Abstract
Neuropeptides are often released into circulatory fluid (hemolymph) to act as circulating hormones and regulate many physiological processes. However, the detection of these low-level peptide hormones in circulation is often complicated by high salt interference and rapid degradation of proteins and peptides in crude hemolymph extracts. In this study, we systematically evaluated three different neuropeptide extraction protocols and developed a simple and effective hemolymph preparation method suitable for MALDI MS profiling of neuropeptides by combining acid-induced abundant protein precipitation/depletion, ultrafiltration, and C18 micro-column desalting. In hemolymph samples collected from crab Cancer borealis several secreted neuropeptides have been detected, including members from at least five neuropeptide families, such as RFamide, allatostatin, orcokinin, tachykinin-related peptide (TRP), and crustacean cardioactive peptide (CCAP). Furthermore, two TRPs were detected in the hemolymph collected from food-deprived animals, suggesting the potential role of these neuropeptides in feeding regulation. In addition, a novel peptide with a Lys-Phe-amide C-terminus was identified and de novo sequenced directly from the Cancer borealis hemolymph sample. To better characterize the hemolymph peptidome, we also identified several abundant peptide signals in C. borealis hemolymph that were assigned to protein degradation products. Collectively, our study describes a simple and effective sample preparation method for neuropeptide analysis directly from crude crustacean hemolymph. Numerous endogenous neuropeptides were detected including both known ones and new peptides whose functions remain to be characterized.
Keywords: Neuropeptides, Hemolymph, Mass spectrometry, MALDI-TOF/TOF, LC-ESI-QTOF, MALDI-FTMS, Quantitation, Crustacean
INTRODUCTION
Neuropeptides are an important class of chemical messengers that modulate nervous system response. These signaling molecules are involved in initiation, modulation and regulation of many physiological processes. Often times these peptides need to be secreted into circulating fluids to exert their hormonal effects on distant organs that participate in a variety of functions such as food intake, pain sensing, stress response, and molting, among many others [1–4]. These peptide hormones have been implicated in regulation of physiological processes, including the control of heart function, hemolymph circulation, and digestion, to name a few [5–7]. Monitoring these released neuropeptides is therefore essential for identification of bioactive neuropeptides and serves as an important step towards understanding their functions. Furthermore, hemolymph serves as an abundant sample source that can be collected without sacrificing the animals. Thus, the analysis of hemolymph neuropeptide profiles offers a great opportunity to monitor peptide secretion changes under different physiological states or in response to different functional manipulations with the usage of the same animal. Consequently, developing effective and highly sensitive methods to examine peptide hormones in circulating hemolymph is an important step toward peptide functional studies.
It has been reported that using traditional immunoassays such as radioactive immunoassay (RIA) and enzyme immunoassay (EIA), several neuropeptides were found to be circulating hormones in crustacean hemolymph including crustacean cardioactive peptide (CCAP) [1], crustacean hyperglycemic hormone (CHH) and molting-inhibiting hormone (MIH) [8–10]. However these methods require antibodies, many of which are not commercially available. Furthermore, they suffer from limitations of cross-reactivity with structurally similar peptides and inability to analyze multiple peptides simultaneously, which greatly impedes the unambiguous identification of neuropeptide isoforms. Since the last two decades, numerous neuropeptides have been discovered in crustacean neuronal tissues using biological mass spectrometry (MS), which provides high speed, great sensitivity and chemical specificity [11–14]. However, compared to the widespread study of neuropeptides using tissue sources, reports on peptide analysis of hemolymph have been quite scarce, which is in part due to the significant analytical challenges for peptide detection in these complex biological fluids. For example, hemolymph contains a large number of abundant proteins including haemocyanin, cryptocyanin, plasma coagulogen, heteroagglutinins, and vitellogenins, among others [15, 16]. These proteins can degrade into small fragments under natural conditions or during sample processing, which not only suppresses the detection of low-abundance peptide hormones, but also complicates data analysis of the hemolymph peptidome. Furthermore, the circulating peptide hormones are present at extremely low concentrations (pM-nM) [17], which is in contrast with the high salt concentrations (~500 mM) inherent to marine organisms that interferes mass spectrometric analysis of neuropeptides. Therefore, developing advanced sample preparation method is a key to improve the sensitivity and selectivity for detection of putative neuropeptides in hemolymph.
Recently, Fastner and coworkers detected pre-ecdysis triggering hormone (PETH) with concentration of 30 to 45 nM during pre-ecdysis in insect hemolymph using heat-induced abundant protein depletion, ultrafiltration and reversed-phase high performance liquid chromatography (RP-HPLC)[17]. In our previous study, using acidified methanol as extraction buffer, Cancer borealis tachykinin-related peptide (CabTRP) Ia was detected in hemolymph of Cancer productus under starvation with accurate mass measurement via MALDI-FTMS [18]. However no MS/MS data was available to confirm the identity of the peptide due to its low concentration in the sample.
This paper provides the first description of effective sample preparation methods for peptidomic profiling and neuropeptide detection in crustacean hemolymph using MS. In this study, we have developed a hybrid sample preparation approach combining ultracentrifugation to remove the high molecular weight species and micro-scale solid-phase extraction to extract and concentrate neuropeptides upon desalting that enables effective detection of peptide hormones in the hemolymph. We systematically investigated three different buffer solutions and two types of protein depletion methods including heat-induced and acid-induced protein denaturation/precipitation. With the effective removal of highly abundant proteins, acidified methanol spiked with EDTA and protease inhibitors produced the best recovery of spiked neuropeptide standards and yielded sensitive detection of neuropeptides from hemolymph samples. CabTRP Ia (m/z 934.5) was detected as a circulating peptide hormone from hemolymph in starved animal with supporting MS/MS information, suggesting a potential role of this peptide in feeding regulation. Several additional neuropeptides were also detected based on accurate mass matching with previously reported neuropeptides. We also de novo sequenced a peptide with C-terminal KFamide, which could be a novel family of neuropeptide(s) whose function remains to be determined. Some common protein degradation products present in hemolymph were also identified with de novo sequencing assisted by Mascot database searching and BLAST.
EXPERIMENTAL
Chemicals and Materials
Methanol, acetonitrile, formic acid (FA), acetic acid and EDTA disodium salt were purchased from Fisher Scientific (Pittsburgh, PA). 2, 5-dihydroxybenzoic acid (DHB) was obtained from MP Biomedicals, Inc. (Solon, OH). α-cyano-4-hydroxy-cinnamic acid (CHCA) was purchased from Sigma-Aldrich (St. Louis, MO). Protease inhibitor Complete-Mini cocktail was purchased from Roche (Indianapolis, IN). C18 micro spin column and 10 kDa molecular weight cutoff tube (MWCO) were purchased from Argos (Elgin, IL). Acidified methanol was prepared using 90% methanol, 9% glacial acetic acid, and 1% water. All water used in this study was deionized water from a Millipore filtration system (Bedford, MA). Peptide standards: angiotensin II (DRVYIHPF, m/z 1046.5), bradykinin (RPPGFSPFR m/z 1060.5), [Arg8] vasopressin (AVP) (CYFQNCPRGamide m/z 1084.4), [Val5] angiotensin I (DRVYVHPFHL m/z 1282.7), angiotensin I (DRVYIHPFHL m/z 1296.7), allatostatin I (APSGAQRLYGFGLamide m/z 1335.7), substance P (RPKPQQFFGLMamide m/z 1347.7) were purchased from the American Peptide Company (Sunnyvale, CA).
Instrumentation
MALDI TOF/TOF
A model 4800 MALDI TOF/TOF analyzer (Applied Biosystems, Framingham, MA) equipped with a 200 Hz, 355 nm Nd:YAG laser was used for both direct peptide profiling and tandem MS fragmentation study for hemolymph samples. Acquisitions were performed in positive ion reflectron mode. Instrument parameters were set using the 4000 Series Explorer software (Applied Biosystems). Mass spectra were obtained by averaging 1000 laser shots covering mass range m/z 500–4000. MS/MS was achieved by 2 kV collision induced dissociation (CID) using air. The peptide sequences were predicted manually with the help of “de novo Explorer” software (Applied Biosystems). A saturated solution of α-cyano-4-hydroxycinnamic acid (CHCA) in 50% (v/v) acetonitrile was used as matrix. For sample spotting, 0.4 μl of sample was spotted on MALDI plate and allowed to dry, followed by application of 0.4 μl matrix solution.
MALDI FTMS
MALDI FTMS experiments were performed on a Varian/IonSpec Fourier transform mass spectrometer (Lake Forest, CA) equipped with a 7.0-T actively shielded superconducting magnet. The FTMS instrument consisted of an external high-pressure MALDI source. A 355-nm Nd:YAG laser (Laser Science, Inc., Franklin, MA) was used to create ions that can be accumulated in the external hexapole storage trap before being transferred through a quadrupole ion guide to the ICR cell. All mass spectra were collected in positive ion mode. The ions were excited prior to detection with an RF sweep beginning at 7050 ms with a width of 4 ms and amplitude of 150 V base to peak. The filament and quadrupole trapping plates were initialized to 15 V, and both were ramped to 1 V from 6500 to 7000 ms to reduce baseline distortion of peaks. Detection was performed in broadband mode from m/z 108.00 to 2500.00.
Capillary LC-ESI-Q-TOF MS/MS
Nanoscale LC-ESI-Q-TOF MS/MS was performed using a Waters capillary LC system coupled to a Q-TOF Micro mass spectrometer (Waters Corp., Milford, MA). Chromatographic separations were performed on a C18 reverse phase capillary column (75 μm internal diameter by 150 mm length, 3 μm particle size; Micro-Tech Scientific Inc., Vista, CA). The mobile phases used were: water with 5% acetonitrile and 0.1% FA (A); acetonitrile with 5% water and 0.1% FA (B); water with 0.1% FA (C). An aliquot of 6.0 μl of desalted hemolymph sample dissolved in 0.1% FA was injected and loaded onto the trap column (PepMap™ C18; 300 μm column internal diameter 1 mm, 5 μm particle size; LC Packings, Sunnyvale, CA, USA) using mobile phase C at a flow rate of 30 μl/min for 3 min. Following this, the stream select module was switched to position the trap column in line with the analytical capillary column, and a linear gradient of mobile phases A and B was initiated. A splitter was added between the mobile phase mixer and the stream select module to reduce the flow rate from 15 μl/min to 200 nl/min. The nanoflow ESI source conditions were set as follows: capillary voltage 3200 V, sample cone voltage 35 V, extraction cone voltage 1 V, source temperature 120°C, cone gas (N2) 10 l/h. Data dependent acquisition was employed for the MS survey scan and the selection of precursor ions and subsequent MS/MS of the selected parent ions. The MS scan range was from m/z 300–2000 and the MS/MS acquisition was from m/z 50–1800. The MS/MS data was converted to peak list file using ProteinLynx software Ver. 2.1 (Waters Corp.) and submitted for Mascot database search against NCBI non-redundant protein database, under taxonomy Metazoa. N-terminal acetylation and C-terminal amidation were specified as variable modifications. Precursor ion mass tolerance was 50 ppm, and product ion mass tolerance 0.3 Da. No enzymatic cleavage specificity was specified.
Animals and Feeding Study
Jonah crabs, Cancer borealis, were shipped from the Fresh Lobster Company (Gloucester, Massachusetts, USA) and maintained without food in an artificial seawater tank at 12–13 °C for a week before used. Crabs were removed from tank and placed on ice for 15 min for cold anesthesia, and then hemolymph sample was collected by inserting a 22-gauge needle attached to a 3-ml plastic syringe through the junction of the thorax and abdomen into the pericardial chamber. After hemolymph collection, the crab was returned to the tank and let recover for two days. Then the same crab was fed with crab meat until it stopped eating, which took 30 to 40 min. Hemolymph sample was collected after packing the crab in ice for 15 min in the same way described above.
Hemolymph Sample Preparation
Different buffer systems were used to extract peptides from freshly obtained hemolymph samples and are listed below:
Method 1
0.75 ml acidified methanol was spiked into 0.75 ml freshly obtained hemolymph to extract peptides and precipitate large proteins.
Method 2
To inhibit protein degradation, half of a protease inhibitor cocktail pill was dissolved in 40 ml of 20 mM EDTA solution. A mixture of 0.45 ml acidified methanol and 0.3 ml protease inhibitor mixed EDTA solution was added to 0.75 ml freshly obtained hemolymph. Protease inhibitor was added to eliminate enzymatic protein degradation during sample preparation and EDTA can prevent cation triggered protein clotting, which has been reported to decrease recovery of putative peptides in the sample [19].
Method 3
0.75 ml protease inhibitor mixed EDTA solution was spiked into 0.75 ml freshly obtained hemolymph. The resulting solution was then boiled for 10 minutes to further inhibit protease activity and precipitate hemolymph proteins.
The sample prepared by each method was centrifuged at 16 000 rcf for 10 minutes then the supernatant was harvested and equally divided into two fractions and filtered through a 10 kDa MWCO tube by centrifuging at 15 000 rcf. The low mass filtrate was concentrated to dryness using a Savant SC 110 SpeedVac concentrator (Thermo Electron Corporation, West Palm Beach, FL) and was resuspended in 80 μl 0.1% FA in water. After sonication for 10 minutes, the sample was desalted by C18 micro spin column according to the product manual and eluted in 6 μl 0.1% FA in 50% acetonitrile (v/v). The sample was then directly used for MALDI MS detection. For LC-ESI-QTOF experiment, the sample was first dried followed by resuspension in 6 μl 0.1% FA in water before injection.
SDS-PAGE
To evaluate the protein removal efficiency of three sample preparation methods, tricine-sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was performed using XCell SureLock Mini-Cell (Invitrogen, Carlsbad, CA) according to the instruction manual. Briefly, both crude hemolymph and processed hemolymph samples were separated with a NuPAGE 10% Bis-Tris Gel and the NuPAGE MES SDS buffer (Invitrogen) at 200V for 30min. After rinsed with water, the gel was then stained with SimplyBlue SafeStain (Invitrogen) for 1hr, and incubated with water over night before visualization.
Formaldehyde Derivatization and Recovery Assay
Stable isotopic labeling with H2/D2 formaldehyde using reductive methylation was performed to measure the concentration of internal standard in the processed hemolymph sample, which can be used to examine the recovery of each method. An aliquot of 6 μl of the final hemolymph sample was mixed with 0.75 μl borane pyridine (120 nM) and 0.5 μl formaldehyde (20%, v/v). Peptide standard mixture solution was labeled in the same way but with deuterium formaldehyde. Both samples were incubated at 37 °C for 20 minutes. The formaldehyde labeled hemolymph sample was then mixed with the peptide standard solution with 1:1 ratio. 0.4 μl of the labeled mixture was spotted and analyzed by MALDI TOF/TOF. The recovery of each internal standard was then calculated based on the ratio of isotopic cluster area from light labeled standards and heavy labeled standards.
The same animal was used in each set of comparison assay to eliminate changes induced by animal variability and different sample background. Internal standards were spiked into fresh hemolymph samples resulting in concentration of 10 or 30 nM. The sample was prepared using three methods described in a previous section. After processing, 0.4 μl of the resulting hemolymph sample was analyzed by MALDI TOF/TOF to compare signal to noise ratio of internal standards prepared by these three methods. Each sample was spotted three times, and every spot was analyzed twice. The signal to noise ratio of internal standard peaks was calculated by averaging six spectra.
RESULTS AND DISCUSSION
Evaluation of Sensitivity and Peptide Coverage of Three Sample Preparation Protocols -- Peptide Detection in Hemolymph with 10 nM Spiked Synthetic Peptides
The sensitivity of MS detection is largely dependent on the sample background. Without sample processing, few peptide peaks can be detected with poor signal-to-noise (S/N) ratios in the crude hemolymph sample due to very high concentration of salts (more than 600 mM) and the presence of large proteins such as clotting factors (Figure S1a). The signal intensity of spiked synthetic peptide standards in crude hemolymph was 100 fold lower compared to standards with the same concentration prepared in pure water (Figures S1b, c). Therefore, removal of salts, abundant proteins, and further concentrating the sample was crucial to detect putative neuropeptides in hemolymph samples.
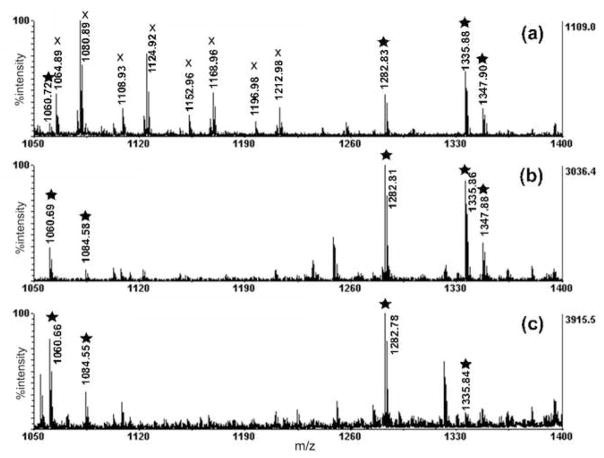
The sensitivity of three different sample preparation protocols described above was evaluated using peptide standards by MALDI-TOF/TOF. Five internal standard peptides were spiked into the original hemolymph sample at a concentration of 10 nM, including bradykinin (m/z 1060.5), [Arg8] vasopressin (AVP, m/z 1084.4), [Val5] angiotensin I (m/z 1282.7), allatostatin I (m/z 1335.7), and substance P (m/z 1347.7). These peptides cover mass range of 500–2000 Da, which is suitable for examining preparation methods for neuropeptide detection. As shown in Figure 1, all five internal standards were detected using Method 2 with good signal intensity, while AVP was not detected using Method 1 and substance P was not detected using Method 3 (n=3). Because acidified methanol is commonly used for neuropeptide extraction, we employed this buffer to denature and precipitate the abundant proteins in hemolymph and extract peptides in Method 1 and Method 2. Methanol provides good solubility for most small peptides, while the high acid concentration of this buffer can denature large proteins including protease to prevent degradation of target peptides during sample preparation. In Method 2, EDTA solution was added to prevent cation trigged protein clotting during the sample preparation process. Additionally, protease inhibitor cocktail pill was added to further eliminate peptide and protein degradation. In Method 3, aqueous EDTA solution was used to extract peptides and abundant proteins were precipitated with heat treatment, resulting in depletion from the hemolymph samples. To evaluate the effectiveness of protein depletion of these three methods, SDS-PAGE gel separation was performed on various hemolymph samples before and after treatment. As seen, all three methods can significantly reduce the abundant protein content in the original hemolymph samples; however the acid induced precipitation is more efficient than the heating induced precipitation (Figure S2). The resulting hemolymph samples can be further simplified by ultrafiltration using a 10 kDa MWCO membrane. This treatment can improve the matrix crystallization, eliminate ion suppression, and may also increase the recovery of peptides from the C18 spin column. Crustacean hemolymph contains a large amount of salt, which greatly interferes with MS detection. After desalting with C18 spin column, better spectral quality can be achieved compared to crude sample (data not shown). In Method 1, the detection of spiked peptide standards was greatly interfered by polymer-like peaks in the mass range from 500 to 1000 Da. Method 3 produced comparable results with Method 2 showing preferential detection of smaller peptides, such as bradykinin and AVP; whereas peptide allatostatin was detected with much lower peak intensity, and substance P was not even detected using this method. This experiment showed that the addition of small amount of organic solvent can facilitate extraction of larger and more hydrophobic peptides. Considering that many neuropeptides are amidated at their C termini, increasing their hydrophobicities, Method 2 should have better recovery than Method 3 for detection of neuropeptides. However, if the peptides of interest are more hydrophilic, Method 3 employing an aqueous extraction buffer can also be considered as an efficient extraction method.
Figure 1.
MALDI TOF/TOF spectral comparison of hemolymph sample spiked with five internal peptide standards at a concentration of 10 nM using three methods. (a) Method 1: pure acidified methanol was used as extraction buffer. (b) Method 2: acidified methanol mixed with 20 mM EDTA and protease inhibitors. (c) Method 3: extract with 20 mM EDTA mixed with protease inhibitor and heat induced protein precipitation/depletion. The internal standard peaks were labeled with stars, while the polymer-like peaks were labeled with x. All peaks shown in the MALDI spectra are singly charged in this study.
Quantitative Evaluation of Recovery of Synthetic Peptides in Hemolymph Sample
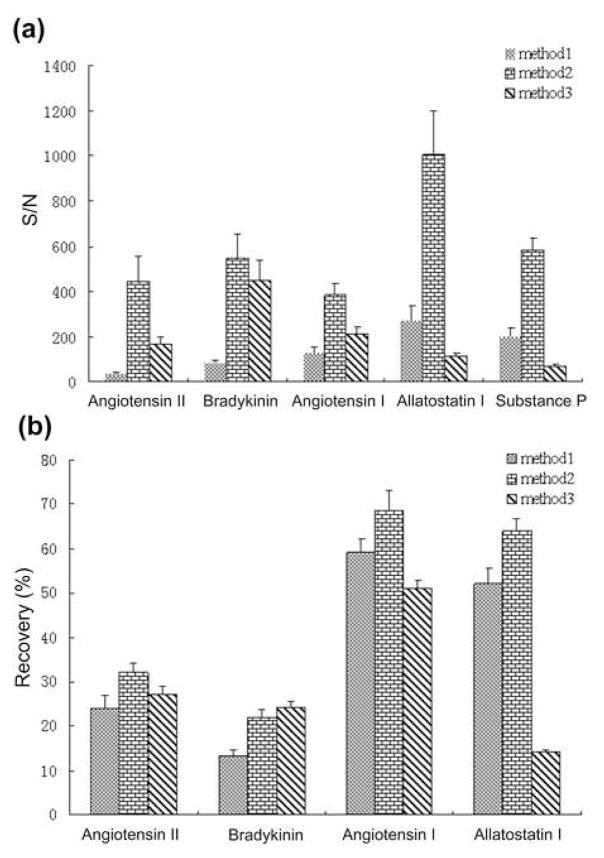
To evaluate the recovery of synthetic peptides by various processing methods, dimethylation using isotopic formaldehyde was performed on processed hemolymph samples. In the 10 nM internal standard spiked experiments, one peptide standard was not detected in samples prepared by either Method 1 or Method 3, hence the concentrations of internal peptide standards were elevated to 30 nM for a more complete and accurate evaluation of recovery rates of these three sample preparation protocols. To calculate the accurate recoveries, the final sample prepared by each method was labeled with formaldehyde and mixed with the same volume of 10−6 M peptide standard mixture labeled with deuterium formaldehyde. This method labels the N-termini of peptides with two methyl groups, resulting in 4 Da mass differences between light labeled and heavy labeled peak pair per incorporated label. It has been reported as a fast and simple reaction that can be applied for differential proteomic and peptidomic analysis [20–22]. Figure 2 shows the result from isotopic labeling experiment of a hemolymph sample processed by method 2. All of the peptide pairs spiked in the sample were detected with good signal intensity. The relative abundances of the internal standards in the final sample can be calculated based on ratios of light labeled peak cluster area and heavy labeled peak cluster area, which were further converted to recoveries. Figure 3 compares the signal-to-noise ratios and recoveries of different peptide standards achieved by three sample preparation methods. Among these three methods, Method 2 provides the best signals for all peptides, and also better recoveries for most cases, suggesting that a combination of acidified methanol, EDTA, and protease inhibitors provides better detection sensitivity than the other two preparation methods. The S/N and recovery values and their relative standard deviation (RSD) for each method are summarized in Table 1 (n=6). Recovery of peptide substance P is not listed, because after formaldehyde labeling this peak is largely interfered by isotopic peaks of m/z 1401.8 present in the hemolymph sample.
Figure 2.
MALDI mass spectrum of formaldehyde labeled hemolymph sample prepared by Method 2. Equimolar ratio of H2-formaldehyde labeled hemolymph sample and D2-formaldehyde labeled 10−6 M peptide standards were mixed and analyzed. The heavy labeled peaks from spiked peptide standards are labeled with open circles, and the light labeled internal standards present in hemolymph sample are indicated with solid circles.
Figure 3.
Quantitative comparisons of three hemolymph preparation methods spiked with 30 nM internal standards. (a) Comparison of signal to noise ratios of five internal peptide standards (N=6). (b) Comparison of overall recovery of four internal peptide standards.
Table 1.
Comparisons of S/N and recoveries of five neuropeptides spiked in the crustacean hemolymph samples prepared using three different methods
| Internal standards |
Method 1 | Method 2 | Method 3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S/N | RSD (%) |
Recovery (%) |
RSD (%) |
S/N | RSD (%) |
Recovery (%) |
RSD (%) |
S/N | RSD (%) |
Recovery (%) |
RSD (%) |
|
| Angiotensin I | 35 | 19 | 24 | 12 | 446 | 25 | 32 | 6 | 165 | 19 | 27 | 7 |
| Bradykinin | 84 | 14 | 13 | 11 | 545 | 20 | 22 | 8 | 448 | 20 | 24 | 5 |
| Angiotensin II | 132 | 17 | 59 | 5 | 385 | 13 | 69 | 6 | 213 | 12 | 51 | 3 |
| Allatostatin I | 276 | 22 | 52 | 7 | 1005 | 20 | 64 | 4 | 113 | 11 | 14.1 | 4 |
| Substance P | 199 | 19 | N/A | N/A | 584 | 9 | N/A | N/A | 69 | 14 | N/A | N/A |
N=6
N/A: the value is not available.
The study of several commonly used peptide standards demonstrates that Method 2 provides acceptable recovery rates ranging from 20% to 70% depending on the properties of specific peptides. A major source of sample loss is due to possible adsorption and interactions of peptides with the protein pellet during protein depletion and precipitation process in all three methods [19]. Filtration through MWCO can also contribute to sample loss, because there is usually solution remains in the high molecular weight portion and the molecular weight cutoff membrane may also absorb peptides. Recovery of Method 1 is comparable with Method 2; however this method yields lower S/N. This is likely because the MS detection was greatly influenced by sample background. As mentioned in the last section, polymer like peaks are commonly detected using Method 1, which can cause poor crystallization and suppress the detection of peptides. The recovery and sensitivity of Method 3 are comparable with Method 2 for the two smaller peptide standards, but much worse for the other three larger peptides, especially allatostatin I and substance P, which have amidation on their C termini, increasing their hydrophobicity. This comparison shows that spiking EDTA aqueous solution and protease inhibitor into acidified methanol is best suited for neuropeptide detection in hemolymph sample.
Profiling and Sequencing of Neuropeptides in Hemolymph
Five groups of hemolymph samples from both fed and unfed Cancer borealis were processed by Method 2 and analyzed by MALDI FTMS and MALDI TOF/TOF. As listed in Table 2, nine known neuropeptides were detected, including two tachykinin-related peptides (TRP, m/z 934.5, m/z 964.5), one A-type allatostatin (m/z 967.5), an orcokinin peptide (m/z 1256.6), the crustacean cardioactive peptide (CCAP, m/z 956.4) and four RFamides (m/z 1104.6, m/z 1005.6, m/z 1105.6, m/z 1019.6). Due to the low intensity of these peaks present in the hemolymph sample, some of the peptides were assigned based on accurate mass measurement alone without MS/MS confirmation. The threshold of mass error was set at 50 ppm. Figure 4 shows representative MALDI mass spectra of two hemolymph samples collected from the same animal under unfed and fed conditions. As shown in Figure 5a, CID fragmentation spectrum of m/z 934.5 was obtained using MALDI TOF/TOF, which indicated the amino acid sequence of tachykinin-related peptide Ia (CabTRP Ia). For the first time, CabTRP Ia was confirmed as a circulating hormone present in the hemolymph, and this peptide was only detected in the hemolymph sampled from unfed animals. Another TRP detected in hemolymph was CabTRP II (m/z 964.5), which was also seen only in the hemolymph collected from a starved animal. This result suggests that TRPs are released into circulating fluid as hormones, and their concentration level is correlated to the feeding status of animals with higher abundance in circulation detected in starved animals. TRPs have previously been reported as bioactive neuropeptides that can activate pyloric motor patterns [18]. They are known to be present extensively in the brain, stomatogastric nervous system (STNS) and sinus gland (a peptide release organ in eyestalk) of several different crustacean species [18, 23, 24]. Based on our recent neuropeptidomic study of neural tissue in Cancer borealis, CabTRP Ia was detected in sinus gland with very low abundance, whereas CabTRP II was detected only in the brain and STNS (unpublished result). Recently both of these two TRPs were detected as gut derived peptides present in midgut epithelium endocrine cells at relatively high abundances [18]. This observation suggests that these two neuropeptides are very likely to be released from the midgut epithelium. However the underlying mechanism governing the release of these peptides and how they are involved with feeding regulation requires further investigation.
Table 2.
List of native neuropeptides detected in Cancer borealis hemolymph
| Peptide sequence | Monoisotopic mass |
TOF/TOF | FTMS | |
|---|---|---|---|---|
| Measured | Theoretical | |||
| Tachykinin-Related Peptides | ||||
| APSGFLGMRamide* | 934.49 | 934.49 | x | |
| TPSGFLGMRamide | ||||
| 964.50 | 964.50 | x | ||
| CCAP | ||||
| PFcNAFTGcamide | 956.44 | 956.38 | x | x |
| RFamide | ||||
| APRNFLRFamide | 1019.56 | 1019.59 | x | |
| GPRNFLRFamide | 1005.61 | 1005.57 | x | |
| GAHKNYLRFamide | 1104.58 | 1104.61 | x | |
| SMPSLRLRFamide | 1105.65 | 1105.63 | x | |
| A type allatostatin | ||||
| ERPYSGLamide | 967.50 | 967.50 | x | |
| Orcokinin | ||||
| NFDEIDRSGFG | 1256.56 | 1256.55 | x | |
| Novel peptide | ||||
| I/LNFTHKFamide* | 905.49 | 905.50 | x | x |
indicates the presence of this peptide is confirmed by MS/MS.
x indicates the presence of this peptide in hemolymph sample as detected by a particular instrument. CCAP is an abbreviation for crustacean cardioactive peptide. Lower case c in the CCAP peptide sequence stands for disulfide-bonding formation. RFamide stands for peptide family ending with Arg-Phe-amide C-terminal sequence.
Figure 4.

Comparisons of the hemolymph peptidomic profiles acquired from the same crab under starved and satiated conditions via MALDI TOF/TOF using Method 2. (a) MALDI TOF/TOF spectrum of hemolymph from crab under starved condition. (b) MALDI TOF/TOF spectrum of hemolymph from the same crab under satiated condition. Neuropeptides and identified protein degradation products are labeled with their sequences or peptide names.
Figure 5.

MALDI TOF/TOF tandem MS sequencing of (a) CabTRP 1a (m/z 934.49) and (b) protein degradation fragment (m/z 1209.7) from the hemolymph sample. The amino acid sequence of each peptide is given above the spectrum. The presence of b and y ions is indicated by horizontal lines above (b ions) or below (y ions) the corresponding amino acid residues in the peptide sequence.
All of the other detected neuropeptides were previously observed in the pericardial organ or sinus gland of Cancer borealis or other crustacean species [12, 13, 24]. It is not surprising that these neuropeptides present in the hemolymph sample, since pericardial organ and sinus gland are major neurohemal organs for releasing peptide hormones into circulating fluid to affect distant stomatogastric motor neurons. A novel peptide with mass of 905.50 Da was also identified in this study using MALDI TOF/TOF, resulting in a deduced sequence of I/LNHTHKFamide. As shown in Figure 6a, the MS/MS fragment ion spectrum of this peptide was obtained by MALDI TOF/TOF directly from processed hemolymph sample. To further confirm the presence of lysine residue in this peptide, the hemolymph sample was derivatized with formaldehyde. The MS/MS spectrum of dimethyl labeled form of this peptide as shown in Figure 6b indicates that there are two labeled sites in this peptide, including N terminus and a lysine residue near the C terminus. The identification and derivation of the amino acid sequence of the new peptide was further confirmed by the tandem MS fragmentation analysis of synthetic peptide standard (Figure 6c) and its dimethylated counterpart (Figure 6d). As shown, identical fragmentation patterns and fragment ions were detected from synthesized standard. Given that the C-terminal amidation is a conserved post-translational modification (PTM) for most neuropeptides and the similarity of the amino acid residue components of this peptide to the well known RFamides, this peptide is likely to belong to a new family of neuropeptide(s). Electrophysiology experiments will be performed in the future to further evaluate and confirm the biological function of this peptide.
Figure 6.

MS/MS sequencing of a novel peptide at m/z 905.50 and its synthetic standard using MALDI TOF/TOF. (a) MS/MS of putative peptide at m/z 905.50 from hemolymph sample with a deduced sequence of I/LNFTHKFamide. (b) MS/MS of synthesized peptide with sequence of LNFTHKFamide at a concentration of 10 μM. (c) MS/MS of formaldehyde derivatized target peptide at m/z 961.56 in hemolymph sample. * indicates the two dimethyl labeled sites, including N terminus and the lysine residue. (d) MS/MS of formaldehyde derivatized synthetic standard peptide with a sequence of LNFTHKFamide at a concentration of 10 μM. * indicates the two dimethyl labeled sites, including N terminus and the lysine residue. The presence of b and y ions is indicated by horizontal lines above (b ions) or below (y ions) the corresponding amino acid residues in the peptide sequence.
Identification of Peptides from Abundant Protein Degradation
With MS/MS information obtained with MALDI TOF/TOF and LC-ESI-QTOF, several abundant peaks in the hemolymph peptidomic profile were identified as protein degradation products (shown in Figure 5b). As shown in Table 3, a series of these protein degradation fragments come from a molting protein cryptocyanin, which has similar structure and sequence as hemocyanin but has no function of oxygen transporting. This protein was characterized in Cancer magister and was reported to be at even higher concentration than hemocyanin at certain molting stages [25]. Huybrechts et al. previously suggested the contamination of these putative cryptocyanin fragments by mass measurements in the analysis of Cancer borealis brain and thoracic ganglia extracts [26]. In this study, MS/MS fragmentation analyses obtained by LC-ESI-QTOF were used to derive the sequences of these peaks (data not shown), and their identities were confirmed with BLAST search. Several of these cryptocyanin degradation fragments have similar masses with some known neuropeptides, such as m/z 1395.7 and m/z 1281.7, which may lead to mis-assignment of these peaks. Careful attention is therefore needed to distinguish protein degradation products from putative neuropeptides with similar masses.
Table 3.
Abundant protein degradation products detected in hemolymph sample
| Proteins | [M+H]+ | Sequence |
|---|---|---|
| Cryptocyanin | 1508.81 | YKIFEPLRESNL |
| 1395.73 | YKIFEPLRESN | |
| 1281.68 | YKIFEPLRES | |
| 1194.65 | YKIFEPLRE | |
| 1065.61 | YKIFEPLR | |
| Beta-actin | 1209.68 | LRVAPEESPVL |
Another protein degradation product was identified with m/z 1209.7 (Figure 5b). This peptide was detected as an abundant peak in fed crab’s hemolymph (Figure 4b). To determine its identity MS/MS fragmentation was obtained on MALDI TOF/TOF shown in Figure 5b. The sequence was deduced manually assisted with “De novo Explorer” software. LC-ESI-QTOF data was submitted for Mascot database search, resulting in the sequence assignment of LRVAPEESPVL, which was shown to be a fragment of actin in several different crustacean species, such as Gecarcinus lateralis and Penaeus monodon. Mascot search with LC-ESI-QTOF data also showed several other fragments from actin, but they were present at lower abundances and were usually not detected in MALDI MS. Enzymatic degradation of proteins can occur under natural conditions or during sample processing, which can be eliminated by immediate denaturing of the degradation enzymes in fresh hemolymph samples using acid extraction and/or adding protease inhibitors. Different degradation products are observed for different sample preparation methods, which maybe due to their different solubilities in different solvents and different enzyme activity in different solvent environments. Due to the lack of sequenced genome for crustacean species the complete characterization of protein degradation products in hemolymph was not attempted.
Although less abundant protein degradation products have been reported using in vivo microdialysis sampling approach where degradation enzymes have been filtered out with molecular weight cutoff membrane [27], the direct analysis of hemolymph samples using methods described in this study represents an attractive alternative strategy that offers simplicity, speed, and throughput. Without the need to perform animal surgery and sophisticated microdialysis probe implantation, direct neuropeptide analysis using crude hemolymph extraction allows collection and concentration of a large volume of hemolymph in a short amount of time, which enables detection and sequencing of neuropeptides present at much lower abundance in circulating fluids. The combination of two different strategies of neuropeptide detection in hemolymph will offer unprecedented information of neuropeptidomes in complex circulating environment under different physiological conditions.
CONCLUSIONS
This study reported and established an effective method of neuropeptide analysis in circulating fluids collected from a marine organism. Through comparison of three different extraction buffer systems and two different abundant protein depletion methods, the use of acidified methanol and protease inhibitor was shown to be efficient for trace-level neuropeptide analysis in hemolymph samples. For the first time, CabTRP Ia was detected directly in crustacean hemolymph sample using MS with confident MS/MS sequence confirmation. Furthermore, the involvement of tachykinin-related peptides in feeding regulation was revealed in this work. A novel peptide with C-terminal KFamide was characterized directly from hemolymph sample. Finally, protein fragments in hemolymph sample were identified with combination of de novo sequencing, database search and BLAST. Collectively, our study represents the first comprehensive peptidomic analysis of Cancer borealis hemolymph. The methodology developed in this study will find applications in expanding peptidomes in many important biological fluids derived from other model systems.
Supplementary Material
Acknowledgments
The authors wish to thank the University of Wisconsin-Biotechnology Center Mass Spectrometry Facility and Drs. Amy Harms and Michael Sussman for access to the MALDI TOF/TOF instrument. This work was supported by a National Science Foundation CAREER Award (CHE-0449991) and the National Institutes of Health through grant 1R01DK071801. L.L. acknowledges an Alfred P. Sloan Research Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Phlippen MK, Webster SG, Chung JS, Dircksen H. Ecdysis of Decapod Crustaceans is Associated with a Dramatic Release of Crustacean Cardioactive Peptide into the Haemolymph. J Exp Biol. 2000;203:521–536. doi: 10.1242/jeb.203.3.521. [DOI] [PubMed] [Google Scholar]
- 2.Turrigiano GG, Selverston AI. A Cholecystokinin-Like Hormone Activates a Feeding-Related Neural Circuit in Lobster. Nature. 1990;344:866–868. doi: 10.1038/344866a0. [DOI] [PubMed] [Google Scholar]
- 3.Bodnar RJ. Endogenous Opiates and Behavior: 2006. Peptides. 2007;28:2435–2513. doi: 10.1016/j.peptides.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marder E, Bucher D. Understanding Circuit Dynamics Using the Stomatogastric Nervous System of Lobsters and Crabs. Annu Rev of Physiol. 2007;69:291–316. doi: 10.1146/annurev.physiol.69.031905.161516. [DOI] [PubMed] [Google Scholar]
- 5.Cruz-Bermudez ND, Marder E. Multiple Modulators Act on the Cardiac Ganglion of the Crab Cancer borealis. J Exp Biol. 2007;210:2873–2884. doi: 10.1242/jeb.002949. [DOI] [PubMed] [Google Scholar]
- 6.McGaw IJ, McMahon BR. The FMRFamide-related Peptides F1 and F2 Alter Hemolymph Distribution and Cardiac Output in the Crab Cancer magister. Biol Bull. 1995;188:186–196. doi: 10.2307/1542084. [DOI] [PubMed] [Google Scholar]
- 7.Zudaire E, Simpson SJ, Montuenga LM. Effects of Food Nutrient Content, Insect Age and Stage in the Feeding Cycle on the FMRFamide Immunoreactivity of Diffuse Endocrine Cells in the Locust Gut. J Exp Biol. 1998;201:2971–2979. doi: 10.1242/jeb.201.21.2971. [DOI] [PubMed] [Google Scholar]
- 8.Chung JS, Webster SG. Dynamics of In Vivo Release of Molt-Inhibiting Hormone and Crustacean Hyperglycemic Hormone in the Shore Crab Carcinus maenas. Endocrinology. 2005;146:5545–5551. doi: 10.1210/en.2005-0859. [DOI] [PubMed] [Google Scholar]
- 9.Lorenzon S, Edomi P, Giulianini PG, Mettulio R, Ferrero EA. Role of Biogenic Amines and cHH in the Crustacean Hyperglycemic Stress Response. J Exp Biol. 2005;208:3341–3347. doi: 10.1242/jeb.01761. [DOI] [PubMed] [Google Scholar]
- 10.Serrano L, Blanvillain G, Soyez D, Charmantier G, Grousset E, Aujoulat F, Spanings-Pierrot C. Putative Involvement of Crustacean Hyperglycemic Hormone Isoforms in the Neuroendocrine Mediation of Osmoregulation in the Crayfish Astacus leptodactylus. J Exp Biol. 2003;206:979–988. doi: 10.1242/jeb.00178. [DOI] [PubMed] [Google Scholar]
- 11.Li L, Kelley WP, Billimoria CP, Christie AE, Pulver SR, Sweedler JV, Marder E. Mass Spectrometric Investigation of the Neuropeptide Complement and Release in the Pericardial Organs of the Crab, Cancer borealis. J Neurochem. 2003;87:642–656. doi: 10.1046/j.1471-4159.2003.02031.x. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Pulver SR, Kelley WP, Thirumalai V, Sweedler JV, Marder E. Orcokinin Peptides in Developing and Adult Crustacean Stomatogastric Nervous Systems and Pericardial Organs. J Comp Neurol. 2002;444:227–244. doi: 10.1002/cne.10139. [DOI] [PubMed] [Google Scholar]
- 13.Fu Q, Goy MF, Li L. Identification of Neuropeptides from the Decapod Crustacean Sinus Glands using Nanoscale Liquid Chromatography Tandem Mass Spectrometry. Biochem Biophys Res Commun. 2005;337:765–778. doi: 10.1016/j.bbrc.2005.09.111. [DOI] [PubMed] [Google Scholar]
- 14.Ma M, Chen R, Sousa GL, Bors EK, Kwiatkowski MA, Goiney CC, Goy MF, Christie AE, Li L. Mass Spectral Characterization of Peptide Transmitters/Hormones in the Nervous System and Neuroendocrine Organs of the American Lobster Homarus americanus. Gen Comp Endocrinol. 2008;156:395–409. doi: 10.1016/j.ygcen.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horn EC, Kerr MS. Hemolymph Protein and Copper Cocnetrations of Adult Blue Crabs (Callinectes sapidus Rathbun) Biol Bull. 1963;125:499–507. [Google Scholar]
- 16.Durliat M. Immunochemical Study of the Proteins of Various Tissues in Crustacea (Decapoda): Nature, Role, Origin. Arch Int Physiol Biochim. 1983;91:297–332. doi: 10.3109/13813458309067977. [DOI] [PubMed] [Google Scholar]
- 17.Fastner S, Predel R, Kahnt J, Schachtner J, Wegener C. A Simple Purification Protocol for the Detection of Peptide Hormones in the Hemolymph of Individual Insects by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry. Rapid Commun Mass Spectrom. 2007;21:23–28. doi: 10.1002/rcm.2800. [DOI] [PubMed] [Google Scholar]
- 18.Christie AE, Kutz-Naber KK, Stemmler EA, Klein A, Messinger DI, Goiney CC, Conterato AJ, Bruns EA, Hsu YWA, Li L, Dickinson PS. Midgut Epithelial Endocrine Cells are a Rich Source of the Neuropeptides APSGFLGMRamide (Cancer borealis Tachykinin-Related Peptide Ia) and GYRKPPFNGSIFamide (Gly1-SIFamide) in the Crabs Cancer borealis, Cancer magister and Cancer productus. J Exp Biol. 2007;210:699–714. doi: 10.1242/jeb.02696. [DOI] [PubMed] [Google Scholar]
- 19.Kwok R, Tobe SS. Hemolymph Clotting in Crustaceans: Implications for Neuropeptide Extraction from Invertebrate Hemolymph. Peptides. 2006;27:590–596. doi: 10.1016/j.peptides.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 20.Hsu JL, Huang SY, Chow NH, Chen SH. Stable-Isotope Dimethyl Labeling for Quantitative Proteomics. Anal Chem. 2003;75:6843–6852. doi: 10.1021/ac0348625. [DOI] [PubMed] [Google Scholar]
- 21.DeKeyser SS, Li L. Matrix-Assisted Laser Desorption/Ionization Fourier Transform Mass Spectrometry Quantitation via in Cell Combination. Analyst. 2006;131:281–290. doi: 10.1039/b510831d. [DOI] [PubMed] [Google Scholar]
- 22.Ji C, Li L. Quantitative Proteome Analysis Using Differential Stable Isotopic Labeling and Microbore LC-MALDI MS and MS/MS. J Proteome Res. 2005;4:734–742. doi: 10.1021/pr049784w. [DOI] [PubMed] [Google Scholar]
- 23.Christie AE, Lundquist CT, Nassel DR, Nusbaum MP. Two Novel Tachykinin-Related Peptides from the Nervous System of the Crab Cancer borealis. J Exp Biol. 1997;200:2279–2294. doi: 10.1242/jeb.200.17.2279. [DOI] [PubMed] [Google Scholar]
- 24.Fu Q, Kutz KK, Schmidt JJ, Hsu YW, Messinger DI, Cain SD, de la Iglesia HO, Christie AE, Li L. Hormone Complement of the Cancer productus Sinus Gland and Pericardial Organ: an Anatomical and Mass Spectrometric Investigation. J Comp Neurol. 2005;493:607–626. doi: 10.1002/cne.20773. [DOI] [PubMed] [Google Scholar]
- 25.Terwilliger NB, Dangott L, Ryan M. Cryptocyanin, a Crustacean Molting Protein: Evolutionary Link with Arthropod Hemocyanins and Insect Hexamerins. PNAS. 1999;96:2013–2018. doi: 10.1073/pnas.96.5.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huybrechts J, Nusbaum MP, Bosch LV, Baggerman G, De Loof A, Schoofs L. Neuropeptidomic Analysis of the Brain and Thoracic Ganglion from the Jonah Crab Cancer borealis. Biochem Biophys Res Commun. 2003;308:535–544. doi: 10.1016/s0006-291x(03)01426-8. [DOI] [PubMed] [Google Scholar]
- 27.Behrens H, Chen R, Li L. Combining Microdialysis, NanoLC-MS, and MALDI-TOF/TOF to Monitor Neuropeptide Secretion in the Crab Cancer borealis. Anal Chem. 2008;80:6949–6958. doi: 10.1021/ac800798h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.