Abstract
The zebrafish ventricular myosin heavy chain (vmhc) gene exhibits restricted expression in the ventricle. However, the molecular mechanism underlying this chamber-specific expression is unclear. Here, we exploited both transient and transgenic technologies to dissect the zebrafish vmhc promoter. We demonstrated that a combination of two transient assays in this animal model quickly identified chamber-specific cis-elements, isolating a 2.2 kb fragment upstream from the vmhc gene that can drive ventricle-specific expression. Furthermore, deletion analysis identified multiple cis-elements that exhibited cardiac-specific expression. To achieve chamber specificity, a distal element was required to coordinate with and suppress a proximal enhancer element. Finally, we discovered that Nkx2.5-binding sites (NKE) were essential for this repressive function. In summary, our study of the zebrafish vmhc promoter suggests that ventricle-specific expression is achieved through an inhibitory mechanism that suppresses expression in the atrium.
Keywords: Vmhc, chamber specificity, promoter analysis, transgenic fish
INTRODUCTION
During vertebrate embryonic development, the heart first forms a simple linear tube and then divides into two types of chambers: the ventricle and atrium (Moorman and Christoffels, 2003). These two chambers not only differ morphologically, but also physiologically with different characteristic rates of contractility (Satin et al., 1988). In order to adapt for these different properties, the ventricle and atrium exhibit distinct chamber-specific gene expression profiles of sarcomeric genes such as myosin heavy chains (MHCs) and myosin light chains (MLCs) (Tabibiazar et al., 2003). There are two cardiac myosin heavy chain genes in mammals, α-and β-MHC (Weiss and Leinwand, 1996; Morkin, 2000), both of which are expressed in the heart tube during early embryogenesis. In the mouse, β-MHC expression is later restricted to the ventricle, while α-MHC expression continues to remain high in the atrium but decreases in the ventricle. Mouse myosin light chain 2v (MLC2v) is continuously detected in the ventricle, whereas myosin light chain 2a (MLC2a) is initially expressed throughout the linear heart tube but later becomes restricted to the atrium (Kubalak et al., 1994; Zammit et al., 2000). In zebrafish, multiple cardiac-specific MHCs have been identified. Atrial myosin heavy chain (amhc)is expressed in the atrium (Berdougo et al., 2003), while ventricular myosin heavy chain (vmhc) is expressed in the ventricle (Yelon et al., 1999). In contrast, the expression of zebrafish myosin light chains does not appear to be restricted to any particular chamber (Chen et al., 2008).
The molecular mechanisms of chamber-restricted expression have been extensively investigated by analyzing promoters of chamber-specific genes (Moorman and Christoffels, 2003; Small and Krieg, 2004). The mouse MLC2v is the most thoroughly studied ventricle-specific gene (Ross et al., 1996; Nguyen-Tran et al., 2003). Promoter dissection identified HF-1a and HF-1b/MEF2 sites, which are responsible for right ventricular expression; however, the specific elements for left ventricular expression still remain unknown. Irx4, a transcription factor that exhibits ventricle-specific expression, has been suggested to play an activating role in determining ventricular specificity. In chickens, Irx4 activates ventricular myosin heavy chain-1 (VMHC1) and suppresses the expression of atrial myosin heavy chain-1 (AMHC1) in the ventricle (Bao et al., 1999). This ventricle-specific gene expression is generally believed to occur through transcriptional activation in specific chambers, while atrium-specific gene expression is achieved by the transcriptional repression of genes in the ventricle (Small and Krieg, 2004). In the quail, slow myosin heavy chain 3 (slMHC3) is initially expressed throughout the linear heart tube, then decreases in the ventricle (Wang et al., 1996). This inhibition requires a vitamin D receptor-binding element (VDRE) and a retinoic acid response element (RARE). The interaction of these factors with Irx4 may be involved in this atrium-specific process (Wang et al., 2001). In addition, analysis of the 5′ regulatory sequences of the mouse MLC2a promoter revealed binding sites for NKX2.5, MEF2, SRF, and retinoic acid. In summary, we conclude that there is no universal molecular mechanism for chamber-specific gene regulation. Rather, additional chamber-specific promoters need to be dissected to further understand the molecular mechanisms mediating this important cardiogenesis event.
Investigation of chamber-specific gene expression requires whole animal models to provide in vivo contexts, in contrast to dissection of cardiac-restricted promoters using adapted cell culture systems. The mouse is currently the most frequently used animal model, but the cost and time to raise numerous transgenic mice prevents thorough dissection of chamber-specific promoters. Thus, we exploited zebrafish to study the molecular mechanism of chamber-restricted gene expression, as they are an economic animal model system to generate transgenic lines and are easily accessible to transient reporter analyses for quantification. Combining both transient and transgenic technologies, we dissected the zebrafish ventricular myosin heavy chain (vmhc) gene promoter. We found that transient injection analysis in this animal model was useful to dissect chamber-specific promoters, and the results were later confirmed by the generation of stable transgenic fish lines. We identified two elements that cooperatively regulated chamber specificity. Interestingly, the distal element functioned as a repressor to suppress atrial expression, suggesting a novel inhibitory mechanism for ventricle-specific gene expression. Finally, our data suggested that the binding sites of cardiogenic transcription factors Nkx2.5 play important roles in this inhibitory function.
RESULTS
A 2.2-kb Fragment Upstream of the vmhc Gene Is Sufficient to Drive Ventricle-Specific Reporter Expression
The vmhc transcript can first be detected in the heart primordium in the anterior lateral plate mesoderm around the 10-somite stage (see Supp. Fig. S1A [arrow], which is available online); this is later than that of titin, which occurs around the 5-somite stage (Seeley et al., 2007), but is earlier than that of essential or regulatory myosin light chains, both of which occur around 16-somites (Chen et al., 2008). Previously, vmhc expression was shown to be immediately restricted to the ventricle after its onset (Yelon et al., 1999), and we consistently detected vmhc expression in the ventricle at each stage of cardiogenesis: cardiac progenitor migration, tube formation, and chamber formation (Supp. Fig. S1A—F). In addition, we detected residual expression in the atrium at 36 hr post-fertilization (hpf) (Supp. Fig. S1E). In addition to cardiac expression, vmhc expression was also detected in the somites starting at 24 hpf (data not shown) and in the extraocular muscles and pharyngeal muscles after 3 days post-fertilization (dpf) (Supp. Fig. S1G).
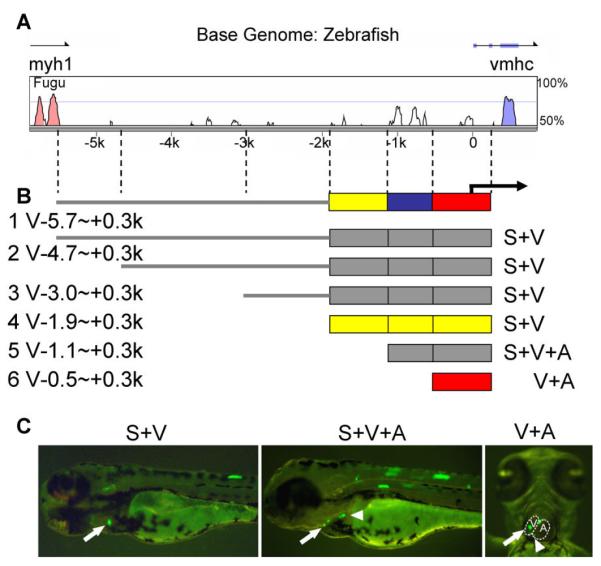
The zebrafish vmhc gene (GenBank accession number: XM_001332905) consists of 39 exons on chromosome 2 and spans 13.3 kb. vmhc is located 6 kb downstream of myh1 (zgc:113832), another MHC homologue that exhibits a ventricle-restricted expression pattern, as revealed by whole mount in situ hybridization (data not shown). By searching the Fugu (Takifugu rubripes) genome, we identified a chromosomal region syntenic to the zebrafish vmhc gene; this region contains a pair of tandemly arranged MHC homologues (Supp. Fig. S2). We aligned and compared the intergenic sequences between these two pairs of mhc genes in the zebrafish (6.7 kb) and Fugu (6.5 kb) using rVista software (Frazer et al., 2004) and were able to identify several conserved regions across both species (Fig. 1A, white peaks), suggesting a regulatory element function for these genes.
Fig. 1.
Identification of a 2.2-kb fragment from a ventricle-specific promoter. A: Sequence comparison of upstream intergenic sequences between the zebrafish vmhc gene with its Fugu homologue. Red and blue peaks represent coding regions for myh1 and vmhc, respectively. White peaks represent inter-species conserved regions. B: Summary of promoter analysis by transient co-injection of naked DNA with an EGFP fragment. The full-length intergenic V-5.7∼+0.3k fragment can drive GFP expression in both the somites and ventricle, as can the V-4.7∼+0.3k, V-3∼+0.3k, and V-1.9∼+0.3k fragments. However, V-1.1∼+0.3k drives GFP expression in both the ventricle and atrium, and V-0.5∼+0.3k drives GFP expression in the heart but not in the somites. The yellow, blue, and red bars on the top line represent fragments required for chamber specificity, somite expression, and cardiac expression, respectively. The yellow or red bars below represent the minimal element sufficient for chamber-specific or cardiac expression, respectively. C: Representative pictures of 3-dpf embryos after co-injection of promoter DNA and the EGFP fragment. Left and middle panels are lateral views; anterior to the left. The right panel is a ventral view; anterior to the top. GFP-positive cells can be detected in single cells or in a group of cells in the somites (left and middle panel) or the heart. The ventricle (indicated by arrows) and the atrium (indicated by arrowheads) could be distinguished due to embryo transparency.
To experimentally dissect the chamber-specific vmhc promoter, we used transient co-injection assays (Muller et al., 1999), which are based on the principle that DNA fragments of different origin usually integrate together into a single breaking point on the chromosome (Bishop and Smith, 1989). Therefore, when a promoter fragment is co-injected with a GFP reporter fragment into one-cell-stage embryos, the GFP expression pattern faithfully reflects promoter activity. We generated a series of promoter fragments derived from the 6.7-kb intergenic region upstream of the zebrafish vmhc gene. All fragments contained the basal promoter as well as 300 bp downstream of the transcription start site. We detected sporadic GFP-positive cells in the heart at 2–3 dpf (Fig. 1C) and in the somites at 3– 4 dpf. It was possible to distinguish GFP-positive cells in the ventricle (arrow) from those in the atrium (arrowhead), due to the transparency of zebrafish embryos. In embryos that were co-injected with the full-length intergenic sequence, V-5.7∼+0.3k (Fig. 1B, line 1), GFP-positive cells were only detected in the somites and ventricle and not in the atrium, suggesting that this 6-kb fragment recapitulated endogenous vmhc expression. This transient co-injection assay appeared to be specific, as a similar ventricle-restricted expression pattern was also observed in embryos co-injected with three other fragments containing a series of 5′ deletions (until—1.9 kb) (Fig. 1B, line 2–4). Further deletions, however, disrupted chamber specificity or ablated expression in the somites. GFP-positive cells were detected in both the atrium and ventricle after co-injection of V-1.1∼+0.3k (Fig. 1B, line 5) or V-0.5∼+0.3k (Fig. 1B, line 6), while GFP-positive cells could be detected in the somites after co-injection of V-1.1∼+0.3k, but not V-0.5∼+0.3k. In summary, these studies indicated that transient co-injection assays are useful for dissecting chamber-specific promoters. Furthermore, we found that a 2.2-kb element was sufficient to recapitulate chamber-specific vmhc expression.
Further Dissection of the vmhc Promoter by Transient Co-Injection Assays
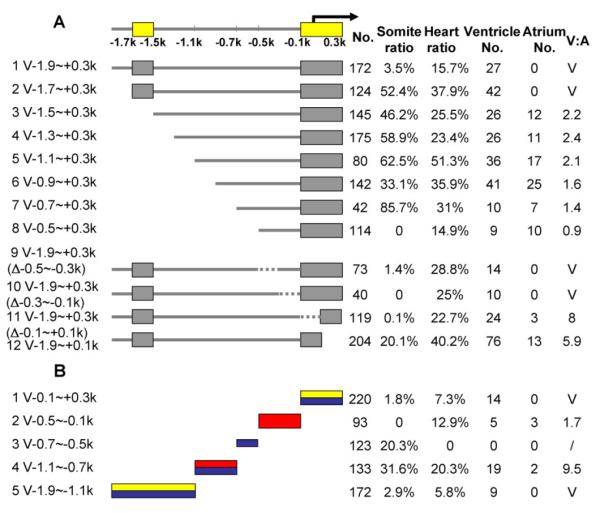
Next, we quantified data from the transient co-injection assays by counting 4-dpf embryos with GFP-positive cells in either the ventricle or atrium. The results were represented as V:A ratios to reflect chamber specificity, where V represents the number of fish with GFP-positive cells in the ventricle and A represents the number of fish with GFP-positive cells in the atrium. As summarized in Figure 2, the V:A ratio for V-1.9∼+0.3k (Fig. 2A, line 1), V-1.1∼+0.3k (Fig. 2A, line 5), and V-0.5∼+0.3k (Fig. 2A, line 8) were V only, 2.4 and 0.9, respectively, suggesting that a repressor located between -1.9 to -1.1 kb was required for chamber specificity. We also calculated the percentage of fish that contained GFP-positive cells in either the somites or the heart (Fig. 2).
Fig. 2.
Dissection of the vmhc promoter using transient co-injection assays. A: Schematic summary of results of serial deletions (lines 1–8) or internal deletions (lines 9–12) to identify minimal cis-elements required for chamber specificity. A distal (V-1.7∼–1.5k) and a proximal (V-0.1∼+0.3k) cis-element were identified and are indicated by yellow bars. B: Schematic summary of results of the minimal cis-elements sufficient for chamber specificity. Both V-0.1∼+0.3k (line 1) and V-1.9∼–1.1k (line 5) drive GFP expression only in the ventricle (yellow bar), while V-0.5∼–0.1k (line 2) and V-1.1∼–0.7k (line 4) drive GFP expression in the entire heart (red bar). Four fragments drive GFP expression in skeletal muscle (blue bar), two are strong enhancers (V-0.7∼–0.5k, line 3 and V-1.1∼–0.7k, line 4) and two are weak enhancers (V-0.1∼+0.3k, line 1 and V-1.9∼–1.1k, line 5). Note that lines 2–5 are fragments lacking the basal promoter. No., number of injected embryos that survived to 4 dpf; Somite/Heart ratio, number of fish with tissue-restricted GFP-positive cells over the total number of fish that survived to 4 dpf; Ventricle No. or Atrium No., number of fish with GFP-positive cells in the ventricle or atrium. A fish with GFP-positive cells in both chambers was counted in both categories. V:A, ratio of GFP-positive cells in the ventricle to that in the atrium.
To identify the minimal cis-elements needed to drive chamber-specific gene expression, we generated a series of fine deletion constructs that deleted every 200 bp from the distal end of the 2.2-kb fragment (Fig. 2A, lines 1–8). Deletion of a 200-bp region located between -1.7 and -1.5 kb resulted in reduction of the V:A ratio from completely ventricle to ∼2 (Fig. 2A, lines 2–3), suggesting that this region represents the minimal distal cis-element required for chamber specificity. We did not generate 5′-deletions from -0.5 kb to +0.3 kb, as the V:A ratio was 0.9 when the V-0.5∼+0.3k fragment was co-injected (Fig. 2A, line 8). Instead, we generated a series of four internal deletion constructs within this proximal region. Deletion of either -0.1 kb to +0.1 kb or +0.1 kb to +0.3 kb resulted in a marginal decrease in the V:A ratio to 8 or 5.9, respectively (compare Fig. 2A, lines 1 and 9–12), suggesting the involvement of a proximal cis-element for chamber-specific vmhc expression.
To identify elements sufficient for chamber-specific gene expression, we generated a series of short fragments within the 2.2-kb region. Most of these fragments (Fig. 2B, lines 2–5) lacked the vmhc basal promoter, but GFP signals could still be detected, possibly due to a stretch of sequences located immediately before the GFP reporter that mimicked the basal promoter function. We found that co-injection of the V-1.9∼–1.1k fragment (Fig. 2B, line 5), which covered the distal element, drove GFP expression only in the ventricle, although co-injection of the V-1.7∼–1.5k fragment did not result in any GFP-positive fish (data not shown). Co-injection of the V-0.1∼+0.3k proximal element (Fig. 2B, line 1) was also able to drive chamber-specific expression. In contrast, co-injection of V-1.1∼–0.7k (Fig. 2B, line 4) or V-0.5∼–0.1k (Fig. 2B, line 2) drove GFP expression in both heart chambers. We also identified four fragments that drove GFP expression in skeletal muscle. V-0.7∼–0.5k (Fig. 2B, line 3) and V-1.1∼–0.7k (Fig. 2B, line 4) drove GFP expression in the somites of 20.3 and 31.6% of embryos, respectively, while V-0.1∼+0.3k (Fig. 2B, line 1) and V-1.9∼–1.1k (Fig. 2B, line 5) drove GFP expression in the somites of 1.8 and 2.9% of embryos, respectively. These data suggest that multiple modular enhancer elements co-exist within the vmhc promoter to cooperatively regulate tissue-specific expression. Thus, both distal and proximal elements are involved in ventricle-specific expression.
Dissection of the vmhc Promoter Using Tol2-Based Transient Assays
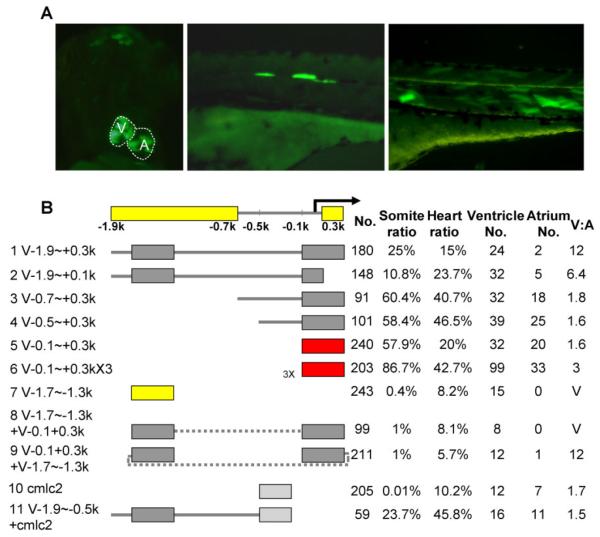
To confirm transient co-injection assay results, we performed classic promoter analysis by cloning a promoter fragment and the GFP reporter gene into a single construct. We used the Tol2 transposon vector system, which was originally identified in Medaka fish (Koga et al., 1996) and later adapted as a vehicle to efficiently integrate ectopic DNA into the zebrafish genome (Kawakami et al., 2000, 2004). Transient injection of Tol2-based plasmids was previously shown to be a valuable tool to analyze tissue-specific promoters in zebrafish (Fisher et al., 2006b; Korzh, 2007). In our hands, adaptation of the Tol2 vector facilitated the identification of GFP-positive fish, due to dramatically increased intensity and larger number of GFP-positive cells within each injected embryo. We easily detected GFP-positive cells in the heart at 2 dpf (Fig. 3A, left panel) and in skeletal muscle at 3 dpf, both of which occurred earlier than in embryos from co-injection assays. Due to increased sensitivity and reduced mosaicism, we could detect GFP in several subsets of muscles, including the extraocular muscles (data not shown), muscle pioneer cells in the body midline (Fig. 3A, middle panel), and myocytes in the whole myotome (Fig. 3A, right panel). Switching to the Tol2 system did not affect the percentage of embryos with GFP-positive cells.
Fig. 3.
Dissection of the vmhc promoter by transient assays using Tol2-based vectors. A: Representative pictures of 3-dpf embryos injected with Tol2 transposon constructs containing vmhc promoter sequences (left and right panel, V-0.5∼+0.3k; middle panel, V-1.9∼+0.3k). Shown in the left panel is a ventral view with anterior to the top; right and middle panels are lateral views with anterior to the left. Multiple GFP-positive cells could be detected in the heart (left) and/or skeletal muscle, including the eye muscle, muscle pioneer cells located in the body midline (middle), and myocytes in the somites (right). B: Schematic summary of results from transient assays using the Tol2 transposon system. A distal (V—1.9∼–0.7k) and a proximal (V+0.1∼+0.3k) element required for the chamber specificity were identified, consistent with results from transient co-injection assays in Figure 2. A shorter distal element (V-1.7∼–1.3k, line 7) is sufficient to drive ventricle-specific expression in this assay. In contrast to results from transient co-injection assays, the proximal element (V-0.1∼+0.3k, line 5) drives GFP expression in the whole heart without chamber specificity in Tol2-based assays. When the distal and proximal elements are linked in tandem (line 8) or in reverse (line 9), the constructs drive GFP expression in the ventricle. However, the distal element cannot alter the expression of the cardiac cmlc2 promoter to be chamber-specific (line 11), which by itself drives GFP expression in the whole heart (line 10). The minimal elements required for chamber specificity (yellow bars, top line), for chamber-restricted expression (yellow bars, line 7) and for cardiac expression (red bars, lines 5–6) are indicated. No., number of injected embryos that survived to 4 dpf; Somite ratio/Heart ratio, number of fish with tissue-restricted GFP-positive cells over total number of fish that survived to 4 dpf; Ventricle No. or Atrium No., number of fish with GFP-positive cells in the ventricle or atrium. A fish with GFP-positive cells in both chambers was counted in both categories. V:A, ratio of GFP-positive cells in the ventricle over that in the atrium.
In contrast to results from the transient co-injection assays, injection of the full-length 2.2-kb fragment (V-1.9∼+0.3k) resulted in a smaller percentage of GFP-positive cells in the atrium (Fig. 3B, line 1). This may be due to the increased sensitivity of the Tol2 system, which more accurately reflects endogenous vmhc expression. As shown in Supp. Figure S1D, residual vmhc mRNA could still be detected in the atrium by in situ hybridization at 36 hpf. Injection of V-0.7∼+0.3k (Fig. 3B, line 3) or V-0.5∼+0.3k (Fig. 3B, line 4), two fragments lacking the distal element, resulted in V:A ratios of less than 2, while injection of V-1.9∼+0.1k, a fragment lacking the proximal region (Fig. 3B, line 2), induced a V:A ratio of 6.4. The results from the Tol2 system experiments confirmed the existence of two cis-elements required for vmhc chamber-specific expression and supported the notion that the distal element plays a stronger role than the proximal element in determining ventricular specificity.
The distal element was sufficient to drive ventricle-specific expression using the Tol2 system, consistent with results from transient co-injection assays. Indeed, injection of the shorter distal V-1.7∼–1.3k element (Fig. 3B, line 7, compared to Fig. 2B, line 5) was sufficient to drive ventricle-specific GFP expression, most likely due to the increased sensitivity of the Tol2 system. However, in contrast to the co-injection assays, injection of the proximal V-0.1∼+0.3k element (Fig. 3B, line 5, compared to Fig. 2B, line 1) resulted in a low V:A ratio of 1.6. One explanation for the inconsistency between the two transient assays could be that variable copies and ratios of the proximal elements and GFP reporter fragments were examined in co-injection assays (Marini et al., 1988; Bishop and Smith, 1989), while the 1:1 ratio of the proximal element and GFP reporter was examined in Tol2-based assays. Consistent with this hypothesis, the injection of a construct containing 3 copies of the proximal element located upstream from a GFP reporter increased the V:A ratio to 3 (Fig. 3B, line 6).
When we combined the minimal distal and proximal elements together in tandem (Fig. 3B, line 8) or in reverse (Fig. 3B, line 9), both constructs were able to drive ventricle-specific expression. This result suggested that the distal element, which may function as a repressor to inhibit gene expression in the atrium, imposed a dominant effect over the proximal element. To test whether the distal element may function as a universal repressor in the atrium, we generated a chimeric construct consisting of V-1.9∼–0.5k, which includes the distal element, and a 300-bp cardiac promoter from the cmlc2 gene (Huang et al., 2003). Much like that of the cmlc2 promoter alone, the chimeric construct exhibited whole heart expression without chamber specificity (Fig. 3B, lines 10 –11). Therefore, we concluded that the repressor function of the distal element is not universal, but functions cooperatively with the proximal vmhc element to achieve chamber-specificity.
Dissection of the vmhc Promoter by Generating Stable Transgenic Fish Lines
To confirm conclusions from transient assays, we generated four transgenic fish lines using Tol2-based constructs. In the Tg(V-1.9k:egfp) line, the GFP reporter was detected only in the ventricle at both the embryonic (Fig. 4C,E,G) and adult stages (Fig. 4I), confirming the chamber specificity of this 2.2-kb fragment. In both the Tg(V-0.7k:egfp) and Tg(V-0.5k:egfp) lines, GFP was detected in the whole heart without chamber specificity (Supp. Figs. S3A, 4D,F,H,J). This result confirmed that the distal element is required for chamber specificity and supported the hypothesis that this element functions as a repressor in the atrium. In the Tg(V-0.1k:egfp) line, GFP was detected in the entire heart in both embryos and in adult fish (Supp. Fig. S3B and data not shown). These results were consistent with results from Tol2-based transient assays, but in contrast to those from co-injection assays. Of note, each transgenic line was expected to contain a single copy of the distal element with a GFP reporter in each insertion locus.
Fig. 4.
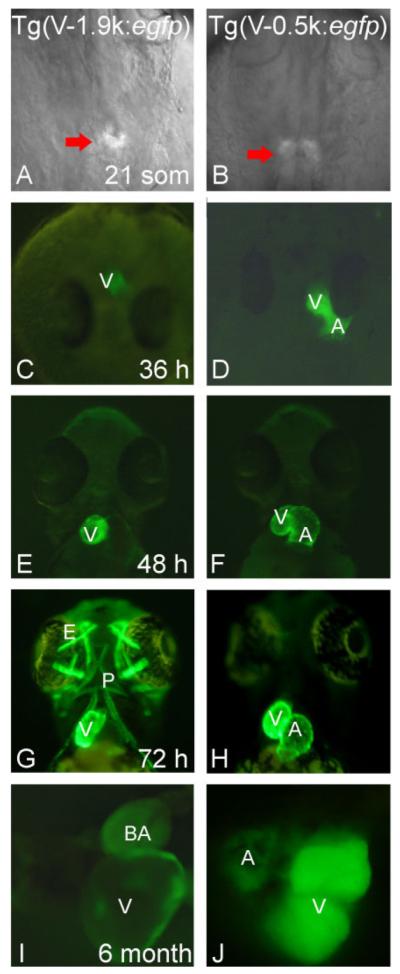
Results from stable transgenic lines are consistent with those from transient assays. A, C, E, G, I: Transgenic fish expressing GFP under the control of the V-1.9∼+0.3k fragment. B, D, F, H, J: Transgenic fish expressing GFP under the control of the V-0.5∼+0.3k fragment. GFP expression begins around the 21-somite stage (A, B). The Tg(V-1.9k:egfp) transgenic line expresses GFP only in the ventricle as well as in both embryonic (C, E, G) and adult stages (I), while the Tg(V-0.5k:egfp) line expresses GFP in the whole heart (D, F, H, J). Tg(V-1.9k:egfp) has an early onset and drives strong GFP expression in the extraocular muscles, pharyngeal muscles, and other muscle types (G; Supp. Fig. S3C, F), while Tg(V-0.5k:egfp) has a late onset and drives weak GFP expression in these muscles types (H; Supp. Fig. S3H). V, ventricle; A, atrium; E, extraocular muscles; P, pharyngeal muscles; BA, bulbus arteriosus. A,B: Dorsal view, anterior to the top. C,D: Head on view. E—H: Ventral view, anterior to the top. I,J: Dissected adult heart.
The fluorescent GFP signal in the heart can be detected in all four transgenic lines starting from the 21-somite stage (Fig. 4A and B), while the GFP transcript can be detected at 16-somites by in situ hybridization (data not shown). In addition to cardiac-specific expression, both the Tg(V-1.9k:egfp) and Tg(V-0.7k:egfp) lines exhibited strong GFP expression in extraocular muscles, pharyngeal muscles, and other muscle types (Fig. 4G; Supp. Fig. S3A,C,D), reaffirming the endogenous vmhc expression pattern. In contrast, GFP signals were barely detectable in skeletal muscles in both of the Tg(V-0.5k:egfp) and Tg(V-0.1k:egfp) lines during early embryogenesis (Fig. 4H; Supp. Fig. S3B). Later, these two lines exhibited weak GFP expression in cephalic musculature and medium-to-strong GFP expression in the trunk musculature (Supp. Fig. S3E). The skeletal muscle expression pattern persisted in adult animals, but was restricted to muscles around the eye, jaw, operculum (Supp. Fig. S3F—H), and subgroups of muscles next to the body border and close to the fins (Supp. Fig. S3H).
Nkx2.5 Binding Sites Are Important for the Chamber-Specific Activity of the vmhc Promoter
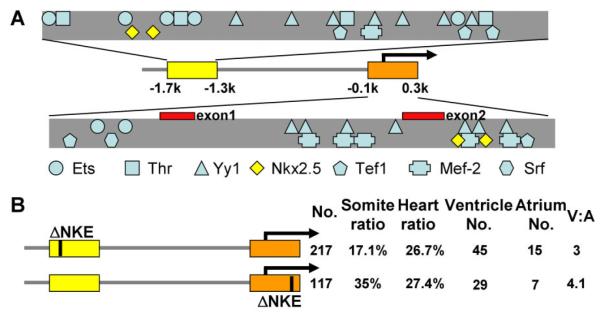
To identify transcription factor(s) involved in regulating chamber-specific vmhc transcription, we used bioinformatics software to predict transcription factor-binding sites within both distal and proximal elements, focusing on those previously reported to be involved in either chamber specificity or MHC gene regulation. We identified multiple binding sites of Ets, Thr, Yy1, Nkx2.5, Tef1, Mef-2, and Srf, but not Tbx, Gata4/5/6, or Irx4 (Knowlton et al., 1995; Ross et al., 1996; Lee et al., 1997; Chen et al., 1998; Wang et al., 2001; Gupta et al., 2003; Small and Krieg, 2003) (Fig. 5A). For the following reasons, we elected to further examine nkx2.5, a member of the NK homeobox gene family and one of the earliest cardiogenic factors. First, the expression of nkx2.5 was reported to be initially restricted to the ventricle at the 7-somite stage during early zebrafish embryogenesis (Schoenebeck et al., 2007). Expression of nkx2.5 then expands into the whole heart at 14-somites, the stage at which vmhc expression begins. Second, nkx2.5 has been previously suggested to be required for ventricular expression of Irx4 (Bruneau et al., 2000). Third, two Nkx2.5-binding sites (NKE) in either the proximal or distal element were identified. An NKE site has also been detected in the corresponding conserved region of the Fugu vmhc promoter (Supp. Fig. S4). We generated fine deletions in the zebrafish 2.2-kb fragment to examine the function of these NKE elements. A 20-bp deletion that eliminated both NKE sites in the distal element of the 2.2-kb fragment reduced the V:A ratio from 12 to 3, and a 15-bp deletion that eliminated both NKE sites in the proximal element resulted in a reduction of the V:A ratio to 4.1 (Fig. 5B). Taken together, this data suggest that NKX-binding sites are required for chamber specificity.
Fig. 5.
nkx2.5 is important for ventricle-specific vmhc gene expression. A: Shown are potential transcriptional factor-binding sites in distal and proximal elements, as predicted by bioinformatic analysis. Yellow diamonds represent the Nkx-binding sites (NKE); red bars represent the first two non-coding exons. B: Schematic summary of results from transient assays using the Tol2 transposon system. Deletion of the two NKE sites in the distal element (∼20 bp) or in the proximal element (∼15 bp) resulted in loss of ventricular specificity. No., number of injected embryos that survived to 4 dpf; Somite ratio/Heart ratio, number of fish with tissue-restricted GFP-positive cells over total number of fish that survived to 4 dpf; Ventricle No. or Atrium No., number of fish with GFP-positive cells in the ventricle or atrium. A fish with GFP-positive cells in both chambers was counted in both categories. V:A, ratio of GFP-positive cells in the ventricle over that in the atrium.
DISCUSSION
A Novel Repressive Mechanism to Achieve Ventricle-Specific Gene Expression
Combining both transient and transgenic techniques in zebrafish, we report here the identification of a 2.2-kb fragment upstream of the vmhc gene that can recapitulate ventricle-restricted gene expression. Further dissection of this fragment suggested a repressor model to explain ventricular chamber specificity. We found that multiple elements set the stage for cardiac-restricted expression, while both a proximal element and a distal element are required for ventricular specificity. The proximal element (located between -0.1 to +0.3kb) encompasses the vmhc transcriptional start site, the first two exons, the first intron, and part of the second intron. Compared to the proximal element, the distal element (-1.7 to -1.3kb) plays a more significant role in chamber specificity. Addition of this 400-bp distal repressor element imposes an inhibitory function to prevent vmhc gene expression in the atrium. This inhibitory function needs to be conferred to a specific basal promoter, such as the one located in the proximal enhancer element of the vmhc gene. It is possible that the basal vmhc promoter contains regulatory components that are likely expressed in the ventricle, as suggested by co-injection assays. Addition of the distal element tips the balance strongly in favor toward ventricle-specific expression. Thus, we describe a novel inhibitory mechanism to achieve ventricular specificity, which differs from the previous concept that ventricle-specific genes are directly controlled by an activating regulatory program, while atrium-specific genes are regulated by a repressor mechanism. It remains to be determined, however, whether the inhibitory mechanism identified here also exists in other ventricle-specific genes and/or in other species.
Nkx2.5 Binding Sites Are Involved in the Repressive Function of the Distal Element to Achieve Ventricular Specificity of the vmhc Gene
Recent investigations using several model organisms have suggested an important function for nkx2.5 in determining cardiac chamber specificity. Mouse knockouts of Nkx2.5 and dHand result in the complete absence of a ventricle, suggesting that Nkx2.5 may function together with dHand to determine ventricle formation (Yamagishi et al., 2001). This activator function of Nkx2.5 was believed to be regulated by Irx4 expression in the ventricle. At least 5 NKEs have been identified upstream of Irx4, and depletion of Nkx2.5 leads to reduction of Irx4 expression (Yamagishi et al., 2001; Small and Krieg, 2004). In Xenopus, nkx2.5 plays an important function in regulating the expression of atrial natriuretic factor (anf) in the atrium. Deletion of the NKE in the anf promoter expanded expression from the atrium into the whole heart, suggesting an inhibitory function of NKE in the ventricle (Small and Krieg, 2003). The repressive function of nkx2.5 has also been reported in Drosophila. Tinman, the Drosophila nkx2.5 orthologue, has been shown to be autoinhibitory, in order to maintain its own expression in cardiac precursors in the dorsal mesoderm (Xu et al., 1998). Depletion of tinman-binding sites resulted in a switch of reporter expression from the dorsal mesoderm to the neighboring dorsal ectoderm. Therefore, it was proposed that Tinman may compete with a repressor for binding sites in cardiac progenitor cells. Indeed, it has been shown that Nkx2.5 interacts with many other transcription factors, including GATA4 (Shiojima et al., 1999), SRF (Chen and Schwartz, 1996), and dHAND (Thattaliyath et al., 2002), to regulate cardiac gene expression. Of particular interest to chamber-specific expression, Tbx2/3 has been proposed to compete with Tbx5 for interaction with Nkx2.5, which would either inhibit or activate chamber differentiation (Hiroi et al., 2001; Christoffels et al., 2004). Tbx2 has also been shown to interact with Nkx2.5 and carry out repressive functions in the atrioventricular canal (Habets et al., 2002).
Here, we dissected the zebrafish vmhc promoter and our result suggested the possible involvement of nkx2.5 in chamber specificity. In contrast to Nkx proteins acting as activators in the ventricle but repressors in the atrium in a recent report concerning cardiomyocyte cell number in these two chambers (Targoff et al., 2008), our genetic data demonstrate that NKEs in either distal or proximal vmhc elements execute repressive functions in both the ventricle and atrium. This paradoxical observation could be explained by a corepressor model, which predicts the following: In general, nkx proteins bind to NKEs and function as transcriptional activators to ensure cardiac-specific expression. However, nkx proteins recruit a repressor complex specifically to NKE sites within the distal and proximal elements of vmhc. This repressive activity of nkx proteins predominates in the atrium but not in the ventricle, likely due to the nature of the repressive partner. As the immediate next step, the identification of this corepressor is important to test this model.
Zebrafish Is a Useful Animal Model to Investigate Chamber-Specific Cardiac Promoters
Here, we dissected a chamber-specific cardiac promoter using three unique techniques in zebrafish. We demonstrated the feasibility of applying transient injection assays to dissect the vmhc cardiac promoter. In addition to the benefit of quick turnaround time, transient assays allow quantification of data, which is not possible from the generation of transgenic fish lines. As demonstrated here, the V:A ratio represents a useful index to determine chamber specificity of cardiac promoters. Based on our data, we propose a three-step methodology to analyze any cardiac- and/or chamber-specific promoter using zebrafish as an in vivo animal model. First, candidate fragments are PCR-amplified and the products directly co-injected with a GFP reporter construct for transient analysis. This assay is very high-throughput, as the tedious steps of molecular cloning are omitted (Muller et al., 1999). This transient technology may be suitable for the initial analysis of strong cardiac promoters, but not for weak cardiac promoters with chamber specificity. In the latter case, large numbers of embryos need to be injected and analyzed, significantly increasing the required workload. Second, promising fragments are cloned into a Tol2-based vector for transient injection assays to dramatically reduce mosaicism, and significantly increase sensitivity. The process of molecular cloning can be accelerated by adapting the recombination-based GATEWAY cloning system (Fisher et al., 2006a; Kwan et al., 2007; Villefranc et al., 2007). In contrast to co-injection assays where varied copy numbers of DNA with physically unlinked GFP fragments integrate, a single copy of a Tol2-based fragment together with the GFP reporter is integrated into a single genomic locus. In addition, it is possible that the Tol2-based fragment may function as an episome, which may contribute to increased sensitivity. Lastly, the results from the transient assays should be confirmed by generating transgenic fish lines for key Tol2-based constructs. The efficiency of Tol2-based transgenesis is approximately 50–70%, making zebrafish the easiest and most economical vertebrate model organism for generating transgenic animals (Kawakami, 2007).
In summary, we have revealed a novel inhibitory mechanism to determine ventricular chamber specificity using the zebrafish animal model. Further dissection of the distal or proximal element, such as cloning of transcription factors by one-hybrid screening, promises to reveal a transcription circuit that involves Nkx2.5 in chamber specificity. Tg(V-1.9k:egfp) is the first transgenic fish line that exhibits chamber-specific gene expression patterns. The ventricle-specific elements identified in this study will further facilitate genetic manipulation of zebrafish. Finally, the methodology can be extended to dissect other chamber-specific promoters in zebrafish and will greatly facilitate our understanding of molecular mechanisms governing chamber-specific gene expression.
EXPERIMENTAL PROCEDURES
Bioinformatics
The vmhc (GenBank accession number AF114427) sequence was used to search the Fugu genomic database to identify any known homologues. The Fugu sequence was then annotated using GENSCAN (http://genes.mit.edu/GENSCAN.html) (Burge and Karlin, 1997) and Augustus (http://augustus.gobics.de) (Stanke et al., 2004) software. Sequence comparison across species was performed using rVista software through the Vista server (http://genome.lbl.gov/vista/index.shtml) (Frazer et al., 2004). Transcription factor-binding sites were predicted using Transcription Element Search System (TESS) software (http://www.cbil.upenn.edu/cgi-bin/tess/tess) (Schug, 2003).
5′ RACE
The GeneRacer Kit (Invitrogen) was used to define the vmhc transcriptional start site through RNA ligase-mediated rapid amplification of 5′ cDNA ends (RLM-RACE). Total RNA was extracted by TRIzol reagent and treated with calf intestinal phosphatase (CIP). Dephosphorylated RNA was then decapped by tobacco acid pyrophosphatase (TAP) and ligated with GeneRacer RNA oligos. After reverse transcription, the 5′ cDNA end was amplified by first-round PCR using the 5′ primer included in the kit and a reverse vmhc primer (5′-TTAATTAGTCAAGCCTACCTTTCTTTTC). This product was then followed by a nested-PCR using the 5′ nested primer included in the kit and a nested reverse vmhc primer (5′-TTGTGCTTCCAGACGCTCTCGATCTGAC). The vmhc transcription start site is the same as nucleotide no. 1 in the GenBank sequence XM_001332905.
In Situ Hybridization
Whole mount in situ hybridization was performed as previously described (Seeley et al., 2007). A vmhc riboprobe was generated by PCR amplification of the cDNA using the following primers: forward primer, 5′-AATGACTTCACAATGCAGAAATC-3′; reverse primer, 5′-ATCTTCATTTGTTCTCAGGGAGT-3′. The PCR product contained a T7 promoter and was transcribed using the AmpliScribe T7-Flash Transcription Kit (Epicenter Bio-technologies).
Generation of a Series of Deletion Constructs
A BAC containing the full-length vmhc genomic region (DKEY-77A20) was ordered from RZPD German Resource Center for Genome Research. The 6-kb upstream sequence of vmhc was PCR-amplified using primers 5′-CAGAGGAATCTGTAAGTGCTG-3′ and 5′-CAAGTATTGCCCACAATTGC-3′, and serial deletion constructs were generated by PCR (primer sequences available upon request). To create an internal deletion product, an asymmetric AvaI site was added to the reverse primer of the distal fragment and the forward primer of the proximal fragment, respectively. The two PCR products were digested with AvaI and linked in tandem by ligase treatment, which was then used as the template for PCR to generate internal deletion constructs.
To generate the chimeric promoter, the cmlc2 promoter was first PCR-amplified (forward primer, 5′-GCATTCATCCATCCTTTTCATC-3′; reverse primer, 5′-TTCACTGTCTGCTTTGCTG-3′) and then cloned into the pD-sRed2-1 vector using SacI/EcoRI sites. The V-1.9∼–0.5k fragment from the vmhc promoter was cloned into the 5′ end of the cmlc2 promoter using XhoI/SacI sites to generate a vmhc-cmlc2 chimeric sequence.
To generate a series of Tol2-based constructs, we first modified the pT2KXIGΔin vector (courtesy of Dr. Koichi Kawakami) by replacing the XhoI/BamHI fragment with a multiple cloning site: TCGAGGTCGACCCGGGCTAGCAAGCTTGAATTCG. vmhc promoter fragments were then cloned into the modified Tol2 vector using XhoI/BamHI sites except V-0.7–+0.3k, which used XhoI/NheI sites and directed a membrane-tagged GFP (unpublished data). Fine deletion constructs eliminating the two NKE sites within the distal and proximal elements were generated by cloning two PCR fragments into the modified Tol2 vector using XhoI/NheI and NheI/BamHI sites, respectively.
Transient Injection Assays
For direct co-injection assays, PCR products were purified with the QIA-quick PCR Purification Kit (Qiagen) and then co-injected with a GFP fragment digested from a pG1 vector using BamHI/NotI. For Tol2-based transient assays, pTol2-vmhc promoter constructs were co-injected with transposase mRNA, which was transcribed from pCS-TP (courtesy of Dr. Koichi Kawakami) using the mMES-SAGE mMACHINE SP6 Kit (Ambion).
Transgenic Zebrafish Lines
Wild-type zebrafish lines (TL and Tu) were maintained in our fish facility and used for in situ hybridization and transient injections. To generate stable transgenic lines, Tol2-based constructs were co-injected with transposase into one-cell staged embryos. GFP-positive embryos were identified at 3 dpf and then transferred into the fish facility until sexual maturity. Individual founder fish were outcrossed with wild-type fish for examination of GFP-positive cells in the offspring. GFP-positive offspring of selected founder lines were raised to establish the F1 generation. The F2 generation was then established by incrossing F1 fish. We have identified at least three different transgenic lines for each construct with expression patterns consistent between different stable lines as well as among different generations.
Supplementary Material
Acknowledgments
Grant sponsor: NIH/NHLBI; Grant number: R01 HL081753.
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Bao ZZ, Bruneau BG, Seidman JG, Seidman CE, Cepko CL. Regulation of chamber-specific gene expression in the developing heart by Irx4. Science. 1999;283:1161–1164. doi: 10.1126/science.283.5405.1161. [DOI] [PubMed] [Google Scholar]
- Berdougo E, Coleman H, Lee DH, Stainier DY, Yelon D. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development. 2003;130:6121–6129. doi: 10.1242/dev.00838. [DOI] [PubMed] [Google Scholar]
- Bishop JO, Smith P. Mechanism of chromosomal integration of microinjected DNA. Mol Biol Med. 1989;6:283–298. [PubMed] [Google Scholar]
- Bruneau BG, Bao ZZ, Tanaka M, Schott JJ, Izumo S, Cepko CL, Seidman JG, Seidman CE. Cardiac expression of the ventricle-specific homeobox gene Irx4 is modulated by Nkx2–5 and dHand. Dev Biol. 2000;217:266–277. doi: 10.1006/dbio.1999.9548. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Chen CY, Schwartz RJ. Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac alpha-actin gene transcription. Mol Cell Biol. 1996;16:6372–6384. doi: 10.1128/mcb.16.11.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kubalak SW, Chien KR. Ventricular muscle-restricted targeting of the RXRalpha gene reveals a non-cell-autonomous requirement in cardiac chamber morphogenesis. Development. 1998;125:1943–1949. doi: 10.1242/dev.125.10.1943. [DOI] [PubMed] [Google Scholar]
- Chen Z, Huang W, Dahme T, Rottbauer W, Ackerman MJ, Xu X. Depletion of zebrafish essential and regulatory myosin light chains reduces cardiac function through distinct mechanisms. Cardiovasc Res. 2008;79:97–108. doi: 10.1093/cvr/cvn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels VM, Hoogaars WM, Tessari A, Clout DE, Moorman AF, Campione M. T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers. Dev Dyn. 2004;229:763–770. doi: 10.1002/dvdy.10487. [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006a;312:276–279. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, McCallion AS. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc. 2006b;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M, Sueblinvong V, Raman J, Jeevanandam V, Gupta MP. Single-stranded DNA-binding proteins PURalpha and PURbeta bind to a purine-rich negative regulatory element of the alpha-myosin heavy chain gene and control transcriptional and translational regulation of the gene expression. Implications in the repression of alpha-myosin heavy chain during heart failure. J Biol Chem. 2003;278:44935–44948. doi: 10.1074/jbc.M307696200. [DOI] [PubMed] [Google Scholar]
- Habets PE, Moorman AF, Clout DE, van Roon MA, Lingbeek M, van Lohuizen M, Campione M, Christoffels VM. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev. 2002;16:1234–1246. doi: 10.1101/gad.222902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi Y, Kudoh S, Monzen K, Ikeda Y, Yazaki Y, Nagai R, Komuro I. Tbx5 associates with Nkx2–5 and synergistically promotes cardiomyocyte differentiation. Nat Genet. 2001;28:276–280. doi: 10.1038/90123. [DOI] [PubMed] [Google Scholar]
- Huang CJ, Tu CT, Hsiao CD, Hsieh FJ, Tsai HJ. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev Dyn. 2003;228:30–40. doi: 10.1002/dvdy.10356. [DOI] [PubMed] [Google Scholar]
- Kawakami K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 2007;8(Suppl 1):S7. doi: 10.1186/gb-2007-8-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci USA. 2000;97:11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Knowlton KU, Rockman HA, Itani M, Vovan A, Seidman CE, Chien KR. Divergent pathways mediate the induction of ANF transgenes in neonatal and hypertrophic ventricular myocardium. J Clin Invest. 1995;96:1311–1318. doi: 10.1172/JCI118166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga A, Suzuki M, Inagaki H, Bessho Y, Hori H. Transposable element in fish. Nature. 1996;383:30. doi: 10.1038/383030a0. [DOI] [PubMed] [Google Scholar]
- Korzh V. Transposons as tools for enhancer trap screens in vertebrates. Genome Biol. 2007;8(Suppl 1):S8. doi: 10.1186/gb-2007-8-s1-s8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubalak SW, Miller-Hance WC, O’Brien TX, Dyson E, Chien KR. Chamber specification of atrial myosin light chain-2 expression precedes septation during murine cardiogenesis. J Biol Chem. 1994;269:16961–16970. [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Lee Y, Nadal-Ginard B, Mahdavi V, Izumo S. Myocyte-specific enhancer factor 2 and thyroid hormone receptor associate and synergistically activate the alpha-cardiac myosin heavy-chain gene. Mol Cell Biol. 1997;17:2745–2755. doi: 10.1128/mcb.17.5.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini NJ, Etkin LD, Benbow RM. Persistence and replication of plasmid DNA microinjected into early embryos of Xenopus laevis. Dev Biol. 1988;127:421–434. doi: 10.1016/0012-1606(88)90328-4. [DOI] [PubMed] [Google Scholar]
- Moorman AF, Christoffels VM. Cardiac chamber formation: development, genes, and evolution. Physiol Rev. 2003;83:1223–1267. doi: 10.1152/physrev.00006.2003. [DOI] [PubMed] [Google Scholar]
- Morkin E. Control of cardiac myosin heavy chain gene expression. Microsc Res Tech. 2000;50:522–531. doi: 10.1002/1097-0029(20000915)50:6<522::AID-JEMT9>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Muller F, Chang B, Albert S, Fischer N, Tora L, Strahle U. Intronic enhancers control expression of zebrafish sonic hedgehog in floor plate and notochord. Development. 1999;126:2103–2116. doi: 10.1242/dev.126.10.2103. [DOI] [PubMed] [Google Scholar]
- Nguyen-Tran VTBCJ, Ruiz-Lozano P, Chien KR. The MLC-2 paradigm for ventricular heart chamber specification, maturation, and morphogenesis. In: Harvey RP, Rosenthal N, editors. Heart development. Academic Press; San Diego, CA: 2003. pp. 255–272. [Google Scholar]
- Ross RS, Navankasattusas S, Harvey RP, Chien KR. An HF-1a/HF-1b/MEF-2 combinatorial element confers cardiac ventricular specificity and established an anterior-posterior gradient of expression. Development. 1996;122:1799–1809. doi: 10.1242/dev.122.6.1799. [DOI] [PubMed] [Google Scholar]
- Satin J, Fujii S, DeHaan RL. Development of cardiac beat rate in early chick embryos is regulated by regional cues. Dev Biol. 1988;129:103–113. doi: 10.1016/0012-1606(88)90165-0. [DOI] [PubMed] [Google Scholar]
- Schoenebeck JJ, Keegan BR, Yelon D. Vessel and blood specification override cardiac potential in anterior mesoderm. Dev Cell. 2007;13:254–267. doi: 10.1016/j.devcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. In: Baxevanis AD, editor. Current protocols in bioinformatics. John Wiley and Sons; Hoboken, NJ: 2003. [DOI] [PubMed] [Google Scholar]
- Seeley M, Huang W, Chen Z, Wolff WO, Lin X, Xu X. Depletion of zebrafish titin reduces cardiac contractility by disrupting the assembly of Z-discs and A-bands. Circ Res. 2007;100:238–245. doi: 10.1161/01.RES.0000255758.69821.b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiojima I, Komuro I, Oka T, Hiroi Y, Mizuno T, Takimoto E, Monzen K, Aikawa R, Akazawa H, Yamazaki T, Kudoh S, Yazaki Y. Context-dependent transcriptional cooperation mediated by cardiac transcription factors Csx/Nkx-2.5 and GATA-4. J Biol Chem. 1999;274:8231–8239. doi: 10.1074/jbc.274.12.8231. [DOI] [PubMed] [Google Scholar]
- Small EM, Krieg PA. Transgenic analysis of the atrialnatriuretic factor (ANF) promoter: Nkx2–5 and GATA-4 binding sites are required for atrial specific expression of ANF. Dev Biol. 2003;261:116–131. doi: 10.1016/s0012-1606(03)00306-3. [DOI] [PubMed] [Google Scholar]
- Small EM, Krieg PA. Molecular regulation of cardiac chamber-specific gene expression. Trends Cardiovasc Med. 2004;14:13–18. doi: 10.1016/j.tcm.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Stanke M, Steinkamp R, Waack S, Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32:W309–312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabibiazar R, Wagner RA, Liao A, Quertermous T. Transcriptional profiling of the heart reveals chamber-specific gene expression patterns. Circ Res. 2003;93:1193–1201. doi: 10.1161/01.RES.0000103171.42654.DD. [DOI] [PubMed] [Google Scholar]
- Targoff KL, Schell T, Yelon D. Nkx genes regulate heart tube extension and exert differential effects on ventricular and atrial cell number. Dev Biol. 2008;322:314–321. doi: 10.1016/j.ydbio.2008.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thattaliyath BD, Firulli BA, Firulli AB. The basic-helix-loop-helix transcription factor HAND2 directly regulates transcription of the atrial naturetic peptide gene. J Mol Cell Cardiol. 2002;34:1335–1344. doi: 10.1006/jmcc.2002.2085. [DOI] [PubMed] [Google Scholar]
- Villefranc JA, Amigo J, Lawson ND. Gateway compatible vectors for analysis of gene function in the zebrafish. Dev Dyn. 2007;236:3077–3087. doi: 10.1002/dvdy.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GF, Nikovits W, Jr., Bao ZZ, Stockdale FE. Irx4 forms an inhibitory complex with the vitamin D and retinoic X receptors to regulate cardiac chamber-specific slow MyHC3 expression. J Biol Chem. 2001;276:28835–28841. doi: 10.1074/jbc.M103716200. [DOI] [PubMed] [Google Scholar]
- Wang GF, Nikovits W, Schleinitz M, Stockdale FE. Atrial chamber-specific expression of the slow myosin heavy chain 3 gene in the embryonic heart. J Biol Chem. 1996;271:19836–19845. doi: 10.1074/jbc.271.33.19836. [DOI] [PubMed] [Google Scholar]
- Weiss A, Leinwand LA. The mammalian myosin heavy chain gene family. Annu Rev Cell Dev Biol. 1996;12:417–439. doi: 10.1146/annurev.cellbio.12.1.417. [DOI] [PubMed] [Google Scholar]
- Xu X, Yin Z, Hudson JB, Ferguson EL, Frasch M. Smad proteins act in combination with synergistic and antagonistic regulators to target Dpp responses to the Drosophila mesoderm. Genes Dev. 1998;12:2354–2370. doi: 10.1101/gad.12.15.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi H, Yamagishi C, Nakagawa O, Harvey RP, Olson EN, Srivastava D. The combinatorial activities of Nkx2.5 and dHAND are essential for cardiac ventricle formation. Dev Biol. 2001;239:190–203. doi: 10.1006/dbio.2001.0417. [DOI] [PubMed] [Google Scholar]
- Yelon D, Horne SA, Stainier DY. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev Biol. 1999;214:23–37. doi: 10.1006/dbio.1999.9406. [DOI] [PubMed] [Google Scholar]
- Zammit PS, Kelly RG, Franco D, Brown N, Moorman AF, Buckingham ME. Suppression of atrial myosin gene expression occurs independently in the left and right ventricles of the developing mouse heart. Dev Dyn. 2000;217:75–85. doi: 10.1002/(SICI)1097-0177(200001)217:1<75::AID-DVDY7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.