (HD42569). M.J.A. is an established investigator of the American Heart Association. M.V. is funded by the National Heart, Lung, and Blood Institute (1R21HL078807 and R21Hl077706) and the Italian Ministry of Education, University and Research (MIUR) D.M. 26.01.2001 #13. J.A.T. is supported by the Texas Children’s Foundation Chair in Cardiac Research grants from the National Institutes of Health (P01), the John Patrick Albright Foundation, and TexGen.
References
- [1].Maron BJ. Hypertrophic cardiomyopathy: a systematic review. JAMA. 2002;287:1308–1320. doi: 10.1001/jama.287.10.1308. [DOI] [PubMed] [Google Scholar]
- [2].Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92:785–789. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- [3].Geisterfer-Lowrance AA, Kass S, Tanigawa G, Vosberg H, McKenna W, Seidman CE, Seidman JG. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- [4].Poetter K, Jiang H, Hassanzadeh S, Master SR, Chang A, Dalakas MC, Rayment I, Sellers JR, Fananapazir L, Epstein ND. Mutations in either the essential or regulatory light chains of myosin are associated with a rare myopathy in human heart and skeletal muscle. Nat. Genet. 1996;13:63–69. doi: 10.1038/ng0596-63. [DOI] [PubMed] [Google Scholar]
- [5].Watkins H, Conner D, Thierfelder L, Jarcho JA, MacRae C, McKenna WJ, Maron BJ, Seidman JG, Seidman CE. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat. Genet. 1995;11:434–437. doi: 10.1038/ng1295-434. [DOI] [PubMed] [Google Scholar]
- [6].Thierfelder L, Watkins H, MacRae C, Lamas R, McKenna W, Vosberg HP, Seidman JG, Seidman CE. Alpha-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell. 1994;77:701–712. doi: 10.1016/0092-8674(94)90054-x. [DOI] [PubMed] [Google Scholar]
- [7].Kimura A, Harada H, Park JE, Nishi H, Satoh M, Takahashi M, Hiroi S, Sasaoka T, Ohbuchi N, Nakamura T, Koyanagi T, Hwang TH, Choo JA, Chung KS, Hasegawa A, Nagai R, Okazaki O, Nakamura H, Matsuzaki M, Sakamoto T, Toshima H, Koga Y, Imaizumi T, Sasazuki T. Mutations in the cardiac troponin I gene associated with hypertrophic cardiomyopathy. Nat. Genet. 1997;16:379–382. doi: 10.1038/ng0897-379. [DOI] [PubMed] [Google Scholar]
- [8].Olson TM, Doan TP, Kishimoto NY, Whitby FG, Ackerman MJ, Fananapazir L. Inherited and de novo mutations in the cardiac actin gene cause hypertrophic cardiomyopathy. J. Mol. Cell. Cardiol. 2000;32:1687–1694. doi: 10.1006/jmcc.2000.1204. [DOI] [PubMed] [Google Scholar]
- [9].Mogensen J, Klausen IC, Pedersen AK, Egeblad H, Bross P, Kruse TA, Gregersen N, Hansen PS, Daandrup U, Borglum AD. Alpha-cardiac actin is a novel disease gene in familial hypertrophic cardiomyopathy. J. Clin. Invest. 1999;103:R39–R43. doi: 10.1172/JCI6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Van Driest SL, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Sarcomeric genotyping in hypertrophic cardiomyopathy. Mayo Clin. Proc. 2005;80:463–469. doi: 10.1016/S0025-6196(11)63196-0. [DOI] [PubMed] [Google Scholar]
- [11].Van Driest SL, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Yield of genetic testing in hypertrophic cardiomyopathy. Mayo Clin. Proc. 2005;80:739–744. doi: 10.1016/S0025-6196(11)61527-9. [DOI] [PubMed] [Google Scholar]
- [12].Genomics of Cardiovascular Development, Adaptation, and Remodelling. NHLBI Program of Genomic Applications, Harvard Medical School; [accessed 09/30/05]. http://www.cardiogenomics.org [Google Scholar]
- [13].Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, Benaiche A, Isnard R, Dubourg O, Burban M, Gueffet JP, Millaire A, Desnos M, Schwartz K, Hainque B, Komajda M. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- [14].Van Driest SL, Vasile VC, Ommen SR, Will ML, Gersh BJ, Nishimura RA, Tajik AJ, Ackerman MJ. Myosin binding protein C mutations and compound herterozygosity in hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2004;44:1903–1910. doi: 10.1016/j.jacc.2004.07.045. [DOI] [PubMed] [Google Scholar]
- [15].Van Driest SL, Jaeger MA, Ommen S, Will ML, Gersh BJ, Tajik AJ, Ackerman MJ. Comprehensive analysis of the beta-myosin heavy chain gene in 389 unrelated patients with hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2004;44:602–610. doi: 10.1016/j.jacc.2004.04.039. [DOI] [PubMed] [Google Scholar]
- [16].Erdmann J, Raible J, Maki-Abadi J, Hummel M, Hammann J, Wollnik B, Frantz E, Fleck E, Hetzer R, Regitz-Zagrosek V. Spectrum of clinical phenotypes and gene variants in cardiac myosin-binding protein C mutation carriers with hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2001;38:322–330. doi: 10.1016/s0735-1097(01)01387-0. [DOI] [PubMed] [Google Scholar]
- [17].Morner S, Richard P, Kazzam E, Hellman U, Hainque B, Schwartz K, Waldenstrom A. Identification of the genotypes causing hypertrophic cardiomyopathy in northern Sweden. J. Mol. Cell. Cardiol. 2003;35:841–849. doi: 10.1016/s0022-2828(03)00146-9. [DOI] [PubMed] [Google Scholar]
- [18].Garcia-Castro M, Reguero JR, Batalla A, Diaz-Molina B, Gonzalez P, Alvarez V, Cortina A, Cubero GI, Coto E. Hypertrophic cardiomyopathy: low frequency of mutations in the beta-myosin heavy chain (MYH7) and cardiac troponin T (TNNT2) genes among Spanish patients. Clin. Chem. 2003;49:1279–1285. doi: 10.1373/49.8.1279. [DOI] [PubMed] [Google Scholar]
- [19].Jaaskelainen P, Soranta M, Miettinen R, Saarinen L, Pihlajamaki J, Silvennoinen K, Tikanoja T, Laakso M, Kuusisto J. The cardiac beta-myosin heavy chain gene is not the predominant gene for hypertrophic cardiomyopathy in the Finnish population. J. Am. Coll. Cardiol. 1998;32:1709–1716. doi: 10.1016/s0735-1097(98)00448-3. [DOI] [PubMed] [Google Scholar]
- [20].Jaaskelainen P, Kuusisto J, Miettinen R, Karkkainen P, Karkkainen S, Heikkinen S, Peltola P, Pihlajamaki J, Vauhkonen I, Laakso M. Mutations in the cardiac myosin-binding protein C gene are the predominant cause of familial hypertrophic cardiomyopathy in eastern Finland. J. Mol. Med. 2002;80:412–422. doi: 10.1007/s00109-002-0323-9. [DOI] [PubMed] [Google Scholar]
- [21].Hayashi T, Arimura T, Itoh-Satoh M, Ueda K, Hohda S, Inagaki N, Takahashi M, Hori H, Yasunami M, Nishi H, Koga Y, Nakamura H, Matsuzaki M, Choi BY, Bae SW, You CW, Han KH, Park JE, Knoll R, Hoshijima M, Chien KR, Kimura A. Tcap gene mutations in hypertrophic cardiomyopathy and dilated cardiomyopathy. J. Am. Coll. Cardiol. 2004;44:2192–2201. doi: 10.1016/j.jacc.2004.08.058. [DOI] [PubMed] [Google Scholar]
- [22].Geier C, Perrot A, Ozcelik C, Binner P, Counsell D, Hoffmann K, Pilz B, Martiniak Y, Gehmlich K, van der Ven PF, Furst DO, Vornwald A, von Hodenberg E, Nurnberg P, Scheffold T, Dietz R, Osterziel KJ. Mutations in the human muscle LIM protein gene in families with hypertrophic cardiomyopathy. Circulation. 2003;107:1390–1395. doi: 10.1161/01.cir.0000056522.82563.5f. [DOI] [PubMed] [Google Scholar]
- [23].Gerull B, Gramlich M, Atherton J, McNabb M, Trombitas K, Sasse-Klaassen S, Seidman JG, Seidman C, Granzier H, Labeit S, Frenneaux M, Thierfelder L. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat. Genet. 2002;30:201–204. doi: 10.1038/ng815. [DOI] [PubMed] [Google Scholar]
- [24].Itoh-Satoh M, Hayashi T, Nishi H, Koga Y, Arimura T, Koyanagi T, Takahashi M, Hohda S, Ueda K, Nouchi T, Hiroe M, Marumo F, Imaizumi T, Yasunami M, Kimura A. Titin mutations as the molecular basis for dilated cardiomyopathy. Biochem. Biophys. Res. Commun. 2002;291:385–393. doi: 10.1006/bbrc.2002.6448. [DOI] [PubMed] [Google Scholar]
- [25].Labeit S, Kolmerer B. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270:293–296. doi: 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- [26].Valle G, Faulkner G, De Antoni A, Pacchioni B, Pallavicini A, Pandolfo D, Tiso N, Toppo S, Trevisan S, Lanfranchi G. Telethonin, a novel sarcomeric protein of heart and skeletal muscle. FEBS Lett. 1997;415:163–168. doi: 10.1016/s0014-5793(97)01108-3. [DOI] [PubMed] [Google Scholar]
- [27].Fung YW, Wang RX, Heng HH, Liew CC. Mapping of a human LIM protein (CLP) to human chromosome 11p15.1 by Xuorescence in situ hybridization. Genomics. 1995;28:602–603. doi: 10.1006/geno.1995.1200. [DOI] [PubMed] [Google Scholar]
- [28].Satoh M, Takahashi M, Sakamoto T, Hiroe M, Marumo F, Kimura A. Structural analysis of the titin gene in hypertrophic cardiomyopathy: identification of a novel disease gene. Biochem. Biophys. Res. Commun. 1999;262:411–417. doi: 10.1006/bbrc.1999.1221. [DOI] [PubMed] [Google Scholar]
- [29].Ackerman MJ, Van Driest SV, Ommen SR, Will ML, Nishimura RA, Tajik AJ, Gersh BJ. Prevalence and age-dependence of malignant mutations in the beta-myosin heavy chain and troponin T gene in hypertrophic cardiomyopathy: a comprehensive outpatient perspective. J. Am. Coll. Cardiol. 2002;39:2042–2048. doi: 10.1016/s0735-1097(02)01900-9. [DOI] [PubMed] [Google Scholar]
- [30].Van Driest SL, Ellsworth EG, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Prevalence and spectrum of thin filament mutations in an outpatient referral population with hypertrophic cardiomyopathy. Circulation. 2003;108:445–451. doi: 10.1161/01.CIR.0000080896.52003.DF. [DOI] [PubMed] [Google Scholar]
- [31].Newman B, Cescon D, Woo A, Rakowski H, Erikkson MJ, Sole M, Wigle ED, Siminovitch KA. W4R variant in CSRP3 encoding muscle LIM protein in a patient with hypertrophic cardiomyopathy. Mol. Genet. Metab. 2005;84:374–375. doi: 10.1016/j.ymgme.2004.11.013. Epub 2005 Jan 22. [DOI] [PubMed] [Google Scholar]
- [32].Hayashi T, Arimura T, Ueda K, Shibata H, Hohda S, Takahashi M, Hori H, Koga Y, Oka N, Imaizumi T, Yasunami M, Kimura A. Identification and functional analysis of a caveolin-3 mutation associated with familial hypertrophic cardiomyopathy. Biochem. Biophys. Res. Commun. 2004;313:178–184. doi: 10.1016/j.bbrc.2003.11.101. [DOI] [PubMed] [Google Scholar]
- [33].Mohapatra B, Jimenez S, Lin JH, Bowles KR, Coveler KJ, Marx JG, Chrisco MA, Murphy RT, Lurie PR, Schwartz RJ, Elliott PM, Vatta M, McKenna W, Towbin JA, Bowles NE. Mutations in the muscle LIM protein and alpha-actinin-2 genes in dilated cardiomyopathy and endocardial fibroelastosis. Mol. Genet. Metab. 2003;80:207–215. doi: 10.1016/s1096-7192(03)00142-2. [DOI] [PubMed] [Google Scholar]
- [34].Knoll R, Hoshijima M, Hoffman HM, Person V, Lorenzen-Schmidt I, Bang ML, Hayashi T, Shiga N, Yasukawa H, Schaper W, McKenna W, Yokoyama M, Schork NJ, Omens JH, McCulloch AD, Kimura A, Gregorio CC, Poller W, Schaper J, Schultheiss HP, Chien KR. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111:943–955. doi: 10.1016/s0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- [35].Van Driest SL, Gakh O, Ommen SR, Isaya G, Ackerman MJ. Molecular and functional characterization of a human frataxin mutation found in hypertrophic cardiomyopathy. Mol. Genet. Metab. 2005:1096–7192. doi: 10.1016/j.ymgme.2005.04.010. [DOI] [PubMed] [Google Scholar]
- [36].Arber S, Hunter JJ, Ross J, Jr., Hongo M, Sansig G, Borg J, Perriard JC, Chien KR, Caroni P. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88:393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]