Abstract
Frontotemporal dementia (FTD) is a clinical term encompassing dementia characterized by the presence of two major phenotypes: 1) behavioral and personality disorder, and 2) language disorder, which includes primary progressive aphasia and semantic dementia. Recently, the gene for familial frontotemporal lobar degeneration (FTLD) with ubiquitin-positive, tau-negative inclusions (FTLD-U) linked to chromosome 17 was cloned. In the present study, 62 unrelated patients from the Washington University Alzheimer's Disease Research Center and the Midwest Consortium for FTD with clinically diagnosed FTD and/or neuropathologically characterized cases of FTLD-U with or without motor neuron disease (MND) were screened for mutations in the progranulin gene (GRN; also PGRN). We discovered two pathogenic mutations in four families: 1) a single-base substitution within the 3′ splice acceptor site of intron 6/exon 7 (g.5913A>G [IVS6–2A>G]) causing skipping of exon 7 and premature termination of the coding sequence (PTC); and 2) a missense mutation in exon 1 (g.4068C>A) introducing a charged amino acid in the hydrophobic core of the signal peptide at residue 9 (p.A9D). Functional analysis in mutation carriers for the splice acceptor site mutation revealed a 50% decrease in GRN mRNA and protein levels, supporting haploinsufficiency. In contrast, there was no significant difference in the total GRN mRNA between cases and controls carrying the p.A9D mutation. Further, subcellular fractionation and confocal microscopy indicate that although the mutant protein is expressed, it is not secreted, and appears to be trapped within an intracellular compartment, possibly resulting in a functional haploinsufficiency.
Keywords: Frontotemporal dementia, FTD, granulin, progranulin, GRN, PGRN
INTRODUCTION
Patients with frontotemporal dementia (FTD) present with one of two types of symptoms: gradual and progressive changes in behavior and personality, or gradual and progressive language dysfunction [Neary et al., 1998; McKhann et al., 2001]. The behavioral form is the most common presentation of FTD, in which there is an early change in social and personal conduct, characterized by difficulty in modulating behavior to the social demands of a situation. The alternative presentation of FTD is early and progressive change in language function. This occurs in the setting of relative preservation of other cognitive domains, such as memory. FTD may also be present with motor neuron disease (FTD-MND). FTD is the second most common form of dementia (12–20%), after Alzheimer's disease (AD), in patients below 65 years of age [Neary et al., 1998]. FTD accounts for about 10% of all dementias with up to 50% of cases demonstrating a positive family history, in which the disease segregates as an autosomal dominant trait.
Gross examination of the frontotemporal lobar degeneration (FTLD) brain typically reveals atrophy of the frontal and temporal lobes. Microscopically, there are stereotypical features: neuronal loss, astrogliosis, and superficial microvacuolation in affected areas. Clinicopathological consensus criteria classify the FTLDs into two main groups: tauopathies (diseases with abnormal cellular aggregates of hyperphosphorylated tau protein) and ubiquitin-positive, tau-negative diseases (disease entities with pathological ubiquitinated inclusions containing TDP-43); and a group of rare diseases including basophilic inclusion body disease, neuronal intermediate filament inclusions disease, and an increasingly rare subset with no detectable inclusion, generically called FTLD and historically referred to as dementia lacking distinctive histopathology [McKhann et al., 2001; Cairns et al., 2007b].
FTD with parkinsonism linked to chromosome 17 (FTDP-17) represents a genetically heterogeneous group of disorders [Hutton et al., 1998]. Many families in this group carry mutations in the gene encoding microtubule-associated protein tau (MAPT) and neuropathological examination reveals neuronal and glial tau-positive inclusions affected areas. Recently, it was shown that several large kindreds linked to chromosome 17 but lacking tau mutations and tau-immunoreactive inclusions represented a subtype of FTLD characterized by neuronal ubiquitin-positive, tau-negative inclusions (FTLD-U) [Mackenzie et al., 2006; Mukherjee et al., 2006; Rosso et al., 2001; Behrens et al., 2007]. Null mutations in the progranulin gene (GRN; MIM# 138945) were reported to cause disease in these families [Baker et al., 2006; Cruts et al., 2006]. A majority of the mutations identified so far cause premature termination of the coding sequence (PTC) leading to degradation of mutant GRN mRNAs by nonsense-mediated decay (NMD) [Gass et al., 2006]. Functional loss of 50% of the GRN protein in these cases causes neurodegeneration as a result of haploinsufficiency [Baker et al., 2006; Cruts et al., 2006]. Interestingly, the DNA binding protein TDP-43 has been shown to be a component of the ubiquitinated inclusions of sporadic and familial FTLD-U and sporadic cases of MND [Neumann et al., 2006, 2007; Cairns et al., 2007a].
GRN belongs to a family of cysteine-rich polypeptide growth factors, and is constitutively expressed in several tissues [Bateman et al., 1990]. The gene encoding GRN is located 1.7 Mb upstream of the MAPT gene. The gene has 12 coding exons encoding a 68.5-kDa precursor glycoprotein, which can be cleaved into seven different granulins (∼6 kDa) [He and Bateman, 2003]. GRN is a secreted protein and brings about its function through cell–cell signaling and signal transduction [Bateman et al., 1990]. GRN is implicated in cell proliferation, wound repair, and as a growth factor [He and Bateman, 1999]. In brain, GRN is expressed in Purkinje and pyramidal cells of the hippocampus and cerebral cortical neurons; however, its role in the central nervous system is poorly understood [Daniel et al., 2000].
In this report, we present the results of our mutation screening of GRN in a set of 62 FTLD patients from the Washington University Alzheimer's Disease Research Center (WUADRC) and the Midwest Consortium for Frontotemporal Dementia (MCFTD), including familial and sporadic FTLD and pathologically confirmed FTLD-U, to estimate the prevalence of GRN mutations in this cohort and establish the molecular mechanism of the mutations identified in this study.
MATERIALS AND METHODS
Subjects
Since 1992, we have ascertained 35 FTD cases through the WUADRC. Additionally, 27 FTLD subjects were obtained from the MCFTD. Table 1 summarizes the clinical characteristics (where available) of the samples used in this study. All procedures were approved by the Washington University School of Medicine Institutional Review Board. Family members older than 18 years of age were included after informed consent was obtained. All individuals were interviewed by an experienced research clinician or nurse, using a modified version of the Family History Interview developed by the Consortium to Establish a Registry for Alzheimer's Disease [Silverman et al., 1994]. For individuals with neurological symptoms, the visit included a clinical assessment, a neurological examination, and a consensus history derived from the individual and/or nearest relatives. The neuropathological diagnoses were determined using the criteria of McKhann et al. [2001] and Cairns et al. [2007b]. When possible, available medical records were also examined. Families HDDD2, FD1, and HDDD1 are three large multiply-affected multigeneration kindreds in this cohort.
TABLE 1.
FTD Subjects Used in This Report*
| WUADRC |
MCFTD |
Total |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Diagnosis | No. of probands | AAO [years] mean (SD) | DI [years] mean (SD) | No. of probands | AAO [years] mean (SD) | DI [years] mean (SD) | No. of probands | AAO [years] mean (SD) | DI [years] mean (SD) |
| Samples with clinical and neuropathological diagnosis: | |||||||||
| FTLD-U | 8 | 56.4 (11.2) | 10.2 (4.5) | 19 | 56.7 (8.3) | 5.7 (3.2) | 27 | 56.6 (8.9) | 7.0 (4.1) |
| FTLD+MND | 1 | 43 | 8 | 8 | 56.7 (7.3) | 3.1 (1.0) | 9 | 54.8 (8.3) | 3.7 (1.9) |
| Samples with clinical diagnosis only: | |||||||||
| FTD | 26 | 64.3 (10.5) | 8.7 (4.1) | 0 | – | – | 26 | 64.3 (10.5) | 8.7 (4.1) |
| Total for all samples | 35 | 62.0 (11.3) | 9.0 (4.1) | 27 | 56.7 (7.8) | 5.0 (3.0) | 62 | 59.7 (10.2) | 7.3 (4.2) |
The patients are stratified according to their diagnosis.
DI, duration of illness.
Family HDDD2
This is an autosomal dominant FTLD-U kindred with characteristic ubiquitin-positive, tau-negative neuronal cytoplasmic and intranuclear inclusions. A detailed clinicopathological description of this family may be found elsewhere [Lendon et al., 1998]. Recently, we reported a pathogenic missense mutation (p.A9D) in the signal peptide of GRN segregating with FTD in this family [Mukherjee et al., 2006].
Family FD1
This is a three generation FTLD-U kindred (Supplementary Fig. S1A; available online at http://www.interscience.wiley.com/jpages/1059-7794/suppmat). Clinical symptoms include disinhibition, progressive dysphasia, and mild memory problems. The mean age at onset (AAO) is 57 years (range 51–68 years). In affected individuals who came to autopsy, there was atrophy of the frontal and temporal lobes. Histology revealed neuronal loss, microvacuolation, and reactive gliosis. There were three sites of ubiquitin-positive and TDP-43-positive aggregates: neuronal cytoplasmic inclusions (NCIs), dystrophic neurites (DNs), and neuronal intranuclear inclusions (NIIs). There were additional coexisting diffuse β-amyloid plaques (Braak amyloid stage = A), but no neuritic plaques or neurofibrillary tangles (Braak neurofibrillary tangle stage = 0).
Family HDDD1
These subjects belong to a three-generation FTLD-U family (Supplementary Fig. S1B). The mean AAO is 62 years (range 56–65 years) with a mean duration of 9.5 years. A detailed clinicopathological description of this kindred is available elsewhere [Morris et al., 1984; Behrens et al., 2007]. Briefly, disease in affected individuals is characterized by progressive language deterioration, mild memory problems, and disinhibition. In cases that came to autopsy, microscopy revealed marked frontal lobe atrophy with dilatation of the lateral ventricles. Histology showed pronounced neuronal loss, status spongiosus, and gliosis in affected areas. Ubiquitin-positive and TDP-43-positive NCIs, DNs, and NIIs were present, and identical to those seen in Families FD1 and HDDD2.
Of the 62 FTD cases studied in this report, 43.6% were neuropathologically confirmed FTLD-U, 14.5% were clinically diagnosed as FTD-MND, and 41.9% were clinically diagnosed as FTD. Table 1 summarizes the clinical profile of subjects used in this study. The mean AAO in the familial FTD group was 60.3±8.6 years with a mean duration of illness of 7±3.7 years. In the sporadic group, the mean AAO was 57.7±11.3 years with a mean duration of illness of 7.5±5.1 years.
Control Samples
A total of 96 Caucasian and 48 African American unrelated controls from the St. Louis metropolitan area were screened for each of the putative pathogenic sequence variants observed in FTD cases in this study. The samples were elderly nondemented controls collected by the WUADRC. The dementia screen in these controls has been described elsewhere [Grupe et al., 2006].
Direct DNA Sequencing
Genomic DNA from affected and unaffected subjects was extracted from peripheral blood and/or brain tissue using standard protocols. All coding exons, flanking intronic sequences, and 5′ & 3′ untranslated exons of GRN were amplified using specific primers and sequenced in both directions according to the manufacturer's instructions (BigDye terminator v3.1 and 1.1 chemistry; Applied Biosystems, Foster City, CA). Reactions were run on an ABI 3100 (Applied Biosystems) and mutation analysis was performed using Sequencher software v4.6 (Gene Codes Corporation, Ann Arbor, MI). Positive calls for mutations were made only if the variant was observed in both reads and absent in a set of 90 controls. Additionally, wherever possible, genetic variants were confirmed using segregation analysis (Families HDDD2, FD1, and HDDD1). All mutation numbering is based on the genomic DNA sequence (EnsEMBL gene ID: ENSG00000030582 [NCBI36:17:39778017:39785996:1]).
Founder Effect Analysis
To determine whether kindred from Families FD1 and HDDD1 might share a common genetic founder, haplotype analysis was performed using microsatellite and SNP markers within and immediately flanking GRN: D17S951 (microsatellite marker), g.5913A>G, g.6048G>A, and D17S934 (microsatellite marker). For microsatellite markers, fluorescently tagged primers were used to amplify the genomic DNA and then fragments were separated using a capillary-based system (ABI 3100). Allele calling was performed using the Genotyper program (Applied Biosystems). SNP genotypes were obtained directly from the DNA sequencing. These SNPs and microsatellite markers were also typed in 96 nondemented Caucasian controls to determine the population frequency of the haplotype background on which the disease mutation occurred.
Analysis of GRN mRNA
To evaluate the stability of GRN mRNA in vivo, lymphoblastoid cells were treated with 100 μg/ml actinomycin D, after which total RNA was isolated at different time intervals. The stability of mRNA was assayed by performing PCR amplification of cDNA using GRN-specific primers.
Total RNA was isolated from brain tissue (frontal cortex) and/or lymphoblastoid cells as per manufacturer's instructions (Rneasy™ total RNA Kit; Qiagen Inc., Valencia, CA). Contaminating DNA was removed from the RNA preparation by digestion with RNase-free deoxyribonuclease (DNase). cDNA was synthesized using Superscript III RT (Invitrogen Life Technologies, Carlsbad, CA) following the manufacturer's protocol. To confirm that the cDNA mixture was free of DNA contamination, a PCR was performed using primer pairs located within the intronic sequence of GRN. No amplification product was observed. Aliquots of cDNA mixture were then subjected to PCR amplification in 50-μl reactions. The amplified cDNA spanned 1,785 bp, covering the entire coding sequence of GRN from start to stop codon.
Transcript Analysis in Individuals Carrying the GRN Splice Acceptor Site Mutation
PCR amplification was performed on cDNA samples obtained from frontal cortex of affected individuals from Family FD1, with primers in exons 6 and 9 (Ex6_FP: GAGGACTAACAGGG CAGTGG); (Ex9_RP:GCCTCTGGGATTGGACAG). RT-PCR products were resolved on a 2% agarose gel to verify the product size. To confirm deletion of exon7 in the GRN cDNA from affected individuals we designed a primer derived from sequences in both exon 6 and exon 8 (in the reverse direction). This primer together with a forward primer in exon 6 will only generate a PCR product if exon 7 has been deleted from the cDNA as predicted by the location of the mutation (Ex6RP: GTCACATTTCACATCCCC CAGTT);(Ex6_FP:GAGGACTAACAGGGCAGTGG). The PCR product obtained in mutation carriers was sequenced to confirm the nature of the PCR product. As expected, no PCR product was generated from individuals with the wild-type GRN sequence.
Analysis of GRN Protein in Lymphoblastoid Cell Lines
Lymphoblastoid cell lines derived from affected and unaffected family members from Families HDDD1, FD1, and HDDD2 were grown in RPMI growth media supplemented with 10% fetalcalf serum (FCS). An equal number of cells were collected and lysed in coimmunoprecipitation (co-IP) lysis buffer (50 mM Tris pH 7.6, 150 mM NaCl, 2 mM EDTA, 1% NP40, 0.5% Triton X100, and a cocktail of protease inhibitors). Total protein was quantified using the bovine serum albumin (BSA) assay (Bio-Rad, Hercules, CA) and an equal amount of total protein (30 μg) was loaded onto a polyacrylamide gel. The amount of media loaded was also adjusted according to protein concentration in the lysates. The samples were run on a 4 to 20% gradient PAGE gel and transferred to a polyvinylidene fluoride (PVDF) membrane. Progranulin was detected by western blotting with a polyclonal anti-GRN antibody (1 in 500 dilution; R&D Systems, Inc., Minneapolis, MN). Themembrane was reblotted with a polyclonal antiactin antibody (1 in 1000 dilution; Sigma, St. Louis, MO) to detect actin as an internal control.
Transfection
The full-length GRN cDNA in pCMV-SPORT6 vector was purchased from Invitrogen. The QuickChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to introduce the p.A9D point mutation into the GRN construct. HEK cells were transiently transfected with wild-type (WT) or p.A9D GRN constructs using the FuGENE 6 transfection reagent (Roche Molecular Biochemicals, Indianapolis, IN). At 24 to 48 hr after transfection, the media was collected and cells were lysed in co-IP lysis buffer. The pellets were washed once with 1 ml co-IP lysis buffer and resuspended in Glycosylation Denaturing Buffer (5% SDS, 0.4 M dithiothreitol [DTT]; New England BioLabs, Ipswich, MA). Deglycosylation was performed on the various fractions (cell lysates, media, and pellets) using PNGase F according to the manufacturer's instructions (New England BioLabs). Both treated and untreated samples from various fractions were resolved on 4 to 20% PAGE as described for the lymphoblastoid cells.
Analysis of GRN Protein in Brain and Cerebrospinal Fluid Samples
Autopsies were performed according to established Alzheimer's Disease Research Center (ADRC) Neuropathology Core protocols [Berg et al., 1998]. Frozen tissue from the frontal lobe was dissected, and the gray and white matter was separated macroscopically and processed individually. Brain tissue was homogenized in 2 ml/g high salt buffer (50 mm Tris pH 7.5, 2 mm EDTA, 0.75 m NaCl, and a cocktail of protease inhibitors), centrifuged at 50,000 rpm at 4°C for 40min, and the supernatant was saved as the high salt fraction. Cerebrospinal fluid (CSF) samples were obtained either by lumbar puncture (LP) or at postmortem (PM) and total protein concentration was determined through Bradford Analysis (BioRad). A total of 20 μg of total CSF protein from each sample were resolved by 10% SDS-PAGE and transferred to PVDF membranes. Membranes were blocked with TBS containing 0.1% Tween-20 and 5% powdered milk, probed with a polyclonal antibody to GRN at a 1:1000 dilution (R&D Systems, Inc.), detected with chemiluminescent reagents (Amersham Biosciences, England, United Kingdom), and exposed to blue X-ray film (Phenix Research).
Immunohistochemistry
Human neuroblastoma cell line, SH-SY5Y (ATCC: CRL-226) was plated at approximately 50% confluency on cover slips in IDEM 10% FCS media supplemented with 100 nM retinoic acid. After cells adhered, 1 μg pCMV-SPORT6 GRN cDNA or pCMV-SPORT6 p.A9D GRN cDNA were transfected following the manufacturers’ protocol (Lipofectomine 2000; Invitrogen Life Technology, Carlsbad, CA). Approximately 12–14 hr posttransfection, cells were washed three times in PBS pH 7.4 and fixed in a 4% paraformaldehyde PBS solution for 20 min. After fixing, cells were washed three times in PBS and blocked for 30 min at room temperature in PBS containing 3% BSA and 0.3% Triton X100. Primary polyclonal anti-GRN antibody (R&D Systems, Inc.) at 1:500 dilution, GRN (Zymed, Inc., San Francisco, CA) at 1:500 dilution or Golgin-97 (Invitrogen) at a 1:200 dilution were incubated on the cells at room temperature for 1 hr. Cells were washed three times in PBS containing 0.3% Triton X100 and incubated with Alexa Fluor (Invitrogen) secondary antibody. Cells were washed as described above, and mounted on slides for confocal imaging. Images were acquired using a Zeiss LSM510 Meta laser scanning confocal microscope (Carl Zeiss Inc., Thornwood, NY) equipped with argon (excitation wavelength 488 nm), HeNe1 (excitation wavelength 543 nm), and HeNe2 (excitation wavelength 633 nm) lasers. A 63 ×, 1.4 numerical aperture Zeiss Plan apochromat oil objective was used. Confocal Z slices of 0.5 μm were obtained using the Zeiss LSM510 software. Experiments were done in triplicate with four fields of view examined with ∼20–35 cells observed per field.
RESULTS
Prevalence of GRN Mutations
DNA sequencing of the coding exons and regulatory regions of GRN in 62 FTD cases resulted in the identification of 10 sequence variants (see Table 2 and Supplementary Table S1). All of the variants identified were single basepair substitutions except for one four-basepair insertion in intron 3. A total of two variants were clearly pathogenic: a single-base substitution within the 3′ splice acceptor site of intron 6 g.5913A>G (IVS6–2A>G) (Fig. 1A); and a missense mutation in exon 1 (g.4068C>A) resulting in an amino acid substitution at residue 9 (p.A9D) [Mukherjee et al., 2006]. Each of these mutations segregated with disease in the families in which they were observed and were absent from 90 Caucasian controls. A missense mutation in exon 10 (g.7011G>A), resulting in a nonsynonymous change at residue 433 (p.R433Q) was also observed in a single African American proband with FTD (Table 2). In addition to screening 90 Caucasian controls, 45 nondemented elderly African American individuals were screened and lacked this variant. However, no family history of FTD was reported for this individual and no other DNA samples were available for segregation analysis, This variant may be a rare polymorphism rather than a pathogenic mutation because another variant at the same codon (p.R433W) has been reported in a neurologically normal individual. Putative pathogenic mutations were detected in five patients, explaining ∼8% of disease predisposition in this cohort (6.4% of disease if the p.R433Q mutation is not pathogenic). The prevalence of GRN mutations within the FTLD-U subset was 14.8% (4/27). The mean AAO for patients with GRN mutations in this dataset was 61.6 (±7.5) years. There was no significant difference in the mean AAO between affected individuals carrying a GRN mutation compared to patients not carrying a GRN mutation; 61.6 vs. 59.8 years. In addition, six sequence variants were identified, which were present in both cases and controls (Supplementary Table S1).
TABLE 2.
Families With Pathogenic Mutations in GRN*
| Subject or family | Pathology | AAO [years] mean (range) (no. of samples or individuals) | Mean duration of illness (years) | Fam Hx | Mutation | Genome | Protein | Location in gene |
|---|---|---|---|---|---|---|---|---|
| HDDD2 | FTLD-U | 62.8 (50–74) (n = 18) | 7.4 | Yes | Missense | g.4068C>A | p.A9D | Ex 1 |
| FD1 | FTLD-U | 57 (50–68) (n = 7) | 11.3 | Yes | Splice site | g.5913A>G (IVS6–2A>G) | p.A237WfsX4 | IVS 6 |
| HDDD1 | FTLD-U | 62 (56–65) (n = 2) | 9.5 | Yes | Splice site | g.5913A>G (IVS6–2A>G) | p.A237WfsX4 | IVS 6 |
| 474 | FTLD-U | 53 (n = 1) | 8 | Yes | Splice site | g.5913A>G (IVS6–2A>G) | p.A237WfsX4 | IVS 6 |
| WUSP1 | FTLD-U | 73 (n = 1) | > 13 | No | Missense | g.7011G>A | p.R433Q | Ex 10 |
All mutation numbering is based on the genomic DNA sequence. EnsEMBL gene ID: ENSG00000030582 (NCBI36:17:39778017:39785996:1).
Fam Hx, family medical history.
FIGURE 1.
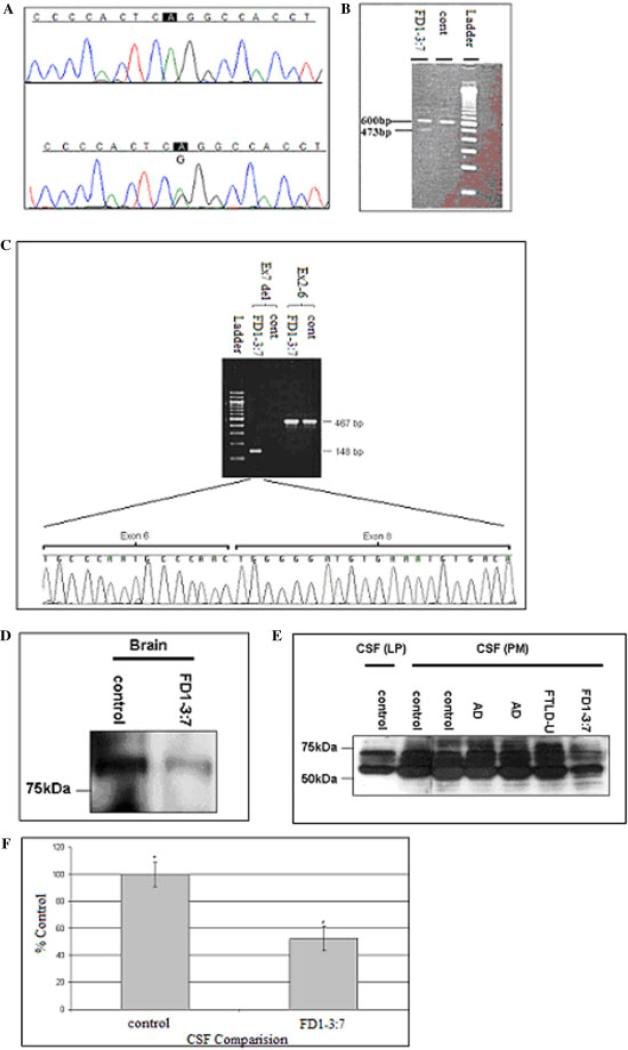
Analysis of the GRN splice site mutation. A: Partial sequence data of the GRN gene in a patient with FTLD-U from Family FD1, harboring a heterozygous splice site mutation (A/G) at IVS6-2 site (lower panel) compared to the normal individual (upper panel). B: Agarose gel electrophoresis of GRN cDNA from an affected individual from Family FD1 (lane 1) showing the mutant allele (473 bp) and the wild-type allele (600 bp). C: GRN cDNA amplified from a case and a control from Family FD1 using two sets of primers (ex6-9 and ex2-6). Sequencing chromatogram showing that exon 7 is missing from the GRN transcript amplified using the ex6-9 primer set from the affected individual from Family FD1. D: Progranulin expression in brain. High salt fraction (each lane contains 40 μg of total protein). E: Progranulin level is lower in CSF of affected individual. Each lane of CSF samples contains 20 μg of total protein. F: Comparison of secreted Progranulin in CSF. Lane 1, control (LP); lanes 2 and 3, controls (PM); lanes 4 and 5, AD); lane 6, FTLD-U sporadic; and lane 7, individual FD1-3:7 Densitometric analysis of samples comparing upper bands (∼70 kDa) set at 100% of secreted progranulin compared to individual FD1-3:7. Compared through repeated measures one-way analysis of variance (ANOVA), Bonferroni post hoc test comparing all samples (n = 6, *P < 0.05). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Evidence for a Common Founder for Families FD1 and HDDD1
Segregation analysis in kindred from Families FD1 and HDDD1 showed that the splice acceptor site mutation (g.5913–2A>G; IVS6–2A>G) segregates with disease (Supplementary Fig. S1). We also identified a nonpathogenic mutation at the +8 position of the 5′ splice donor site of intron 7 (IVS7+8G>A), which is present on the disease haplotype of both Family FD1 and Family HDDD1 kindreds. The splice acceptor site mutation (g.5913–2A>G; IVS6–2A>G) was identified in another sample (sample 474) (Table 2) lacking the IVS7+8G>A mutation. Segregation analysis was not possible in this proband due to the unavailability of DNA from other family members, However, this result demonstrates that the disease-causing mutation (g.5913–2A>G; IVS6–2A>G) has occurred independently on at least two occasions and that Families FD1 and HDDD1 may be related.
To determine whether the kindred from Families FD1 and HDDD1 share a common founder, haplotype analysis was performed using two microsatellite markers (D17S951 and D17S934) flanking GRN as well as two SNPs found within the gene: IVS6–2A>Gand IVS7+8G>A. Haplotype analysis showed an unambiguous disease haplotype between D17S951 and D17S934, shared by the affected members of both kindreds (Supplementary Fig. S1C). The disease haplotype was observed in all affected individuals and did not show a difference between the various subphenotypes (Supplementary Fig. S1C). The frequency of the haplotype on which the disease mutation occurred was computed to be 3% by typing the markers in a set of 93 matched controls.
Molecular Analysis of the Intron 6 Splice Acceptor Site Mutation
Brain tissue from the frontal cortices of two individuals (3:7 and 3:22) from Family FD1 kindred was used to isolate total RNA and analyze the GRN transcript. Amplification of GRN cDNA using forward and reverse primers in exons 6 and 9, respectively, generated a cDNA fragment of 473 bp in addition to the wild-type transcript fragment of 600 bp (Fig. 1B). The size of the smaller fragment was consistent with that predicted if the mutation resulted in skipping of exon 7. To confirm the nature of this fragment, we designed another PCR assay that would only produce a product if exon 6 were spliced directly to exon 8. PCR amplification resulted in a 148-bp product in the affected individual, which was confirmed to contain exon 6 spliced to exon 8 by sequencing (Fig. 1C). Skipping of exon 7 results in premature termination of the transcript because of a novel stop codon in exon 8 when spliced to exon 6. This mutant transcript is likely to be degraded by NMD, as has been reported for other similar mutations in GRN, leading to a 50% loss in GRN protein. Western blot analysis confirmed that this mutation causes a significant reduction in GRN protein compared to control samples (Fig. 1D). Since GRN is a secreted protein, we postulated that GRN would be present in human CSF and that the mutation carriers in Family FD1 would have 50% less GRN. GRN was easily detected in CSF obtained either at postmortem or by LP, indicating that GRN is highly abundant in CSF. The major fragments detectable by western blot were smaller than those observed in cell lysates. Comparison of the CSF from a member of Family FD1 clearly shows the same fragments of GRN but at much lower levels than the two normal individuals (Fig. 1E and F).
The Molecular Mechanism of the p.A9D Mutation
In silico experiments (Signal IP Software; http://www.cbs.dtu.dk/services/SignalP/) show that the p.A9D mutation reduces the signal peptide probability score from a normal value of 0.95 to 0.79, suggesting that this mutation would significantly impair the functioning of the signal peptide (Fig. 2A).
FIGURE 2.
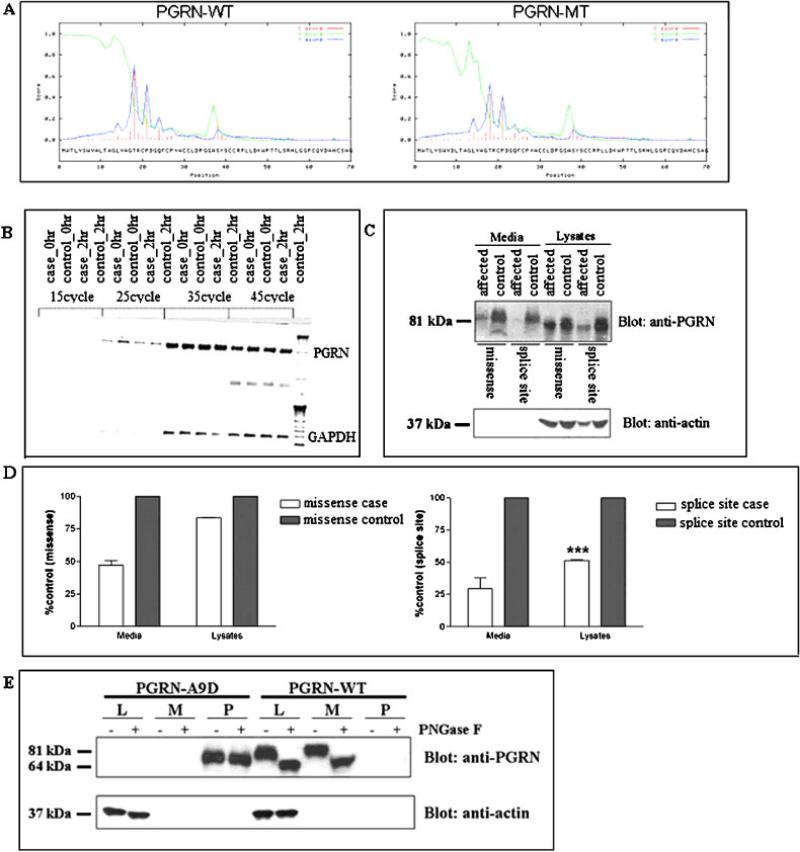
Analysis of the p.A9D mutation. A: Output of the Signal IP program, demonstrating a significant decrease in the prediction score for the signal peptide between the wild-type GRN and mutant GRN. B: Agarose gel electrophoresis of GRN product of RT-PCR showing the stability of GRN mRNA. GRN cDNA was amplified using gene-specific primers using a range of cycles. There was no significant difference between the case and control band intensity. C: Western blot analysis of the conditioned media and cell lysates from lymphoblastoid cell lines using anti-GRN polyclonal antibody against full-length GRN (R&D Systems). A representative of three independent experiments is shown. D: Semiquantitative densitometric analysis. The densities of the GRN bands were quantified using Quantity One Software (Bio-Rad) and normalized to actin levels. The levels of GRN protein in cases from Family HDDD2 (left panel) and Family FD1 (right panel) are presented as a percentage of their respective familial controls. GRN levels in the lysates were significantly higher in cases with the missense mutation compared to cases with the splice site mutation (***P < 0.001), whereas GRN levels in the media were similar (P = 0.203), as determined by a two-tailed Student's t-test. E: The p.A9D mutant protein is located in detergent-insoluble nuclear pellets in an overexpression system. HEK cells were transfected with WT or p.A9D constructs. Three fractions: lysates (L), media (M), and detergent-insoluble pellets (P) were taken, half of each sample was subjected to PNGase treatment and then all fractions were separated by SDS-PAGE; GRN was detected by western blotting as described in Materials and Methods. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
To investigate whether the p.A9D mutation affects GRN mRNA or protein expression, we established lymphoblastoid cell lines from affected and unaffected individuals from the kindred of Family HDDD2. We initially investigated whether the p.A9D mutation affected GRN mRNA levels. To evaluate the stability of GRN mRNA in vivo, lymphoblastoid cells were treated with 100 μg/ml actinomycin D, after which total RNA was isolated at different time intervals. The stability of mRNA was assayed by performing PCR amplification of cDNA obtained by reverse transcribing total RNA isolated from lymphoblastoid cell lines treated with actinomycin D. Semiquantitative analysis of total cDNA did not show any difference in the amount of GRN mRNA in cases when compared to controls from Family HDDD2 (Fig. 2B). This result demonstrates that the p.A9D mutation does not result in NMD, suggesting that it must affect GRN function through a different mechanism.
To investigate whether the p.A9D mutation affects GRN at the protein level we assessed steady-state levels of GRN protein in the lymphoblastoid lines. Both cell lysates and cell growth media were analyzed (Fig. 2C). Semiquantitative analysis revealed that the steady-state levels of GRN in lysates from individuals carrying the intron 6 splice acceptor site mutation was about 50% of the levels observed in a control individual (Fig. 2D, right panel), confirming that this mutation results in a 50% loss of GRN protein. In contrast, the steady-state level of GRN in the lysates from Family HDDD2 cases was about 83% compared to the controls (Fig. 2D, left panel). However, secreted GRN protein assayed using the media fraction showed that p.A9D mutation carriers had GRN protein levels that were 50% of those seen in the control cell line (Fig. 2D, left panel). While we cannot distinguish between mutant and wild-type GRN, the decreased proportion of secreted GRN in affected individuals from Family HDDD2 suggests that the mutant protein is not secreted. We also observed that the secreted GRN protein runs slightly higher than the majority of the GRN protein in the lysates (Fig. 2C). We postulate that the secreted protein is fully glycosylated, while the lysates contain a mixture of species undergoing glycosylation, with some being fully glycosylated and others being only partially glycosylated.
To unambiguously detect the mutant protein, we introduced the p.A9D mutation into the GRN cDNA and overexpressed mutant and wild-type constructs in HEK cells. While the wild-type protein was highly expressed and detected in both the lysates and culture media, the mutant was only detectable in the detergent-insoluble pellet (Fig. 2E), a fraction containing no detectable wild-type protein. Deglycosylation experiments indicated that the mutant protein was not glycosylated (Fig. 2E).
Subcellular Localization of GRN
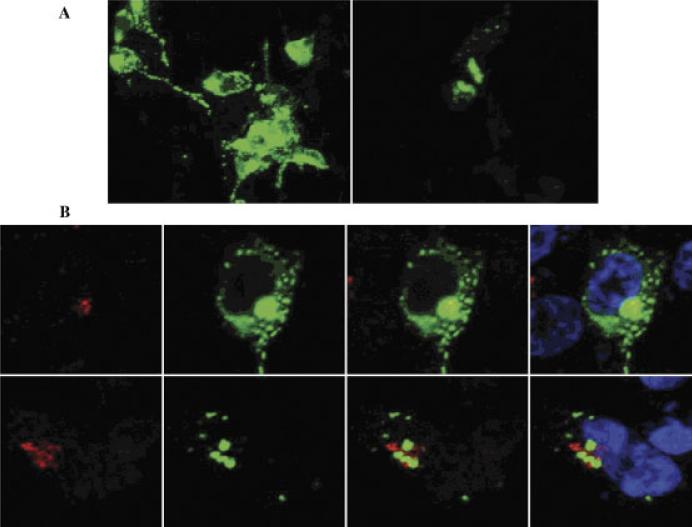
Western blot analyses suggest that the mutant p.A9D protein is not being trafficked through the secretory pathway, resulting in the mutant protein being mislocalized within a membranous compartment within the cell. To investigate this more fully, we performed immunocytochemistry in differentiated neuroblastoma cells (SHSY-5Y) transfected with either mutant or wild-type GRN cDNA. Confocal microscopy of living cells revealed that over-expressed wild-type GRN is distributed throughout the secretory pathway (Fig. 3A). However, overexpressed p.A9D GRN localizes within the Golgi network and cytosol (Fig. 3B). This defect in trafficking, leading to an absence of secreted mutant protein, most likely leads to a functional haploinsufficiency in mutation carriers.
FIGURE 3.
Subcellular localization of GRN missense mutation (p.A9D). Progranulin p.A9D accumulates within the Golgi network and cytosol of transfected SHSY-5Ycells. A: Compressed Z-stack displays apical to basal distribution of p.A9D and wild-type progranulin in SHSY-5Y cells. The p.A9D progranulin appears to partially colocalize within the trans-Golgi network and cytosol. B: Mutant progranulin accumulates within the Golgi network and cytosol. Upper row (wild-type), lower row (p.A9D); trans-Golgi marker, Golgin-97 (red) antiprogranulin (green), andTo-PRO3 nuclear stain (blue).
DISCUSSION
Recent findings have demonstrated that GRN mutations cause FTLD-U linked to 17q21. Based on brain and lymphoblastoid GRN cDNA analysis, most of the mutations described so far are predicted to cause functional loss of GRN through NMD [Baker et al., 2006; Cruts et al., 2006; Masellis et al., 2006].
In this study, we report mutation screening of GRN in a series of clinical and neuropathologically well-characterized cases of FTD identified by WUADRC and MCFTD. To establish the prevalence and specificity of GRN mutations, we screened 62 patients (26 had a clinical diagnosis of FTD and of those where neuropathology was performed, 27 had FTLD-U and nine had FTLD-MND) (Table 1). Most of the putative pathogenic variants were identified in familial FTLD-U samples (4/33 vs. 1/30). More than one-half of the patients (66.7%) presented with a language deficit, and 26.1% of these subjects carried a mutation in GRN. One patient, carrying the R433Q mutation, was an exception, with no reported family history of FTD or language dysfunction. The evidence that this mutation is pathogenic is weaker than the evidence for the other mutations. Additionally, none of the patients within the FTLD-MND subgroup harbored a genetic variant in GRN in this screen. This observation is similar to an earlier report [Gass et al., 2006].
The splice acceptor site mutation (IVS6–2A>G) was found in three cases. Haplotype analysis using two intragenic SNPs and two microsatellite markers flanking GRN suggests that kindred from Families FD1 and HDDD1 may share a common founder. Study of GRN mRNA revealed two alleles in mutation carriers of kindred from Family FD1, representing the wild-type and the mutant RNA. The size of the mutant mRNA fragment corresponds with the expected size of a transcript in which exon 7 is excluded. Deletion of the 127-bp exon 7 is predicted to cause a change in the reading frame, resulting in an aberrant transcript and premature termination of the protein. This was further supported by western blot analysis, which showed a significant reduction in the amount of GRN protein in cases compared to controls.
In this study we show that the missense mutation in Family HDDD2, which replaces Ala with Asp (p.A9D) within the hydrophobic core of the signal peptide, is expressed. Both the western blotting of cellular extracts and the confocal microscopy of transfected cells indicate that mutant protein is located in an abnormal cellular compartment. This result is in contrast to an earlier study that reported loss of GRN function in individuals carrying the p.A9D mutation possibly through the degradation of mutant mRNA [Gass et al., 2006]. We have performed a detailed analysis of the mutation at the DNA, RNA, and protein levels using samples from both brain and lymphoblastoid cell lines from affected and normal individuals as well as expression of cDNA constructs. Our data shows that the p.A9D protein is synthesized but gets trapped partially within the Golgi apparatus and accumulates within the cytosol. Since GRN is normally a secreted protein, trapping of the mutant GRN protein in the trans Golgi apparatus and within the cytosol most likely explains the disease phenotype since this results in a functional haploinsufficiency.
Our mutation screening and functional analysis support the original hypothesis that GRN mutations causing FTLD-U result in haploinsufficiency. However, as our functional data demonstrate, this haploinsufficiency can result either from a 50% reduction in GRN mRNA or from a similar decrease in secreted GRN protein. The loss of one copy of GRN, as a pathogenic mechanism appears to be in contrast to all other identified neurodegenerative disease mutations, wherein the disease onset is due to accumulation of a toxic protein. However, several recent studies have demonstrated that the ubiquitin positive inclusions in FTLD-U contain TDP-43. It may therefore be that loss of GRN expression leads to an accumulation of aggregated TDP-43, which may be toxic to the cell and lead to neurodegeneration. Although the exact role of GRN in the central nervous system is not completely understood, increasing the level of expression of GRN in these patients, possibly by stabilizing the wild-type protein may delay onset or prevent disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank the participants for their collaboration in the project. We also acknowledge the support of the Clinical, Psychometrics, and the Genetics Cores of the WUADRC, and the expert technical assistance of Deborah Carter, Katherine Paulsmeyer, and Jeffrey Strider of the WUADRC Neuropathology Core and the Betty Martz Laboratory for Neurodegenerative Research. This work was supported by grants from the National Institute on Aging (NIA) (AG05681 and AG03991 [to J.C.M.], AG12300 [to K.J.H. and C.L.W.], AG13854 [to E.B.], AG16976 [to E.B., N.C., K.J.H., and C.L.W.]); McDonnell Center for Molecular and Cellular Neurobiology (to N.C.); Barnes Jewish Foundation (to A.G.); the Washington University Genome Sequencing Center (to A.G.); the Winspear Family Center for Research on the Neuropathology of Alzheimer's Disease (to K.J.H. and C.L.W.); and the McCune Foundation (UTSW). O.M. is a Fogarty International Postdoctoral fellow (grant #TW 0511-05) and J.S.K.K. is a Ford Foundation Predoctoral Fellow.
Grant sponsors: McDonnell Center for Molecular and Cellular Neurobiology; Barnes Jewish Foundation; Washington University Genome Sequencing Center; Winspear Family Center for Research on the Neuropathology of Alzheimer's Disease; McCune Foundation (UTSW); National Institute on Aging (NIA); Grant numbers: AG05681; AG03991; AG12300; AG13854; and AG16976.
Footnotes
Communicated by Christine Van Broeckhoven
The Supplementary Material referred to in this article can be accessed at http://www.interscience.wiley.com/jpages/1059-7794/suppmat.
REFERENCES
- Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S, Cannon A, Dwosh E, Neary D, Melquist S, Richardson A, Dickson D, Berger Z, Eriksen J, Robinson T, Zehr C, Dickey CA, Crook R, McGowan E, Mann D, Boeve B, Feldman H, Hutton M. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- Bateman A, Belcourt D, Bennett H, Lazure C, Solomon S. Granulins, a novel class of peptide from leukocytes. Biochem Biophys Res Commun. 1990;173:1161–1168. doi: 10.1016/s0006-291x(05)80908-8. [DOI] [PubMed] [Google Scholar]
- Behrens MI, Mukherjee O, Tu PH, Liscic RM, Grinberg LT, Carter D, Paulsmeyer K, Taylor-Reinwald L, Gitcho M, Norton JB, Chakraverty S, Goate AM, Morris JC, Cairns NJ. Neuropathologic heterogeneity in HDDD1: a familial frontotemporal lobar degeneration with ubiquitin-positive inclusions and progranulin mutation. Alzheimer Dis Assoc Disord. 2007;21:1–7. doi: 10.1097/WAD.0b013e31803083f2. [DOI] [PubMed] [Google Scholar]
- Berg L, McKeel DW, Jr, Miller JP, Storandt M, Rubin EH, Morris JC, Baty J, Coats M, Norton J, Goate AM, Price JL, Gearing M, Mirra SS, Saunders AM. Clinicopathologic studies in cognitively healthy aging and Alzheimer's disease: relation of histologic markers to dementia severity, age, sex, and apolipoprotein E genotype. Arch Neurol. 1998;55:326–335. doi: 10.1001/archneur.55.3.326. [DOI] [PubMed] [Google Scholar]
- Cairns NJ, Neumann M, Bigio EH, Holm IE, Troost D, Hatanpaa KJ, Foong C, White CL, 3rd, Schneider JA, Kretzschmar HA, Carter D, Taylor-Reinwald L, Paulsmeyer K, Strider J, Gitcho M, Goate AM, Morris JC, Mishra M, Kwong LK, Stieber A, Xu Y, Forman MS, Trojanowski JQ, Lee VM, Mackenzie IR. TDP-43 in familial and sporadic frontotemporal lobar degeneration with ubiquitin inclusions. Am J Pathol. 2007a;171:227–240. doi: 10.2353/ajpath.2007.070182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns NJ, Bigio EH, Mackenzie IR, Neumann M, Lee VM, Hatanpaa KJ, White CL, 3rd, Schneider JA, Grinberg LT, Halliday G, Duyckaerts C, Lowe JS, Holm IE, Tolnay M, Okamoto K, Yokoo H, Murayama S, Woulfe J, Munoz DG, Dickson DW, Ince PG, Trojanowski JQ, Mann DM. Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the Consortium for Frontotemporal Lobar Degeneration. Acta Neuropathol. 2007b;114:5–11422. doi: 10.1007/s00401-007-0237-2. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, Rademakers R, Vandenberghe R, Dermaut B, Martin JJ, van Duijn C, Peeters K, Sciot R, Santens P, De Pooter T, Mattheijssens M, Van den Broeck M, Cuijt I, Vennekens K, De Deyn PP, Kumar-Singh S, Van Broeckhoven C. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- Daniel R, He Z, Carmichael KP, Halper J. Cellular localization of gene expression for progranulin. J Histochem Cytochem. 2000;48:999–1009. doi: 10.1177/002215540004800713. [DOI] [PubMed] [Google Scholar]
- Gass J, Cannon A, Mackenzie IR, Boeve B, Baker M, Adamson J, Crook R, Melquist S, Kuntz K, Petersen R, Josephs K, Pickering-Brown SM, Graff-Radford N, Uitti R, Dickson D, Wszolek Z, Gonzalez J, Beach TG, Bigio E, Johnson N, Weintraub S, Mesulam M, White CL, 3rd, Woodruff B, Caselli R, Hsiung GY, Feldman H, Knopman D, Hutton M, Rademakers R. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum Mol Genet. 2006;15:2988–3001. doi: 10.1093/hmg/ddl241. [DOI] [PubMed] [Google Scholar]
- Grupe A, Li Y, Rowland C, Nowotny P, Hinrichs AL, Smemo S, Kauwe JS, Maxwell TJ, Cherny S, Doil L, Tacey K, van Luchene R, Myers A, Wavrant-De Vrieze F, Kaleem M, Hollingworth P, Jehu L, Foy C, Archer N, Hamilton G, Holmans P, Morris CM, Catanese J, Sninsky J, White TJ, Powell J, Hardy J, O'Donovan M, Lovestone S, Jones L, Morris JC, Thal L, Owen M, Williams J, Goate A. A scan of chromosome 10 identifies a novel locus showing strong association with late-onset Alzheimer disease. Am J Hum Genet. 2006;78:78–88. doi: 10.1086/498851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Bateman A. Progranulin gene expression regulates epithelial cell growth and promotes tumor growth in vivo. Cancer Res. 1999;59:3222–3229. [PubMed] [Google Scholar]
- He Z, Bateman A. Progranulin (granulin-epithelin precursor, PC-cell-derived growth factor, acrogranin) mediates tissue repair and tumorigenesis. J Mol Med. 2003;81:600–612. doi: 10.1007/s00109-003-0474-3. [DOI] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, Hackett J, Adamson J, Lincoln S, Dickson D, Davies P, Petersen RC, Stevens M, de Graaff E, Wauters E, van Baren J, Hillebrand M, Joosse M, Kwon JM, Nowotny P, Che LK, Norton J, Morris JC, Reed LA, Trojanowski J, Basun H, Lannfelt L, Neystat M, Fahn S, Dark F, Tannenberg T, Dodd PR, Hayward N, Kwok JB, Schofield PR, Andreadis A, Snowden J, Craufurd D, Neary D, Owen F, Oostra BA, Hardy J, Goate A, van Swieten J, Mann D, Lynch T, Heutink P. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. 8. [DOI] [PubMed] [Google Scholar]
- Lendon CL, Lynch T, Norton J, McKeel DW, Jr, Busfield F, Craddock N, Chakraverty S, Gopalakrishnan G, Shears SD, Grimmett W, Wilhelmsen KC, Hansen L, Morris JC, Goate AM. Hereditary dysphasic disinhibition dementia: a frontotemporal dementia linked to 17q21–22. Neurology. 1998;50:1546–1555. doi: 10.1212/wnl.50.6.1546. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Butland SL, Devon RS, Dwosh E, Feldman H, Lindholm C, Neal SJ, Ouellette BF, Leavitt BR. Familial frontotemporal dementia with neuronal intranuclear inclusions is not a polyglutamine expansion disease. BMC Neurol. 2006;6:32. doi: 10.1186/1471-2377-6-32. 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masellis M, Momeni P, Meschino W, Heffner R, Jr, Elder J, Sato C, Liang Y, St George-Hyslop P, Hardy J, Bilbao J, Black S, Rogaeva E. Novel splicing mutation in the progranulin gene causing familial corticobasal syndrome. Brain. 2006;129(Pt 11):3115–3123. doi: 10.1093/brain/awl276. [DOI] [PubMed] [Google Scholar]
- McKhann GM, Albert MS, Grossman M, Miller B, Dickson D, Trojanowski JQ. Work group on frontotemporal dementia and Pick's disease. Arch Neurol. 2001;58:1803–1809. doi: 10.1001/archneur.58.11.1803. [DOI] [PubMed] [Google Scholar]
- Morris JC, Cole M, Banker BQ, Wright D. Hereditary dysphasic dementia and the Pick-Alzheimer spectrum. Ann Neurol. 1984;16:455–466. doi: 10.1002/ana.410160407. [DOI] [PubMed] [Google Scholar]
- Mukherjee O, Pastor P, Cairns NJ, Chakraverty S, Kauwe JS, Shears S, Behrens MI, Budde J, Hinrichs AL, Norton J, Levitch D, Taylor-Reinwald L, Gitcho M, Tu PH, Tenenholz Grinberg L, Liscic RM, Armendariz J, Morris JC, Goate AM. HDDD2 is a familial frontotemporal lobar degeneration with ubiquitin-positive, tau-negative inclusions caused by a missense mutation in the signal peptide of progranulin. Ann Neurol. 2006;60:314–322. doi: 10.1002/ana.20963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neary D, Snowden JS, Gustafson L, Passant U, Stuss D, Black S, Freedman M, Kertesz A, Robert PH, Albert M, Boone K, Miller BL, Cummings J, Benson DF. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–1554. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:13–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Neumann M, Mackenzie IR, Cairns NJ, Boyer PJ, Markesbery WR, Smith CD, Paul Taylor J, Kretzschmar HA, Kimonis V, Forman MS. TDP-43 in the ubiquitin pathology of frontotemporal dementia with VCP mutations. J Neuropathol Exp Neurol. 2007;66:152–157. doi: 10.1097/nen.0b013e31803020b9. [DOI] [PubMed] [Google Scholar]
- Rosso SM, Kamphorst W, de Graaf B, Willemsen R, Ravid R, Niermeijer MF, Spillantini MG, Heutink P, van Swieten JC. Familial frontotemporal dementia with ubiquitin-positive inclusions is linked to chromosome 17q21–22. Brain. 2001;124(Pt 10):1948–1957. doi: 10.1093/brain/124.10.1948. [DOI] [PubMed] [Google Scholar]
- Silverman JM, Raiford K, Edland S, Fillenbaum G, Morris JC, Clark CM, Kukull W, Heyman A. The consortium to establish a registry for AD (CERAD). Part VI. Family history assessment: a multicenter study of first-degree relatives of Alzheimer's disease probands and nondemented spouse controls. Neurology. 1994;44:1253–1259. doi: 10.1212/wnl.44.7.1253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.