Abstract
Quantitative interpretation and prediction of Hofmeister ion effects on protein processes, including folding and crystallization, have been elusive goals of a century of research. Here, a quantitative thermodynamic analysis, developed to treat noncoulombic interactions of solutes with biopolymer surface and recently extended to analyze the effects of Hofmeister salts on the surface tension of water, is applied to literature solubility data for small hydrocarbons and model peptides. This analysis allows us to obtain a minimum estimate of the hydration b1 (H2O Å−2) of hydrocarbon surface and partition coefficients Kp characterizing the distribution of salts and salt ions between this hydration water and bulk water. Assuming that Na+ and ions of Na2SO4 (the salt giving the largest reduction in hydrocarbon solubility as well as the largest increase in surface tension) are fully excluded from the hydration water at the hydrocarbon surface, we obtain the same b1 as for air-water surface (∼0.18 H2O Å−2). Rank orders of cation and anion partition coefficients for nonpolar surface follow the Hofmeister series for protein processes, but are strongly offset for cations in the direction of exclusion (preferential hydration). Assuming a coarse-grained decomposition of water accessible surface area (ASA) into nonpolar, polar amide, and other polar ASA and the same hydration b1 to interpret peptide solubility increments, we determine salt partition coefficients for amide surface. These partition coefficients are separated into single-ion contributions based on the observation that both Cl− and Na+ (also K+) occupy neutral positions in the middle of the anion and cation Hofmeister series for protein processes. Independent of this assignment, we find that all cations investigated are strongly accumulated at amide surface while most anions are excluded. Ion effects are independent and additive, allowing successful prediction of Hofmeister salt effects on micelle formation and other processes from structural information (ASA).
1 Introduction
Virtually all noncovalent biopolymer processes are significantly affected by salt con-centration.1–5 At moderate to high salt concentrations (> 0.1 M), osmotic effects (dependent on the number of ions per formula unit but otherwise nonspecific) and ion-specific effects (traditionally called Hofmeister effects) are most significant, while Coulombic effects (valence-specific but otherwise nonspecific) dominate at relatively low salt concentrations. Processes that expose biopolymer surface to the aqueous salt solution (e.g. unfolding, disassembling, dissociating, or dissolving the biopolymer) are typically driven by an increase in concentration of salts with a guanidinium cation and/or a thiocyanate, perchlorate or iodide anion. The reverse direction of these processes is favored by addition of sodium or potassium salts with either a carboxylate, sulfate, phosphate, or fluoride anion. The general conclusion from thermodynamic studies with polymers and model compounds1,3,6,7,7,9 is that interactions of salts with nonpolar surface are unfavorable (“salting-out”) and salt-specific, following the Hofmeister order, while salt-amide interactions are favorable (“salting-in”) and may or may not be salt-specific.3,7,7,10,11 Recently, surface spectroscopists and computation-alists have focused attention on differences in distributions of different Hofmeister ions in the vicinity of the air-water surface, with an emphasis on atmospheric implications.12 What is needed now, and what our work addresses, is a thermodynamic characterization of interactions of Hofmeister ions with biochemically-relevant molecular surfaces so that prediction of salt effects on the thermodynamics of biopolymer processes, which expose or bury surface, is possible.
We recently analyzed and interpreted surface tension increments of a spectrum of electrolytes, including Hofmeister salts, to quantify the partitioning of the cation and anion between bulk water and the air-water interface.13,14 Partition coefficients Kp of ions, defined as ratios of the local concentration of the ion in surface water to the bulk concentration, are found to be independent of salt concentration and of the nature of the companion ion. Alkali metal cations and sulfate and carbonate anions are found to be highly excluded from the air-water interface (local concentration much lower than bulk; Kp << 1).15 By contrast, thiocyanate, perchlorate, and iodide anions are present at concentrations at the air-water interface which are comparable to or exceed their bulk concentrations (Kp > 1), and guanidinium is much less excluded than other cations. Rank orders of partition coefficients of cations and anions generally both correspond to the protein-based Hofmeister ion series, but are offset in the direction of exclusion relative to those inferred for the protein series.
In this paper, we apply a parallel analysis to model compound solubility free energy increments quantifying the effects of Hofmeister salts on solubility of hydrocarbons and oligoamides. From this analysis we obtain single ion partition coefficients Kp,i quantifying the accumulation or exclusion of the spectrum of Hofmeister cations and anions in the vicinity of molecular hydrocarbon and amide surface. As illustrated in this article, these values of Kp,i are all that is needed to predict the thermodynamic consequences of burying or exposing any amount of hydrocarbon and/or amide surface in any process. For example, the process of micelle formation from a nonionic surfactant buries nonpolar (primarily hydrocarbon) surface. From the amount of nonpolar surface buried per monomer incorporated and the Kp,i for salt ions and hydrocarbon surface, we successfully predict the effect of a given concentration of any Hofmeister salt on the critical monomer concentration (CMC), the reciprocal of the equilibrium constant for micelle formation. In unfolding (denaturation) of a globular protein, approximately 80–90% of the newly-exposed protein surface is hydrocarbon and amide surface.16 Values of Kp,i for interactions of salt ions with hydrocarbon and amide surfaces therefore should be sufficient to predict the Hofmeister (i.e. noncoulombic) part of the salt dependence of the stability of a folded protein to denaturation.Ongoing work in our laboratory is separating coulombic and Hofmeister effects of salts on protein stability17 and other protein and nucleic acid processes18 to test these predictions.
2 Thermodynamic background and analysis
2.1 Analysis of salt effects on model compound solubility using the Solute (Salt ion) Partitioning Model (SPM)
Specific, often-large effects of various salts (designated component 3) on solubility of nonpolar and sparingly soluble polar model compounds (designated component 2), called “salting out” or “salting in”, as well as salt effects on distributions of these nonelectrolytes between hydrocarbon solvents and water, generally follow the empirical Setschenow equation:19 the negative logarithm of the solubility S2 or the distribution coefficient D2 of component 2 is a linear function of molar salt concentration C3 with slope ks, designated the Setschenow coefficient of that salt (cf. Fig. 1). The transfer of a model compound to water from a nonaqueous phase involves the formation of a hydration layer of water at the surface of the compound. The Salt ion Partitioning Model (SPM), developed and applied previously to analyze effects of salt concentration on surface tension,13,14 and noncoulombic effects of guanidinium chloride (GuHCl) and potassium glutamate (KGlu), relative to KCl, on protein processes,18,20–22 predicts that the addition of a salt will affect the thermodynamics of the model compound transfer process if the sum of the local cation and anion concentrations in the hydration water of the model compound differs from the sum of their bulk concentrations. If the sum of the local concentrations of salt ions in the hydration water is higher (lower) than in bulk water, then the salt favors (disfavors) the transfer process and increases (decreases) the solubility of the model compound.
Fig. 1.
(A) Effects of sodium salts on benzene solubility. The logarithm of the ratio of solubilities of benzene in the absence and presence of sodium salts (So/S) is plotted as a function of salt concentration; Na2SO4 is the most effective “salting-out” agent, and NaI has the least effect on solubility. Data are from McDevit and Long3 with error bars determined from reported accuracy of solubility measurements. (B) Effects of three sodium salts on solubility of blocked amino acids with unbranched, aliphatic side chains. The side chains contain 0, 1, 3, or 4 carbon atoms (triangle, closed circle, square, and open circle, respectively). Figure has been redrawn from Nandi and Robinson.33
Analysis of the effect of the salt Mν+ Xν− on the solubility of a (nonelectrolyte) model compound (as quantified by the solubility free energy increment (SFEI), which is proportional to the Setschenow coefficient) using the SPM yields:23–25
| (1) |
In Eq. 1, the partition coefficient Kp,3 of the electroneutral salt component Mν+Xν− is related stoichiometrically to individual partition coefficients of the cation (Kp,+) and anion (Kp,−):13,14
| (2) |
where ν=ν+ + ν− is the number of ions per formula unit of the salt. In Eq. 2, the single-ion partition coefficients, Kp,i, are defined as where and are the local and bulk molal concentrations of the salt ion (cation or anion) and In Eq. 1, the hydration of the model compound (i.e. the number of bound water molecules) is b1(ASA), where ASA is its water-accessible surface area, calculated using Surface Racer 3.0,26 with a probe radius of 1.4 Å and accepted values of van der Waals radii (set I27) and b1 is the number of water molecules per unit area in the hydration layer(s). Also in Eq. 1, is the water molality (55.5 mol/kg), and ϵ± is the nonideality correction dlnf±/dlnm3.14,28
Equation 1 predicts that the nonideality corrected SFEI of any uncharged solute in any salt solution is determined only by its hydration b1(ASA) and by the quantity ν(Kp,3−1)=ν+(Kp,+−1)+ν−(Kp,−−1), which quantifies the net contribution of Hofmeister accumulation/exclusion and osmotic effects of salt ions. (If the model compound were an electrolyte, the SFEI would also include a coulombic effect of the salt.) If a salt ion is completely excluded (Kp,ion=0) from the water of hydration b1(ASA) of the solute, then changing its concentration makes a purely osmotic contribution to the SFEI;15 if both cation and anion of the salt are completely excluded, then ν(Kp,3−1)=–ν. Such a salt (e.g. Na2SO4 in the case of hydrocarbon solutes, see below) is the best “salting out” salt because addition of this salt only changes the water activity, thereby disfavoring hydration of the dissolved solute and so reducing its solubility. For solute-salt combinations where (Kp,3−1)= 0, addition of the salt has no effect on solubility (SFEI = 0). This can result from a situation in which Kp,+=1 and Kp,−=1, where no local concentration gradient exists for either cation or anion. More generally (Kp,3−1)= 0 whenever (ν+Kp,++ν−Kp,−)/ν=1, which can occur if one ion is accumulated and the other is excluded (e.g. Kp,− > 1, Kp,+ <1). In either case, if the local concentration of salt ions is equal to its bulk, complete compensation is achieved between Hofmeister (Kp,i) and osmotic effects of that salt. For solute-salt combinations where Kp,3 >1, the model compound is “salted in”, even if one ion is locally excluded as long as the other ion is sufficiently accumulated in the local hydration water of the solute.
3 Results and Discussion
3.1 Hydrocarbon model compounds
Table 1 compares solubility free energy increments (SFEI=–RTdlnS2/dC3; Eq. 1) for effects of selected Hofmeister salts (and HCl) on solubility of aliphatic and aromatic hydrocarbons (for the complete data set, see Table S1);2–7,33,35 representative data for benzene are shown in Fig. 1A. Also tabulated in Table 1 are the corresponding surface tension increments13,14 and the intrinsic (composite) thermodynamic quantities b1(Kp,3 – 1) characterizing the net accumulation or exclusion of these salts for both molecular hydrocarbon and air-water surfaces (Eq. 1). The four hydrocarbons (benzene, toluene, propane, and butane) differ significantly in ASA and in SFEI for a given salt, but the calculated values of b1(Kp,3 – 1) for each salt are strikingly similar. With the exception of GuHCl, which is less excluded from butane than from benzene or toluene, all salts exhibit the same value of b1(Kp,3 – 1) for aliphatic and aromatic surfaces. For example, SFEI characterizing the effect of KCl on solubility of benzene, toluene, and butane yield values of b1(Kp,3 – 1) of −0.056, −0.058, and −0.057, respectively. Since all of these salts “salt out” hydrocarbons, values of b1(Kp,3 – 1) are all negative, indicating net exclusion of salt ions from molecular hydrocarbon surface (i.e. Kp,3 < 1). Values for sulfate (2:1) salts are similar for both hydrocarbon and air-water surfaces, whereas the 1:1 salts investigated yield values of b1(Kp,3 – 1) for molecular hydrocarbon surface which are approximately half as large in magnitude as those determined for the air-water surface.
Table 1.
Composite b1(Kp,3–1) values for selected electrolytes at hydrocarbon and air-water surfaces
| benzene | toluene | propane | butane | air-water | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (ASA=212Å2) | (ASA=249Å2) | (ASA=207Å2) | (ASA=236Å2) | |||||||
| electrolyte | SFEIa | b1(Kp-1) | SFEI | b1(Kp-1) | SFEI | b1(Kp-1) | SFEI | b1(Kp-1) | STIa | b1(Kp-1) |
| Na2S04 | 747 | −0.172 | 909 | −0.177b | - | - | - | - | 3.99 | −0.194 |
| NaCl | 266 | −0.064 | 299 | −0.061 | 273 | −0.067 | 304 | −0.065 | 2.49 | −0.126 |
| Nal | 130 | −0.030 | - | - | - | - | - | - | 1.64 | −0.079 |
| KCl | 226 | −0.056 | 275 | −0.058 | - | - | 255 | −0.057 | 2.29 | −0.121 |
| KI | - | - | - | - | 147 | −0.036 | 138 | −0.030 | 1.66 | −0.085 |
| LiCl | 192 | −0.043 | 260 | −0.050 | 214 | −0.049 | 241 | −0.048 | 2.37 | −0.113 |
| HCl | 65.4 | −0.014 | - | - | - | - | 76.3 | −0.015 | −0.40 | 0.019 |
| GuHCl | −72.3 | 0.020 | −104 | 0.024 | - | - | 20.6 | −0.005 | 1.08 | −0.062 |
The solubility free energy increments are determined from the reported Setschenow coefficients1–6 and are in units of cal mol−1 molar−1, and the surface tension increments13,14 have the same units, normalized by surface area (i.e. cal mol−1 Å−2 molar−1).
Propagation of the reported 0.5% accuracy of solubility measurements3 results in uncertainties in b1(Kp-1) of 0.001–0.003. We propose a more conservative estimate of 0.005 for all b1(Kp-1) values reported here. For a b1 value of 0.177, this represents an uncertainty in Kp of 0.03.
In the case of toluene, the SFEI for Na2SO4 yields the most negative b1(Kp,3 – 1) value (i.e. Na2SO4 is the most excluded from the water of hydration of toluene). Assuming that Na+ and are completely excluded from this hydration water, we obtain a minimum estimate of 0.18 H2O Å−2 for the hydration b1. The same assumption applied to surface tension increments (i.e. complete exclusion of Na+ and from the air-water surface) yielded a similar b1 (0.19 H2O Å−2) for the number of water molecules per unit area at the air-water surface.13,14 To obtain composite partition coefficients Kp,3 characterizing electrolyte-model compound interactions, we assume that b1 is a molecular property of the model compound hydration layer, independent of the concentration and nature of the electrolyte. Values of Kp,3 are then analyzed to obtain individual ion Kp,i using Eq. 2 and the assumption that at molecular hydrocarbon surface. The partition coefficients obtained in this way are presented in Table 2.
Table 2.
Single ion partition coefficients (Kp,i) for hydrocarbon molecular surfacec; Comparison with air-water surfaced.
| Ion | toluene | benzene | n-butane | n-propane | average Kp,hc | air-water Kp |
|---|---|---|---|---|---|---|
| Na+ | 0.00 | 0.05 | - | - | 0.03 ± 0.04 (2)e | 0.00 |
| K+ | 0.09 | 0.14 | 0.12 | 0.05 | 0.10 ± 0.04 (4) | 0.12 |
| Li+ | 0.27 | 0.29 | 0.22 | 0.23 | 0.25 ± 0.08 (5) | 0.08 |
| 0.56 | 0.42 | - | 0.28 | 0.41 ± 0.12 (4) | 0.25 | |
| H+ | - | 0.61 | 0.59 | - | 0.60 ± 0.01 (2) | 1.50 |
| GuH+ | 0.99 | 0.99 | 0.70 | - | - | 0.67 |
| 0.00 | 0.00 | - | - | 0.00 | 0.00 | |
| F− | 0.88 | 0.96 | - | - | 0.92 ± 0.05 (2) | 0.53 |
| HO− | - | 1.03 | - | - | 1.00 | 0.58 |
| Cl− | 1.22 | 1.23 | 1.24 | 1.22 | 1.23 ± 0.08 (6) | 0.69 |
| Br− | 1.47 | 1.40 | - | - | 1.44 ± 0.05 (4) | 0.86 |
| 1.48 | 1.47 | - | - | 1.47 ± 0.01 (2) | 0.98 | |
| 1.56 | 1.54 | - | - | 1.55 ± 0.01 (3) | 1.77 | |
| I− | - | 1.62 | 1.54 | - | 1.59 ± 0.04 (3) | 1.18 |
| SCN− | - | 1.80 | - | - | 1.80 | 1.64 |
From data of Table 1
14
Numbers in parentheses denote the number of determinations of Kp for a particular ion.
As seen in Table 2, the trends observed for molecular hydrocarbon surface parallel those previously reported for ion partitioning at the air-water surface;13,14 comparisons of ion distributions near these surfaces, obtained from molecular dynamics simulations, yield a similar conclusion.36 Alkali metal cations are strongly excluded, GuH+ is much less excluded than the other cations, and the rank order of partitioning of anions matches their placement in the traditional biopolymer Hofmeister series. (The proton, not part of the traditional Hofmeister series, is relatively highly accumulated at both surfaces, though much less so at the hydrocarbon molecular surface (Kp=0.6) than at the air-water surface (Kp=1.5).) A comparison of Kp values for the hydrocarbons with those obtained for the air-water surface reveals a proportionality (Kp,SFEI ∼1.6Kp,STI, cf. Fig. 2) for all cations and anions (with the possible exception of GuH+) which are excluded from the air-water surface (Kp,STI ≲ 1). The series of anions that accumulate at the air-water surface (with Kp increasing from 1.2 to 1.8) do not exhibit this proportionality; these anions all accumulate to the same extent at molecular hydrocarbon surface (Kp,SFEI=1.6±0.1).
Fig. 2.
Comparison of ion partition coefficients calculated from hydrocarbon solubility free energy increments and from surface tension increments (ordinate and abscissa, respectively). For ions which are excluded from the air-water interface (Kp,STI < 1), there is a clear proportionality, given by Kp,SFEI = 1.6Kp,STI. However, anions that accumulate at the air-water surface (Kp,STI > 1) all partition similarly at hydrocarbon molecular surface (Kp,SFEI ∼ 1.6).
3.2 Coarse-grained surface decomposition of model peptides and small amide molecules
Nandi and Robinson performed extensive series of solubility and distribution studies on various end-blocked model peptides in salt solutions.7,33,37 In contrast to the situation for hydrocarbons, where all salts examined except GuHCl decrease solubility in water, some salts (e.g. NaSCN, NaClO4) increase the solubility of some model peptides (cf. Fig. 1B and Table 3 for a subset of this data). This has been interpreted as a balance between the salting-out of nonpolar groups and the salting-in of the peptide group. From the trends observed for the N-acetyl (glycine)n ethyl ester series (I, where n=1–4),
Table 3.
Solubility free energy increments (SFEI)f characterizing effects of Hofmeister salts on solubilty of the peptide AG4E and salt partition coefficients for total surface and polar amide surface g obtained from those SFEI.
| salt | SFEI | ||
|---|---|---|---|
| Na2SO4 | 709 | 0.69 | 2.22 |
| KF | 314 | 0.85 | 1.67 |
| NaCl | 68.2 | 0.97 | 1.71 |
| KCl | 68.2 | 0.97 | 1.66 |
| LiCl | 13.6 | 0.99 | 1.59 |
| NaBr | 0.00 | 1.00 | 1.56 |
| NaI | −300 | 1.13 | 1.73 |
| GuHCl | −351 | 1.19 | 1.68 |
| NaSCN | −354 | 1.16 | 1.74 |
| NaClO4 | −436 | 1.21 | 2.11 |
The SFEI are calculated from the reported Setschenow coefficients7 and are in units of cal mol−1 molar−1.
Nandi and Robinson concluded that salt effects on peptide groups are additive and estimated a salting-in constant for this group (consisting of the methylene carbon as well as the amide functional group) which was somewhat salt-specific.7 The glycine peptide series provides a set of model compounds with varying ASA composition; specifically, the polar amide (N,O) surface contribution increases from 13% (AG1E) to 28% (AG4E).
In order to extend our ASA-based analysis (Eq. 1) to these compounds and determine salt-amide partition coefficients, we propose that the contributions from different types of surface are independent and additive, and that the same hydration b1 characterizes the entire molecular surface.38,39 (This hypothesis of additivity has proved successful in SPM-based analyses of interactions of urea and osmolytes with model compounds and proteins.16,25,38) In other words, the partition coefficient (from Eq. 1) characterizing the distribution of uncharged solute component 3 (or a particular salt ion; Eq. 2) between the hydration water of the model compound (component 2) and bulk water is the sum of contributions from different types of surface on the model compound. As in our previous analyses, we use a coarse-grained decomposition of the ASA of component 2 into fractional nonpolar (hydrocarbon; np), polar amide (N, O; pa) and other polar (here, polar ester oxygen; eo) surface:
| (3) |
Salt effects on solubility of ethyl acetate7–9,42 were used to determine for the interaction of each salt with the ester oxygen surface (Table S2). These values, along with those determined for hydrocarbon surface and the coarse-grained surface areas for each acetylglycine ethyl ester (Table 4) allow us to obtain component partition coefficients for Hofmeister salts at polar amide surface from Eq. 3 (Table S2). These salt-amide Kp values and Kp values for ester oxygen surface are summarized in Fig. 3. Uncertainties shown for the values are the standard deviations from the mean for the four members of the acetyl glycine ethyl ester series (Table S2).
Table 4.
Coarse-grained decomposition of the water-accessible surface area (ASA) of ethyl acetate, secondary amides (models for peptide backbone amides), and a representative globular protein.
| C | amide O | amide N | other polar | |||
|---|---|---|---|---|---|---|
| model compound | ASAtotal (Å2) | ASA (%) | ASA (%) | ASA (%) | ASA (%) | |
| ethyl acetate | 267.6 | 221.5 (83%) | - | - | 46.1 (17%) | |
| AG1E | 354.8 | 267.2 (75%) | 35.3 (10%) | 11.1 (3%) | 41.2 (12%) | |
| AG2E | 442.2 | 313.0 (71%) | 65.8 (15%) | 22.2 (5%) | 41.2 (9%) | |
| AG3E | 529.4 | 358.7 (68%) | 96.3 (18%) | 33.3 (6%) | 41.2 (8%) | |
| AG4E | 616.7 | 404.5 (66%) | 126.8 (21%) | 44.3 (7%) | 41.2 (7%) | |
| acylphosphatase (F)h | 5616.9 | 2938.6 (52%) | 612.7 (11%) | 425.1 (8%) | 1640.5 (29%) | |
| extended state (U) | 14306.3 | 8438.0 (59%) | 1846.8 (13%) | 865.3 (6%) | 3156.2 (22%) | |
| ΔASAF→U | 7154.2 | 5499.3 (63%) | 1234.1 (14%) | 440.3 (5%) | 1515.8 (17%) | |
The ASA values presented are for horse muscle acylphosphatase, which exposes 83% nonpolar and polar amide surface on unfolding, close to the average (85%) obtained from an ASA analysis of 29 monomeric proteins.16
Fig. 3.
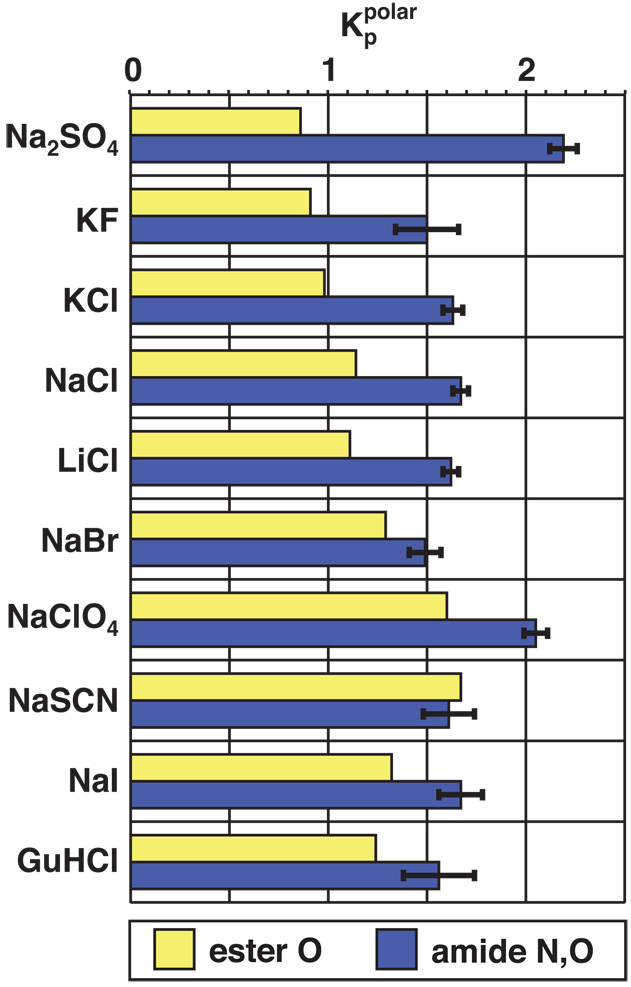
Salt partition coefficients for polar surfaces. Solubility/distribution data for ethyl acetate and the AGnE series were used to calculate salt partition coefficients for ester oxygen (yellow) and amide nitrogen and oxygen (blue) surfaces (see text). The error bars for polar amide surface are the standard deviations from the mean for the four peptides; all salts are moderately accumulated at polar amide surface, and NaClO4 and Na2SO4 are more strongly accumulated than the other salts.
For most of the salts investigated, excellent agreement is obtained between values of calculated (Eq. 1) from solubility free energy increments of the different members of the glycine peptide series.43 Consistent with the previous conclusion that all salts “salt in” the amide group,3,33 we find that, as a net effect of the behavior of the individual cation and anion, all salts investigated accumulate at polar amide surface. In particular, we find that alkali halide salts, GuHCl, and NaSCN all accumulate to similar extents (with ranging from 1.47 to 1.67). Interestingly, Na2SO4 and NaClO4 are somewhat more accumulated at polar amide surface ( and 2.05, respectively). In contrast to the results of the hydrocarbon and air-water surface analyses, the traditional Hofmeister order is not observed for salt-amide partition coefficients (Lack of specificity of salt-amide interactions had previously been inferred from solubility data for N-methylacetamide and N-methylpropionamide7 as well as from recycling chromatography studies.11) In particular, we find that Na2SO4, a strong protein stabilizer and precipitant and the most excluded salt at air-water and hydrocarbon surfaces, is the most accumulated salt at polar amide surface. The partition coefficients of salts for polar (ester) oxygen surface comprise a somewhat compressed Hofmeister ordering (as compared to the hydrocarbon case), ranging from slight exclusion (Na2SO4, KF) to moderate accumulation (NaI, NaClO4).
3.2.1 Strategy for obtaining single-ion contributions to polar amide partition coefficients
To obtain partition coefficients characterizing accumulation or exclusion of individual ions at air-water and hydrocarbon-water surfaces, we chose Na2SO4, the most excluded salt, as a reference and assumed that both Na+ and are completely excluded from the water of hydration of these surfaces (i.e. ). No analogous approach is possible for amide surface, however, because neither Na2SO4 nor any other salt investigated is excluded from amide surface (Fig. 3). Separation of the individual contributions of cation and anion to salt-amide partition coefficients is therefore arbitrary. As a basis for separating salt-amide partition coefficients into single-ion contributions, we use the classical observation that NaCl and KCl typically exhibit little if any Hofmeister-osmotic (i.e. non-coulombic) effect on unfolding transitions of globular proteins, and that Cl−, Na+, and K+ions are also centrally positioned in individual anion and cation series derived from unfolding data.3,6 From these semiquantitative results, we conclude that the NaCl and KCl component partition coefficients for the surface exposed in unfolding the typical globular protein must be near unity and that the single-ion partition coefficients of these ions must be similar to one another and close to unity as well. The peptide AG4E has a similar surface composition as that exposed in unfolding the typical globular protein (2/3 nonpolar, 1/3 polar; Table 4), and indeed the overall NaCl and KCl component partition coefficients for AG4E surface are equal and close to unity (Kp,NaCl=Kp,KCl=0.97; Table 3). Moreover, the effects observed for limited series of NaX (and KX) and MCl investigated parallel those observed for protein unfolding, and are consistent with a central positioning of the individual Na+ (and K+) and Cl− ions. We therefore assign overall partition coefficients of Na+, K+, and Cl− for AG4E surface to be equal to one another and to the observed value (0.97). Simulations may be able to test and/or refine this reasonable but arbitrary assignment.44
With the overall partitioning behavior of Na+, K+, and Cl− for the surface of AG4E as a reference, single ion partition coefficients for these ions and the polar amide surface of AG4E were calculated: and 45 The remaining singleion partition coefficients were calculated from these reference values and the values for AG4E (Table 3). As shown in Fig. 4, cations are highly accumulated (), halide anions, and SCN− are moderately to highly excluded and ClO4 is slightly accumulated (Differences in cation and anion values are small, with the exception of and may not be outside of experimental and propagated error.) The qualitative conclusion of net cation accumulation at, and anion exclusion from, polar amide surface is supported by two key observations: 1) on average, 75% of the polar amide surface in these end-blocked amino acids (and in the backbone amide group of an unfolded protein, cf. Table 4) is contributed by the partially negative oxygen, and 2) Na2SO4 (for which Na+ contributes twice as much to the salt component Kp,3 as it does for a 1:1 salt) is the most accumulated salt at this surface. Also shown in Fig. 4 are the single-ion partition coefficients determined for nonpolar hydrocarbon surface by assuming complete exclusion of Na+ and Placements of cations and anions on the horizontal axis are based on their positions in the traditional biopolymer Hofmeister series. It is clear from the trends shown in Fig. 4 that the partitioning behavior of ions at nonpolar surface is the origin of the range of Hofmeister salt effects observed for biopolymer processes.
Fig. 4.
Predicted single-ion partition coefficients for nonpolar and internal (backbone) polar amide surface, obtained from salt component partition coefficients as described in the text. The rank orders of cations and anions are independent of the details of the separation of the component Kp. For nonpolar surface, the conventional Hofmeister order is observed for both cations and anions. In particular, GuH+ is much less excluded than the alkali metal cations. For the alkali metal cations, strong accumulation at backbone polar amide surface counterbalances the strong exclusion from nonpolar surface. Because GuH+ is only slightly excluded from nonpolar surface, its accumulation at polar amide surface dominates and results in its denaturing abilities.
3.3 Comparison of predicted and observed effects of Hofmeister salts on solubility (or distribution coefficients) of other pep-tides and amides
From the partition coefficients of individual Hofmeister salt ions for hydrocarbon, amide and ester oxygen surface determined in previous sections, noncoulombic effects of any salt on any process changing the exposure of these types of surface to water can be predicted. Effects of Hofmeister salts on solubility (or distribution coefficients) of various N-acetyl amino acid ethyl esters (end-blocked peptides; II) with hydrocarbon side chains have been determined.33,37,42
(Compounds investigated are designated AAE, AFE, AVE, AnVE, ALE, AnLE. In these acronyms the first and last letters stand for “acetyl” and “ethyl ester”, respectively; the central letters A, F, V and L stand for alanine, phenylalanine, valine, and leucine, and nV and nL represent norvaline and norleucine.) A coarse-grained treatment of the ASA of these compounds yields the results in Table S3 for nonpolar (hydrocarbon), polar amide, and polar oxygen surface areas. These peptides are 80–87% hydrocarbon, 7–11% polar amide, and 6–10% polar ester oxygen surface, making them a good test of the applicability of the hydrocarbon and amide (AGnE) data sets to predict Hofmeister salt effects.
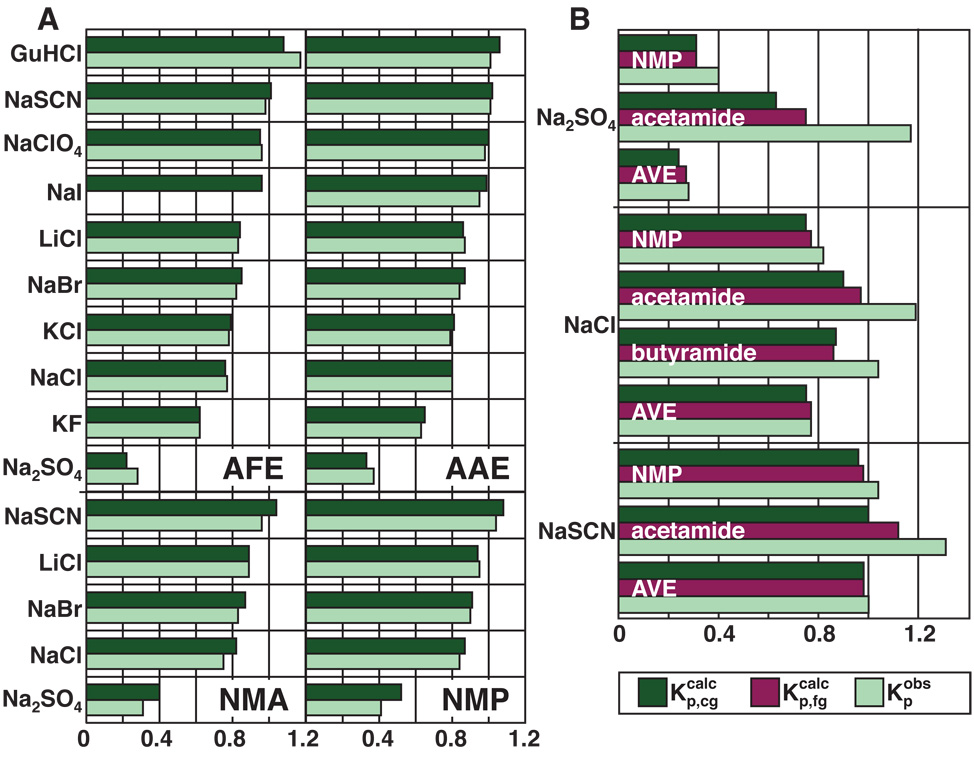
Use of amide ASAs (Table S3) together with the characteristic ionic Kp values (Table 2 and Fig. 4) allows us to predict from Eq. 3 the overall partition coefficient of each salt investigated for each of these compounds, and compare with that determined from the experimental Setschenow coefficient (Eq. 1). Figure 5A compares predicted and observed (from solubility/distribution) values of Kp for salts and two end-blocked peptides from the extremes of the series (AAE and AFE). Agreement for other members of the series is equally good. Also compared in Fig. 5A are predicted and observed values of Kp for salts and two secondary amides (N-methyl acetamide (NMA) and N-methyl propionamide (NMP), obtained by analysis of distribution data.7 In all cases, the agreement between observed and predicted values of Kp is very good; the average discrepancy is 5% in Kp (for all salts) and 13% in observed and predicted Setschenow coefficients (for salts with ks > |0.03|). Additional experiments are needed to determine whether the major source of these discrepancies is experimental error or failures of one or more of the assumptions of coarse-graining, additivity and context-independence of the contributions to the calculated salt partition coefficient.
Fig. 5.
(A) Comparison of observed salt partition coefficients (, light green), obtained from application of Eq. 1 to solubility/distribution increments, with those predicted from coarse-grained (cg) surface area decomposition (, dark green) for AFE, AAE, NMA, and NMP (see text). (B) Comparison of observed salt partition coefficients with those predicted by coarse-grained and fine-grained amide ASA decompositions for NMP, acetamide, butyramide, and AVE. Agreement between observed and calculated partition coefficients for terminal amides is dramatically improved when the amide ASA is deconstructed into individual contributions of the nitrogen and oxygen . The three salts Na2SO4, NaCl, and NaSCN are predicted to accumulate at the oxygen surface (Kp=2.3, 1.8, and 1.5, respectively). Both Na2SO4 and NaCl are moderately excluded from the amide nitrogen surface (Kp=0.7 and 0.9), while NaSCN is slightly accumulated at this surface (Kp=1.2). Since most protein amide groups (peptide backbone) have similar fractional exposures of the amide nitrogen and oxygen to those observed for the end-blocked peptides, the partition coefficients determined for polar amide (N,O) surface should be sufficient for predicting effects on protein processes.
The model compounds shown in Fig. 5A all have internal (secondary) amides, for which the water-accessible surface area (ASA) of the amide oxygen (20–35Å2; see Table 4 and Table S3) is on average three times that of the amide nitrogen (8–13Å2). This 3:1 ASA ratio is also characteristic of amide ASA in folded and unfolded proteins, most of which is from backbone (secondary) amides. For terminal (primary) amides, similar to the side chain amides of asparagine and glutamine, where the ASA of amide nitrogen (62Å2; see Table 5) is larger than that of oxygen (∼37–51Å2), the agreement between and is poor. We solved for the interaction coefficients of three salts with the nitrogen (N) and oxygen (O) using two compounds with different surface compositions (formamide and NMA, with O:N surface area ratios of 0.7 and 1.8, respectively). When this fine-grained treatment of amide surface area is applied to terminal amides (i.e. acetamide and butyramide), the agreement between the calculated and observed partition coefficients ( and ) is dramatically improved (Fig. 5B). The largest improvement is observed for acetamide, which has a similar O:N ASA ratio (0.7) to the amide groups on asparagine or glutamine (0.5–0.6; see Table 5) but a very different ratio than that for the end-blocked peptides and peptide groups in proteins (∼ 3; Table 4 and Table S3). Indeed, little to no improvement is observed for the end-blocked amino acids (cf. AVE in Fig. 5B) upon application of the fine-grained ASA decomposition. The finer-grained ASA analysis is necessary for applications to side chain amides, but the majority of protein amides are internal (backbone), where the coarse-grained (determined for the full spectrum of salts) should be sufficient.
Table 5.
Coarse-grained decomposition of the water-accessible surface area (ASA) of primary amides including protein side chain amides.
| C | amide O | amide N | ||
|---|---|---|---|---|
| model compound | ASAtotal (Å2) | ASA (%) | ASA (%) | ASA (%) |
| formamide | 161.8 | 40.2 (25%) | 51.3 (32%) | 70.2 (43%) |
| acetamide | 200.7 | 94.0 (47%) | 44.9 (22%) | 61.8 (31%) |
| butyramide | 256.4 | 157.8 (62%) | 36.9 (14%) | 61.7 (24%) |
| pentanamide | 285.3 | 186.7 (65%) | 36.9 (13%) | 61.7 (22%) |
| hexanamide | 314.2 | 216.0 (69%) | 36.9 (12%) | 61.7 (20%) |
| asparagine side chain | 129.5 | 38.1 (29%) | 29.8 (23%) | 61.6 (48%) |
| glutamine side chain | 151.7 | 46.6 (31%) | 40.0 (26%) | 65.1 (43%) |
3.4 Comparison of Predicted and Observed Effects of Hofmeister Salts on the Critical Concentration (CMC) for Formation of a Nonionic Micelle
Formation of micelles from amphiphilic monomers in aqueous solution is a highly cooperative self-assembly process driven largely by burial of nonpolar (hydrocarbon) surface on each monomer in the interior of the micelle (the hydrophobic effect46). The reciprocal of the experimentally-determined critical monomer concentration (CMC−1) is the equilibrium constant for the process of transferring a monomer from the solution to the micelle. Salt effects on the CMC, like any other observed equilibrium constant, can be coulombic and/or Hofmeister-osmotic effects;5 for assembly of uncharged monomers, no coulombic effects are expected. In a classic work, Ray and Nemethy reported a particularly extensive data set characterizing the effects of the full spectrum of Hofmeister salts (from GuHSCN to Na2SO4) on the CMC of micelle formation by nonionic OPE (OctylphenoxyPolyEthoxy) surfactants (compound III).47,48
Here we analyze initial slopes –dlnCMC/dC3 using Eq. 1 (with S2 replaced by CMC). For these OPE monomers, only nonpolar surface area (ASA) is buried in micelle formation; most of this is hydrocarbon ASA, with a small contribution from ether oxygens. Hence, we propose that SPM partition coefficients characterizing the accumulation or excluson of individual salt ions in the vicinity of nonpolar (hydrocarbon) surface (Table 2) are sufficient to predict the effects of any Hofmeister salt on the CMC.
Since all salt cations are excluded from hydrocarbon surface (Kp < 1), almost all salts reduce the solubility of hydrocarbons in water (cf. Table 1); these same salts are predicted to favor micelle formation, since this reduces the water-accessible nonpolar surface area of the monomer, and therefore should lower the CMC. Since alkali metal cations are almost completely excluded from hydrocarbon surface in water, differences in effects of their salts on the CMC are predicted to be determined mostly by the extent of anion accumulation at the nonpolar surface of the free OPE monomer which is buried in the micelle. Alkali fluorides are predicted to strongly favor micelle formation (lower the CMC) because of strong exclusion of the alkali metal cation and slight exclusion of F− from nonpolar surface. On the other end of the Hofmeister series, alkali iodides and thiocyanates are predicted to exert only small effects on the CMC because of largely compensating effects of anion accumulation and cation exclusion at nonpolar surface. Because GuH+ exhibits very different partitioning behavior from alkali metal cations at hydrocarbon surface, being only modestly excluded (Kp = 0.7) from aliphatic surface and neither excluded from nor accumulated at aromatic surface (Kp=1.0), salts which are combinations of GuH+ and an accumulated anion like I−or SCN− are predicted to favor micelle disassembly and therefore increase the CMC. On the other hand, (GuH)2SO4 is predicted to strongly favor micelle formation and reduce the CMC because of strong sulfate exclusion (Kp = 0) and modest exclusion of the two GuH+ cations.
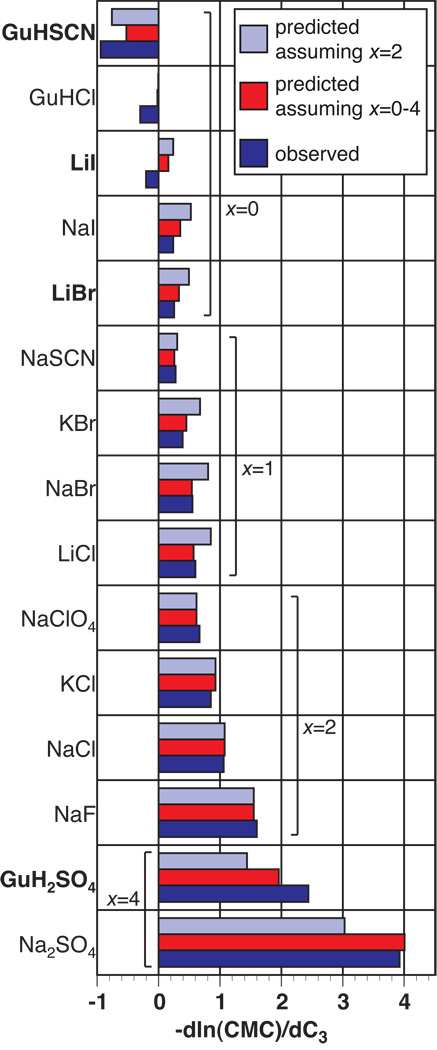
Quantitative predictions of the SPM (Eq. 1) for the low-salt value of the derivative –dlnCMC/dC3 are compared with the corresponding quantities obtained from the data of Ray and Nemethy in Fig. 6. Dark blue bars are experimental values and light blue bars are predictions based on the single ion Kp data in Table 2, assuming that the p-tert-octylphenoxy group and two ethoxy groups of each monomer are buried in the micelle (cf. Table S5). (The nonpolar ether oxygens of the buried ethoxy groups (∼5% of the total ΔASA) are treated as equivalent to hydrocarbon surface.) The agreement between the experimental data and the model predictions is remarkably good given the simplicity of the model and the possibility that the choice and/or concentration of salt perturbs the structure of the micelle.49 Even better agreement between predicted and observed values of –dlnCMC/dC3 is obtained if the number of buried ethoxy groups is systematically increased from 0 for the most accumulated salts to 4 for the most excluded salts, suggesting a modest effect of the choice of Hofmeister salt on the structure of the micelle. While there is no independent verification of the prediction that 2 ± 2 ethoxy groups are buried, the finding that the salt derivative of the experimental CMC is unaffected as the number of ethoxy groups is increased from 9–10 to 30 places an upper bound of 9 ethoxy groups that become water-inaccessible in the micelle. Additionally, NMR studies on OPEn (n=9–10, 12–13, 30) in aqueous solution (above and below the CMC), in bulk surfactant, and in hydrocarbon solvents have been performed; comparisons of the proton chemical shifts in the different environments led the authors to conclude that the interior of the micelle was not in contact with water and that the ethoxy groups nearest the phenyl ring were “less solvated” than those closer to the hydroxyl group (see compound I).50
Fig. 6.
Comparison of observed and predicted effects of Hofmeister salts on micelle formation. Experimental values of the equilibrium constant derivative —dlnCMC/dC3 for effects of Hofmeister salts on formation of a p-tert-octylphenoxy(polyethoxy)ethanol micelle47 are represented by the dark blue bars. Light blue and red bars represent predicted values based on the SPM/ASA analysis. The best agreement of the calculated results to the experimental data for all salts is obtained if it is assumed that two ethoxy groups are buried along with the p-tert-octylphenoxy moiety (light blue). Even better agreement is attained if the number of ethoxy groups buried is varied from 0 for salts with Kp >>1 to 4 for salts with Kp <<1(red; see text). Salts designated in bold letters are those for which additivity of independently determined anion and cation Kp,i was used to predict —dlnCMC/dC3.
3.5 Prediction of Hofmeister salt effects on protein folding and other biopolymer assembly processes
Folding of a polypeptide chain into the native globular structure of a single-domain enzyme or regulatory protein requires dehydration and burial of thousands of Å2 of previously water-accessible molecular surface of the peptide backbone and amino acid side chains. The composition of the previously water accessible surface area (ASA) which is buried upon folding is very similar for all globular proteins: approximately 2/3 nonpolar (hydrocarbon) and 1/3 polar. Little if any charged surface is buried in the folded protein. Of the polar ASA, approximately half is polar amide surface (predominantly the secondary amides of the peptide backbone but also primary amides of glutamine and asparagine side chains); the remainder of the polar ASA originates from other polar side chain groups (e.g. hydroxyls of serine, threonine and tyrosine). Although the buried surface is heterogeneous at the atomic level, with interspersed nonpolar and polar atoms, precedent exists for the application of an ASA-based thermodynamic analysis to predict or interpret the consequences of changing the exposure of these types of surface to water in protein processes. In particular, heat capacity changes (ΔCp) for protein folding and for interactions of proteins with ligands and other proteins agree well with those predicted from an ASA analysis based on experimental values of the ΔCp for dissolving small hydrocarbons and amides.51 Although the surface buried in protein folding is largely uncharged, interpretation of salt effects on protein folding requires the separation of coulombic salt effects (arising from changes in the surface density of charged groups in folding) from Hofmeister-osmotic effects of salts. This quantitative analysis is in progress; here we make and discuss some qualitative predictions based on the SPM analysis and Hofmeister ion partition coefficients for hydrocarbon and amide surface obtained above.
Figure 4 illustrates the novel explanation, obtained from this SPM analysis, for the observation that GuHCl is a protein denaturant, but KCl is not (equivalent to the observation that GuHCl solubilizes AG4E and KCl does not). The ASA exposed in unfolding of a globular protein is mostly nonpolar but includes a significant polar amide contribution.16 Both GuH+ and K+ are predicted to accumulate (strongly and to similar extents) at polar amide surface (e.g. the peptide backbone); however, K+ is predicted to be strongly excluded from nonpolar surface, while GuH+ is only weakly excluded from this surface. When weighted by surface composition, these opposing effects for K+ appear to largely compensate and we therefore propose an overall Kp near unity. (Similar compensation for Cl− also appears to occur, resulting in a proposed overall Kp ≃ 1 for the surface exposed in unfolding.) For GuH+, however, accumulation at amide surface is not compensated by exclusion from nonpolar surface, yielding net accumulation at the protein surface exposed in unfolding (Kp,GuH+=1.4) as also observed for urea (Kp,urea=1.1).21,25 Hence, both qualitative points of view in the literature regarding why GuHCl denatures proteins (but KCl does not) are partially correct; namely, the somewhat successful competition of GuH+ with water for nonpolar surface (resulting in only moderate exclusion) and favorable interactions with polar amide surface (resulting in strong accumulation) together make GuHCl a denaturant. Additionally, consistent with experiment, GuHSCN is predicted to be a much stronger denaturant (overall Kp,GuHSCN=1.4) than GuHCl (Kp,GuH Cl=1.2) because of the increased accumulation of SCN− at nonpolar surface (relative to Cl−). On the other hand, while the ions of (GuH)2SO4 are as strongly accumulated at polar amide surface as those of GuHSCN, the strong exclusion of from nonpolar surface provides a compensating contribution to the overall Kp characterizing the interaction of this salt with the surface exposed in unfolding (overall and results in the observed weak stabilizing effect (similar to KCl).3,6
Other processes, including formation of a polypeptide alpha helix or a DNA double helix and binding of proteins to DNA, bury surfaces that are far more polar or charged than that buried in folding a globular protein. In these situations, since a smaller fraction of the buried surface is nonpolar, we predict that few if any stabilizing Hofmeister effects will be observed and that differences between effects of different Hofmeister salts will be suppressed relative to the effects on folding of globular proteins. These expectations are currently being tested.
Supplementary Material
Tables containing solubility data, the composite thermodynamic quantity b 1(Kp 3, – 1) for each salt, and surface area breakdowns for the model compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
This work was supported by NIH grant GM47022.
References
- 1.von Hippel PH, Schleich T. Accounts of Chemical Research. 1969;2:257–265. [Google Scholar]
- 2.Lohman TM. CRC Crit Rev Biochem. 1986;19:191–245. doi: 10.3109/10409238609084656. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin RL. Biophys. J. 1996;71:2056–2063. doi: 10.1016/S0006-3495(96)79404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Timasheff SN. Adv Protein Chem. 1998;51:355–432. doi: 10.1016/s0065-3233(08)60656-7. [DOI] [PubMed] [Google Scholar]
- 5.Record MT, Jr., Zhang W, Anderson CF. Adv. Protein Chem. 1998;51:281–353. doi: 10.1016/s0065-3233(08)60655-5. [DOI] [PubMed] [Google Scholar]
- 6.von Hippel PH, Wong KY. Science. 1964;145:577–580. doi: 10.1126/science.145.3632.577. [DOI] [PubMed] [Google Scholar]
- 7.Schrier EE, Schrier EB. J. Phys. Chem. 1967;71:1851–1860. doi: 10.1021/j100865a049. [DOI] [PubMed] [Google Scholar]
- 8.Nandi PK, Robinson DR. J. Am. Chem. Soc. 1972;94:1299–1308. doi: 10.1021/ja00759a042. [DOI] [PubMed] [Google Scholar]
- 9.Arakawa T, Timasheff SN. Biochemistry. 1982;21:6545–6552. doi: 10.1021/bi00268a034. [DOI] [PubMed] [Google Scholar]
- 10.von Hippel PH, Peticolas V, Schack L, Karlson L. Biochemistry. 1973;12:1256–1264. doi: 10.1021/bi00731a003. [DOI] [PubMed] [Google Scholar]
- 11.Hamabata A, von Hippel PH. Biochemistry. 1973;12:1264–1271. doi: 10.1021/bi00731a004. [DOI] [PubMed] [Google Scholar]
- 12.Petersen PB, Saykally R. J. Annu. Rev. Phys. Chem. 2006;57:333–364. doi: 10.1146/annurev.physchem.57.032905.104609. [DOI] [PubMed] [Google Scholar]
- 13.Pegram LM, Record MT. J.Proc Natl Acad Sci U S A. 2006;103:14278–14281. doi: 10.1073/pnas.0606256103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pegram LM, Record MTJ. J Phys Chem B. 2007;111:5411–5417. doi: 10.1021/jp070245z. [DOI] [PubMed] [Google Scholar]
- 15.Salts of these ions increase surface tension by a bulk osmotic effect analogous to freezing point depression (i.e. an increase in [salt] increases the thermodynamic (entropic) cost of unmixing bulk water from a salt solution to form pure water at the air-water interface, analogous to unmixing bulk water to form pure ice in the case of freezing point depression).
- 16.Hong J, Capp MW, Saecker RM, Record MTJ. J. Biochemistry. 2005;44:16896–16911. doi: 10.1021/bi0515218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pegram LM, Jr., R MT. in preparation. 2008 [Google Scholar]
- 18.Meulen KV, Saecker RM, Jr., R MT. J. Mol. Biol. 2008;377:9–27. doi: 10.1016/j.jmb.2007.11.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long FA, McDevit WF. Chem. Rev. 1952;51:119–169. [Google Scholar]
- 20.Record MT, Jr., Anderson CF. Biophys. J. 1995;68:786–794. doi: 10.1016/S0006-3495(95)80254-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Courtenay ES, Capp MW, Saecker RM, Record MT., Jr. Proteins: Struct., Funct., and Genetics. 2000;41:72–85. doi: 10.1002/1097-0134(2000)41:4+<72::aid-prot70>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 22.Courtenay ES, Capp MW, Record MT., Jr. Protein Sci. 2001;10:2485–2497. doi: 10.1110/ps.ps.20801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Equation 1 for the SFEI is completely analogous to earlier analyses of thermodynamic effects of solutes on biopolymer processes. The SPM is a two-domain (local, bulk) interpretation of preferential interaction coefficients Γ, fundamental thermodynamic quantities (integrations of the distribution of solute around a biopolymer) that describe the competition of solute and water for a surface. Equation 1 should only be applied for model compounds with low aqueous solubility and/or small salt effects (i.e. free energy increments). Rigorous application requires various corrections determined from isopiestic distillation data for two- and three-component solutions. As this data is not available, and the low-solubility requirement is met in most cases, the internally consistent analysis developed here should be applicable to salt effects on biopolymer processes.
- 24.Anderson CF, Felitsky DJ, Hong J, Record MT. Biophys. Chem. 2002:101–102. doi: 10.1016/s0301-4622(02)00159-x. 497–511. [DOI] [PubMed] [Google Scholar]
- 25.Cannon JG, Anderson CF, Record MT., Jr. J. Phys. Chem. B. 2007;111:9675–9685. doi: 10.1021/jp072037c. [DOI] [PubMed] [Google Scholar]
- 26.Tsodikov OV, Record MT, Jr., Sergeev YV. J. Comput. Chem. 2002;23:600–609. doi: 10.1002/jcc.10061. [DOI] [PubMed] [Google Scholar]
- 27.Livingstone JR, Spolar RS, Record MT. J. Biochemistry. 1991;30:4237–4244. doi: 10.1021/bi00231a019. [DOI] [PubMed] [Google Scholar]
- 28.The solute partitioning model fundamentally treats salt effects as a function of salt activity. This requires the nonideality correction term (1 + ϵ±) on the right side of Eq. 1, which is directly related to the concentration dependence of the osmolality, i.e. (1 + ϵ±) ≡ (dOsm/dm3)/v. The osmolality, Osm ≡ vϕm3, is determined from the reported molal osmotic coefficients ϕ, and dOsm/dm3 is approximated as linear in the concentration range 0.1–0.7m. For GuHCl, when curvature in the solubility data and in dOsm/dm3 is taken into account, Kp,3 typically changes <2% in this concentration range.
- 29.Long FA, McDevit WF. J. Am. Chem. Soc. 1952;74:1773–1777. [Google Scholar]
- 30.Saylor JH, Whitten AI, Claiborne I, Gross PM. J. Am. Chem. Soc. 1952;74:1778–1781. [Google Scholar]
- 31.Poulson SR, Harrington RR, Drever JI. Talanta. 1999;48:633–641. doi: 10.1016/s0039-9140(98)00292-6. [DOI] [PubMed] [Google Scholar]
- 32.Hovorka S, Dohnal V, Carrillo-Nava E, Costas M. J. Chem. Thermo. 2000;32:1683–1705. [Google Scholar]
- 33.Nandi PK, Robinson DR. J. Am. Chem. Soc. 1972;94:1308–1315. doi: 10.1021/ja00759a043. [DOI] [PubMed] [Google Scholar]
- 34.Wetlaufer DB, Malik SK, Stoller L, Coffin RL. J. Am. Chem. Soc. 1964;86:508–514. [Google Scholar]
- 35.Most solubility and distribution measurements were performed at 25°C; benzene solubility in a few salt solutions was measured at 30°C. This temperature difference is insignificant because the temperature dependence of solubility free energy increments for a particular solute, where measured, is found to be small.
- 36.Horinek D, Netz R. Phys Rev Lett. 2007;99:226104/1–226104/4. doi: 10.1103/PhysRevLett.99.226104. [DOI] [PubMed] [Google Scholar]
- 37.Nandi PK, Robinson DR. Biochemistry. 1984;23:6661–6668. doi: 10.1021/bi00321a058. [DOI] [PubMed] [Google Scholar]
- 38.Felitsky DJ, Cannon JG, Capp MW, Hong J, Van Wynsberghe AW, Anderson CF, Record MT. J. Biochemistry. 2004;43:14732–14743. doi: 10.1021/bi049115w. [DOI] [PubMed] [Google Scholar]
- 39.If surface water has the same density as bulk water, then the value of b1 determined from the complete exclusion of Na2SO4 from hydrocarbon surface (b1=0.18) corresponds to an interfacial region approximately 5 Å thick (somewhat less than two water layers). Previous work has determined that glycine betaine is fully excluded from two layers of water surrounding anionic surface (i.e. b1=0.22). The similarity of the b1 values for the disparate surfaces (hydrocarbon and negatively charged) provides justification for the assumption of a universal value of b1.
- 40.Altshuller AP, Everson HE. J. Am. Chem. Soc. 1953;75:4823–4827. [Google Scholar]
- 41.Glasstone S, Dimond DW, Jones EC. J. Chem. Soc. 1926:2935–2939. [Google Scholar]
- 42.Robinson DR, Jencks WP. J Am Chem Soc. 1965;87:2462–2470. doi: 10.1021/ja01089a028. [DOI] [PubMed] [Google Scholar]
- 43.Since solubility of ethyl acetate is very high (∼ 0.9M), we have also determined Kppa by analyzing the SFEI for the three equivalent ASA differences (i.e. AGnE–AGn-1E), thus avoiding the need to account for the ester oxygen contribution. Results are qualitatively the same although somewhat larger values of Kppa are obtained for the 1:1 salts. Since these values are obtained from small differences (both in the SFEI and in ΔASA), there is more variation in the results for a particular salt. For this reason, we have elected to determine Kppa by using ethyl acetate to correct for the small contribution of the ester oxygens to the AGnE series.
- 44.Baynes BM, Trout BL. J. Phys. Chem. B. 2003;107:14058–14067. [Google Scholar]
- 45.Partition coefficients for ions at amide surface are actually determined for the total polar surface (28% polar amide and 7% ester oxygen). The single-ion contribution from the ester oxygen surface is predicted to be small, but these Kpvalues should be taken as approximate.
- 46.Tanford C. J Phys Chem. 1974;78:2469–2479. [Google Scholar]
- 47.Ray A, Némethy G. J. Am. Chem. Soc. 1971;93:6787–6793. doi: 10.1021/ja00754a014. [DOI] [PubMed] [Google Scholar]
- 48.Gratzer WB, Beaven GH. J Phys Chem. 1969;73:2270–2273. doi: 10.1021/j100727a028. [DOI] [PubMed] [Google Scholar]
- 49.Deguchi K, Meguro K. J. Colloid and Interface Sci. 1975;50:223–227. [Google Scholar]
- 50.Podo F, Ray A, Némethy G. J Am Chem Soc. 1973;95:6164–6171. [Google Scholar]
- 51.Spolar RS, Livingstone JR, Record MT. J. Biochemistry. 1992;31:3947–3955. doi: 10.1021/bi00131a009. [DOI] [PubMed] [Google Scholar]
- 52.Morrison TJ, Billett F. J. Chem. Soc. 1952:3819–3822. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables containing solubility data, the composite thermodynamic quantity b 1(Kp 3, – 1) for each salt, and surface area breakdowns for the model compounds. This material is available free of charge via the Internet at http://pubs.acs.org.