Abstract
In mammals, DNA is methylated at cytosines within CpG dinucleotides. Properly regulated methylation is crucial for normal development1,2. Inappropriate methylation may contribute to tumorigenesis by silencing tumor-suppressor genes3-10 or by activating growth-stimulating genes11-13. Although many genes have been identified that acquire methylation and whose expression is methylation-sensitive14,15, little is known about how DNA methylation is controlled16. We have identified a DNA sequence that regulates establishment of DNA methylation in the male germ line at Rasgrf1. In mice, the imprinted Rasgrf1 locus is methylated on the paternal allele within a differentially methylated domain (DMD) 30 kbp 5′ of the promoter. Expression is exclusively from the paternal allele in neonatal brain17. Methylation is regulated by a repeated sequence, consisting of a 41-mer repeated 40 times, found immediately 3′ of the DMD. This sequence is present in organisms in which Rasgrf1 is imprinted18. In addition, DMD methylation is required for imprinted Rasgrf1 expression. Together the DMD and repeat element constitute a binary switch that regulates imprinting at the locus.
We generated mice in which the Rasgrf1 repeats were replaced by a single loxP site. This allele of Rasgrf1 was designated Rasgrf1tm1Pds (Fig. 1). To evaluate whether the mutation affected methylation of the DMD, we subjected tail DNA from mice inheriting the repeat deletion from their mother or father to Southern-blot analysis using PstI and the methylation-sensitive enzyme NotI. A single NotI site is present in the DMD, and its methylation state in DNAs from all somatic tissues tested is diagnostic of the methylation state of the DMD17. PstI-digested DNA from wildtype mice (Rasgrf1+/+) gave rise to an 8.0-kbp band on Southern blots hybridized to the probe shown in Fig. 2a. The unmethylated maternal fragment was further cleaved by NotI, generating a 2.8-kbp band (Fig. 2a,b) as previously reported17,19. In heterozygous mice inheriting the mutated allele from the mother (Rasgrf1tm1Pds/+), there was no change in the methylation status of either allele. PstI digestion generated a 3.0-kbp fragment from the mutated maternal allele and an 8.0-kbp fragment from the wildtype paternal allele. Digestion with both PstI and NotI further cleaved the 3.0-kbp maternal fragment to 2.8 kbp. In contrast, animals bearing a paternally transmitted repeat deletion (Rasgrf1+/tm1Pds) lost methylation of the paternal allele. PstI digestion of DNA gave rise to an 8.0-kbp wildtype maternal allele and a 3.0-kbp mutated paternal allele band. Both were reduced to 2.8 kbp after NotI digestion, indicating that neither allele was methylated at the NotI site (Fig. 2b). Bisulfite sequencing revealed that loss of methylation was extensive throughout the DMD on the mutant paternal allele (Table 1). This indicated that the Rasgrf1 repeats are needed for paternal allele–specific DNA methylation patterns seen at the locus.
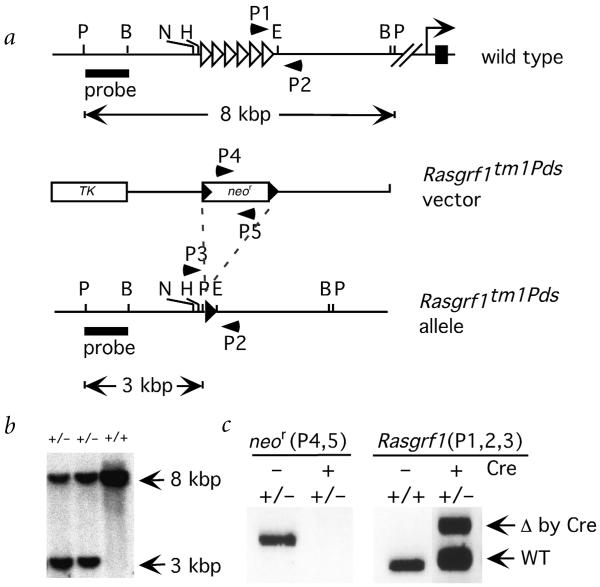
Fig. 1.
Targeting the Rasgrf1 repeats. a, The wildtype Rasgrf1 locus, targeting vector and correctly targeted locus are shown. Open rightward-pointing triangles represent the Rasgrf1 repeat element. The rightward-pointing arrow and black rectangle to the right of it represent the transcriptional start site and first exon of the gene, respectively, which are located approximately 30 kbp 3′ of the repeats. Filled rightward-pointing triangles represent loxP sites on the targeting vector and targeted allele. b, Southern-blot hybridization of PstI-digested DNA from mice using the probe shown in a produced band sizes of 8.0 kbp and 3.0 kbp for the wildtype and mutated alleles, respectively. c, PCR analysis was carried out on tail DNA from progeny of heterozygous male Rasgrf1tm1Pds mice crossed with females containing (+) or lacking (−) the Zp3cre transgene. Primers, depicted as arrowheads numbered P1–P5 in a, detected the neor cassette (left panel, P4 and P5), the wildtype Rasgrf1 allele (WT; right panel, P1 and P2) and the Rasgrf1tm1Pds allele (Δ by Cre rightpanel, P2 and P3). Restriction sites are PstI (P), BamHI (B), NotI (N), HindIII (H) and EcoRV (E).
Fig. 2.
NotI site methylation is lost upon paternal transmission of the Rasgrf1 repeat mutation. a, The wildtype maternal and mutated paternal alleles are shown along with the possible PstI/NotI (P/N) restriction fragments arising from methylated or unmethylated NotI sites. b, Tail DNA from wildtype (+/+) or heterozygous mice with a maternally (−/+) or paternally (+/−) transmitted Rasgrf1tm1Pds allele was digested with PstI and/or NotI, and Southern blots were hybridized with the probe indicated. Mice were on a mixed C57BL/6 and 129S4/SvJae background.
Table 1.
Results of bisulfite sequencing
| CpG | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NotI | N | |||||||||||||||||||||||||
| HhaI | H | H | H | H | H | |||||||||||||||||||||
| Tail wildtype paternal allele | ||||||||||||||||||||||||||
| X | O | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | |||||||||
| X | O | X | X | X | X | X | X | X | X | O | X | X | X | X | X | X | X | |||||||||
| X | X | X | X | X | X | X | O | X | X | X | X | X | X | X | X | X | X | |||||||||
| X | X | X | X | X | X | X | O | X | X | X | X | X | X | X | X | X | X | |||||||||
| X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | |||||||||
| X | X | X | X | O | O | X | O | X | X | X | X | O | X | X | X | X | X | |||||||||
| O | O | X | X | X | X | X | O | O | O | X | X | X | O | X | X | X | O | |||||||||
| O | O | X | X | X | X | X | O | O | O | X | X | X | O | X | X | X | O | |||||||||
| Tail wildtype maternal allele | ||||||||||||||||||||||||||
| O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | |||||||||
| O | O | O | O | O | X | O | O | O | O | O | O | O | O | O | O | X | O | |||||||||
| O | O | O | O | O | O | O | X | O | O | O | O | O | O | O | O | O | O | |||||||||
| O | O | O | O | O | O | O | X | O | O | O | O | O | O | O | O | O | O | |||||||||
| O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | |||||||||
| Tail mutant paternal allele | ||||||||||||||||||||||||||
| O | O | O | X | O | O | O | O | O | O | O | O | O | O | X | O | O | O | O | O | O | O | X | O | O | O | |
| O | O | O | O | O | O | O | X | O | O | O | O | O | O | O | O | O | X | O | O | O | O | O | O | O | O | |
| O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | |
| O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | |
| O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | |
| O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | |
| O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | |
| Sperm wildtype allele | ||||||||||||||||||||||||||
| X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | O | ||
| X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | O | ||
| X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | O | ||
| X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | ||
| X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | ||
| X | X | X | X | X | X | X | X | X | X | O | X | X | X | X | X | X | X | X | X | X | X | X | X | O | ||
| X | X | X | X | X | X | X | X | X | X | O | X | X | X | X | X | X | X | X | X | X | X | X | X | O | ||
| X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | ||
| X | X | X | X | X | X | X | X | X | X | O | X | X | X | X | X | X | X | X | X | X | X | X | X | O | ||
| X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | ||
| Sperm mutated allele | ||||||||||||||||||||||||||
| O | O | O | O | O | O | O | O | O | O | O | O | O | O | X | X | O | O | O | O | O | O | O | O | O | O | |
| O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | |
| O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | |
| X | O | X | X | X | X | X | X | X | X | X | X | X | X | O | X | X | O | O | X | X | O | X | X | X | X | |
| O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | |
| O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | |
| O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | |
| O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | |
| O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | O | |
Each line represents the sequence of independent clones of DMD PCR products generated by amplification of bisulfite-treated templates prepared from the indicated tissue and mice. Clones representing the mutated allele were derived from three independent mice for both the tail and sperm DNA samples. Identification of the parental alleles from wildtype tail DNA was based on the methylation state of the NotI site (N). The relative positions of up to 26 CpGs that were assayed (1–26) are shown. Those contained in the NotI (N) and HhaI (H) sites are indicated. O, unmethylated CpG; X, methylated CpG.
The loss of paternal allele–specific methylation may have resulted from a failure to establish the normal paternal methylation pattern in the male germ line or failure of the established pattern to be maintained in the somatic tissue of the progeny. To distinguish between these possibilities, we assayed the methylation status of the Rasgrf1 DMD in sperm DNA from males heterozygous for the repeat mutation, using three separate methods. The first assay was a Southern blot using the same digests and probes used in Fig. 2. Results showed a substantial reduction in methylation of the mutated allele (Fig. 3a). Previous studies have suggested that although the NotI site methylation state is diagnostic for paternal allele–specific methylation in somatic tissue, the germline methylation imprint may be more accurately reflected by the methylation of the five HhaI sites within the Rasgrf1 DMD19. We therefore assessed the methylation status of these sites in sperm DNA using PCR reactions specific for the mutated and wildtype alleles, with primers that spanned all five HhaI sites. Fig. 3b shows the mutant and wild-type loci and the location of primer pairs used in this analysis. Sperm DNA was digested with HhaI or left undigested, before PCR analysis, using the two allele-specific assays. We carried out a third control PCR reaction using a primer pair that spanned a region of Rasgrf1 lacking HhaI sites. In all DNA samples, the wildtype allele was amplified using primers P1 and P2 (Fig. 3c, upper gel), indicating that the HhaI sites were detectable in the methylated state on the wildtype alleles. When we amplified uncut DNA from sperm of wildtype or heterozygous mice using mutated allele–specific primers P3 and P4, a band of the predicted size was generated only in the reactions containing heterozygous DNA, indicating that the product was indeed allele-specific. However, the band was substantially diminished in reactions containing sperm DNA from heterozygotes that had been pre-digested with HhaI. This indicated that one or more of the five HhaI sites were hypomethylated at a high frequency on the mutated allele. This result was seen regardless of whether the mutated allele was paternally or maternally transmitted (data not shown for maternal transmission). An internal control PCR reaction using primers P5 and P6 showed that band loss was not due to a failure in the amplification reaction. We carried out bisulfite sequencing to evaluate whether the loss of methylation affected just one or more of the five HhaI sites and to determine whether CpGs other than those in the HhaI and NotI sites were affected. The results indicated that loss of methylation on the mutated allele of sperm DNA affected CpGs across the entire DMD and not just those in the NotI and HhaI sites (Table 1). In addition, the disruption of DMD methylation was comparable in both somatic and sperm DNA templates. These three independent means of analysis showed that loss of methylation caused by the repeat deletion was a common event affecting many CpG residues within the DMD at a time when methylation of the locus was normally established in the male germ line.
Fig. 3.
Loss of paternal allele methylation is due to a failure to establish Rasgrf1 methylation in the male germ line. a, The methylation status of the NotI site in DNA isolated from sperm and tail of heterozygous Rasgrf1+/tm1Pds (+/−) mice was analyzed by Southern blot as described in Fig. 2. b, Structure of the locus in heterozygotes and location of primers used in PCR analysis of HhaI site methylation. c, Sperm DNA was amplified by PCR after digestion with HhaI (H) or without previous restriction enzyme treatment (U). PCR reactions included primers in b specific for the wildtype allele (P1 and P2) or the mutated allele (P3 and P4), or spanned a control sequence devoid of HhaI sites, which served as a control for successful PCR amplification (P5 and P6). Mice were on a mixed C57BL/6 and 129S4/SvJae background.
Rasgrf1 is expressed predominantly from the paternal allele in many tissues; however, expression in neonatal brain is exclusively from the paternal allele17. To determine whether the Rasgrf1tm1Pds mutation had any effect on imprinted expression of Rasgrf1, we analyzed RNA from the brain of neonatal progeny of mutant animals crossed with wildtype C57BL/6 or PWK mates. The cross facilitated separate expression analysis of the two parental Rasgrf1 alleles because of an exonic polymorphism that exists between 129S4/SvJae, the strain in which the mutation was made, and PWK17. Maternal transmission of the repeat deletion did not affect the normal expression pattern of the locus—expression was still from the paternal allele only on all strain backgrounds in amounts comparable to those seen in wild-type mice (data not shown). However, paternal transmission of the Rasgrf1tm1Pds allele resulted in substantial silencing of the normally active allele in heterozygous progeny of C57BL/6 mothers (Fig. 4, +B6/−). Notably, the silencing of the mutated paternal allele was variable in progeny of PWK mothers (Fig. 4, +PWK/−). In those animals, Rasgrf1 expression levels ranged from those seen in +B6/− mice (fourth panel from the top) to amounts seen in +B6/+ and +PWK/+ animals (bottom panel). In +PWK/− mice with wildtype levels of expression, the DMD showed extensive methylation despite the presence of a paternally transmitted Rasgrf1tm1Pds allele, whereas in animals with substantially silenced Rasgrf1 expression the paternal allele was unmethylated (Fig. 4, right panel). This indicated that it is the methylation state of the DMD rather than the presence of the repeats that has the most direct influence on expression of the locus, ruling out the trivial explanation that the repeats contain a promoter or enhancer element needed for Rasgrf1 expression. Moreover, the Rasgrf1 DMD behaves like a methylation-sensitive enhancer-blocking element, as is the case for the DMD at H19/Igf2 (refs 12,13,20; B.J. Yoon et al., unpublished data). The variable methylation seen among progeny of PWK mothers also suggested that there are strain-specific modifiers of the methylation phenotype. Analysis of the nine +PWK/− progeny revealed three animals with residual methylation on the mutated paternal allele (data not shown). Methylation analysis of a much larger number of mice on a mixed C57BL/6 ×129S4/SvJae background revealed no such variability in methylation of the mutated paternal allele. Mice on this background were used in the experiments reported in Fig. 3.
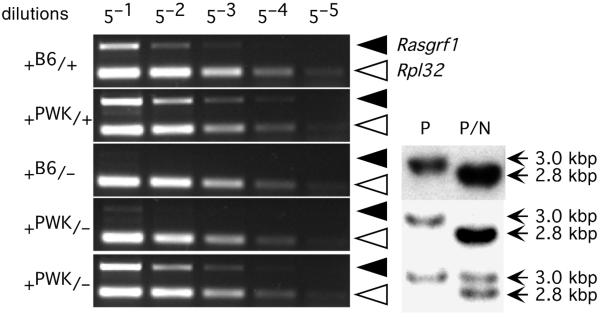
Fig. 4.
Loss of DMD methylation leads to Rasgrf1 silencing. Rasgrf1 expression levels in neonatal brain were measured by RT–PCR (left panel). The mice included wildtype progeny of C57BL/6 (+B6/+) or PWK mothers (+PWK/+) as well as animals heterozygous for a paternally transmitted Rasgrf1tm1Pds allele, whose mothers were also either C57BL/6 (+B6/−) or PWK (+PWK/−). cDNAs were serially diluted in fivefold increments before PCR amplification. Primer pairs detected Rasgrf1 (solid triangles) or Rpl32 (open triangles), which served as an internal control. The methylation status of the Rasgrf1 DMD in heterozygous animals was assayed by the Southern-blot hybridization method used in Figs 1 and 2 (right panel). In DNA digested with both PstI (P) and NotI (N), the 2.8-kbp band is diagnostic for an unmethylated DMD, whereas the 3.0-kb band is diagnostic for a methylated DMD.
The presence of transgene repeats has been correlated with changes in local DNA methylation and gene expression in diverse systems21-23. The repetitive nature of the Rasgrf1 repeats, the presence of 39 CpGs within the 1,611-nt repeat region, or other structural or sequence motifs may be required for the Rasgrf1 repeats to regulate establishment of DNA methylation in the male germ line. These motifs may serve as sites of interaction with collaborating trans-acting factors that themselves dictate local methylation patterns. The variable loss of methylation and correlation between methylation and expression seen in mutant progeny of PWK mothers was reminiscent of previous studies of H19 imprinting24 and may be due to strain-specific differences in loci encoding such factors. Expression of these factors must be specific to the male or female germ line for Rasgrf1 and other imprinted loci to undergo allele-specific methylation.
Studies of methylation regulation at other loci have been reported. The maternally-expressed H19/Igf2 locus contains a DMD that is hypermethylated at CpGs within and adjacent to the promoter on the paternal allele25. A targeted deletion of the DMD abolished paternal allele–specific DNA methylation and Igf2 silencing when transmitted paternally26. As is the case for the Rasgrf1 DMD, the H19 DMD is an enhancer blocker (refs 12,13; B.J. Yoon et al., unpublished). However, in contrast with the Rasgrf1 repeat mutation, which caused loss of methylation establishment, the H19 mutation interfered with the maintenance or persistence of methylation patterns after their normal establishment. Although it is not known how methylation is established at the H19 locus, repeats seem not to be required. A 461-nt G-rich repetitive element at H19 was shown to be dispensable for establishment of paternal-allele methylation of an H19 transgene27. It is possible that regulation of transgene methylation differs from methylation of endogenous loci, that redundant mechanisms regulate methylation at H19 or that mechanisms regulating methylation at imprinted loci vary. These possibilities are not mutually exclusive.
At the Igf2r locus, a 3-kbp intronic sequence regulates methylation of the maternal allele before fertilization28. Transient transgenesis experiments identified a 6-nt allele-discrimination sequence (ADS) that protects the paternal allele from methylation and an 8-nt de novo methylation sequence (DNS) that facilitates methylation of the unprotected maternal allele. The ADS and DNS sequences interact with factors present in androgenetic and gynogenetic ES cells, respectively16. Dot-plot analysis shows that the Igf2r imprinting control region contains some repeated elements; however, it is not clear if these are needed for the ADS or DNS to function. Although a single copy of the 6-nt ADS is found within the Rasgrf1 repeats, it is not likely to be functionally significant, as in Igf2r transgenes the ADS sequence protects the paternal allele from methylation. Because the paternal allele is methylated at Rasgrf1, the ADS cannot be protecting the locus from methylation.
Methods
Generation of mutant mice
We prepared the Rasgrf1-targeting construct (pBJR3) which replaced a 2-kbp repeat–containing fragment with a PGKneor cassette flanked by loxP sites as follows. The 5′ vector arm consisted of a 2-kbp fragment immediately 5′ of the repeat that we PCR-amplified using Rasgrf1 reverse and forward primers derived from the plasmid pSPL3, which contained the 5′ Rasgrf1 fragment. The reverse primer modified four nucleotides of the wildtype Rasgrf1 sequence to generate new PstI and HindIII sites immediately 5′ to the repeat. We cloned the PstI fragment from the amplification into the PstI site of pBluescriptII (Stratagene). The 5′ arm, isolated as a SacII-HindIII fragment from the resulting clone, was inserted into the SacII and HindIII sites of pTK-NEOF which carried the loxP-neor-loxP and HSVTK markers (D. Lam and P. Aplan, National Cancer Institute). We inserted the 3-kb homologous arm, consisting of an EcoRV-SmaI fragment located 3′ of the repeats, into the SalI site of pTKNEOF to generate the pBJR3 targeting vector. We carried out ES cell culture and blastocyst microinjections according to the standard protocols. We excised the neor selectable marker by crossing germline progeny with Zp3Cre transgenic mice29.
Sperm DNA isolation
We prepared sperm DNA from 8-wk or older heterozygous male mice inheriting a mutated allele from their fathers, mothers or wildtype control littermates by allowing sperm to swim out from the cauda epididymis into 500 μl phosphate-buffered saline (PBS). We added SDS (1.5 μl of 10% w/v) and incubated the sperm at room temperature for 15 min. Cells were centrifuged (3 min, 16,000 × g), resuspended in 100 μl PBS, diluted with an additional 900 μl PBS and 100 μl β-mercaptoethanol (1 M), and then incubated at 37 °C for 1 h. Cells were centrifuged (3 min, 16,000 × g), resuspended in 100 μl PBS, centrifuged again (5 min, 16,000 × g), resuspended in 200 ml PBS, diluted with an equal volume of 2 × lysis buffer (200 mM Tris pH 8.5, 200 mM EDTA, 400 mM NaCl, 2% SDS) and proteinase K added (400 μg/ml−1), followed by an overnight incubation at 55 °C with shaking. We then isolated DNA by phenol chloroform extraction and ethanol precipitation.
Methylation analysis
We carried out assays for NotI site methylation using Southern-blot hybridization as described17 and assays for HhaI site methylation in sperm as described19, with modifications. Using sperm DNA that was digested with HhaI or left uncut, we carried out PCR amplification. PCR product sizes were as follows: P1−P2, 263 bp; P3−P4, 228 bp; P5−P6, 148 bp.
We carried out bisulfite sequencing as described30 with some modifications, using freshly prepared solutions. Briefly, 1.0 μg of sheared DNA in 50 μl water was denatured by addition of 5 μl fresh 3 M NaOH, then incubated at 37 °C for 10 min. After adding 30 μl 10 mM hydroquinone and 520 μl 3 M sodium bisulfite, we allowed deamination to proceed at 50 °C for 16 h. We purified treated DNA using a Qiaquick gel extraction kit according to the manufacturer's instructions (Qiagen) and eluted DNA from the kit column in 50 μl elution buffer. The bisulfite reaction was completed by the addition of 5 μl fresh 3 M NaOH followed by a 5-min incubation at RT. We purified DNA again using a Qiaquick gel extraction kit and eluted it in 30 μl. For PCR, we amplified 1 μl of DNA (primers available upon request). We gel-purified all PCR products, cloned them using a TA or TopoII cloning kit (Invitrogen) and then sequenced individual clones.
Expression analysis by RT–PCR
We measured expression of Rasgrf1 using a quantitative PCR assay. We perpared cDNA using oligo(dT) primers and 0.5 μg total RNA. A fraction of this material (10%) was serially-diluted and used for PCR. A reaction mixture containing four primer pairs was prepared and added to each cDNA dilution. One set of the primers amplified a 378-bp product that spanned a Rasgrf1 intron–exon boundary, and another set amplified a 128-bp product specific for Rpl32 which served as an internal control.
PCR primers
Available upon request.
Acknowledgments
This work was made possible through grants from the NIH and the Roswell Park Alliance to P.D.S., C.P. and to the Roswell Park Cancer Institute. The authors dedicate this work to the memory of V. Chapman.
References
- 1.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 2.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 3.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv. Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 4.Ohtani-Fujita N, et al. CpG methylation inactivates the promoter activity of the human retinoblastoma tumor-suppressor gene. Oncogene. 1993;8:1063–1067. [PubMed] [Google Scholar]
- 5.Herman JG, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc. Natl Acad. Sci. USA. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herman JG, Jen J, Merlo A, Baylin SB. Hypermethylation-associated inactivation indicates a tumor suppressor role for p15INK4B. Cancer Res. 1996;56:722–727. [PubMed] [Google Scholar]
- 7.Yoshiura K, et al. Silencing of the E-cadherin invasion-suppressor gene by CpG methylation in human carcinomas. Proc. Natl Acad. Sci. USA. 1995;92:7416–7419. doi: 10.1073/pnas.92.16.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graff JR, et al. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995;55:5195–5199. [PubMed] [Google Scholar]
- 9.Esteller M, et al. hMLH1 promoter hypermethylation is an early event in human endometrial tumorigenesis. Am. J. Pathol. 1999;155:1767–1772. doi: 10.1016/S0002-9440(10)65492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herman JG, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc. Natl Acad. Sci. USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Issa JP, Vertino PM, Boehm CD, Newsham IF, Baylin SB. Switch from monoallelic to biallelic human IGF2 promoter methylation during aging and carcinogenesis. Proc. Natl Acad. Sci. USA. 1996;93:11757–11762. doi: 10.1073/pnas.93.21.11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 13.Hark AT, et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 14.Costello JF, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nature Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 15.Jackson-Grusby L, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nature Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 16.Birger Y, Shemer R, Perk J, Razin A. The imprinting box of the mouse Igf2r gene. Nature. 1999;397:84–88. doi: 10.1038/16291. [DOI] [PubMed] [Google Scholar]
- 17.Plass C, et al. Identification of Grf1 on mouse chromosome 9 as an imprinted gene by RLGS-M. Nature Genet. 1996;14:106–109. doi: 10.1038/ng0996-106. [DOI] [PubMed] [Google Scholar]
- 18.Pearsall RS, et al. A direct repeat sequence at the Rasgrf1 locus and imprinted expression. Genomics. 1999;55:194–201. doi: 10.1006/geno.1998.5660. [DOI] [PubMed] [Google Scholar]
- 19.Shibata H, et al. A methylation imprint mark in the mouse imprinted gene Grf1/Cdc25Mm locus shares a common feature with the U2afbp-rs gene: an association with a short tandem repeat and a hypermethylated region. Genomics. 1998;49:30–37. doi: 10.1006/geno.1998.5218. [DOI] [PubMed] [Google Scholar]
- 20.Kanduri C, et al. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 21.Linn F, Heidmann I, Saedler H, Meyer P. Epigenetic changes in the expression of the maize A1 gene in Petunia hybrida: role of numbers of integrated gene copies and state of methylation. Mol. Gen. Genet. 1990;222:329–336. doi: 10.1007/BF00633837. [DOI] [PubMed] [Google Scholar]
- 22.Goyon C, Barry C, Gregoire A, Faugeron G, Rossignol JL. Methylation of DNA repeats of decreasing sizes in Ascobolus immersus. Mol. Cell. Biol. 1996;16:3054–3065. doi: 10.1128/mcb.16.6.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene silencing in mammals. Nature Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 24.Jones BK, Levorse JM, Tilghman SM. Igf2 imprinting does not require its own DNA methylation or H19 RNA. Genes Dev. 1998;12:2200–2207. doi: 10.1101/gad.12.14.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tremblay KD, Saam JR, Ingram RS, Tilghman SM, Bartolomei MS. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nature Genet. 1995;9:407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- 26.Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stadnick MP, et al. Role of a 461-bp G-rich repetitive element in H19 transgene imprinting. Dev. Genes Evol. 1999;209:239–248. doi: 10.1007/s004270050248. [DOI] [PubMed] [Google Scholar]
- 28.Stoger R, et al. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993;73:61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- 29.Lewandoski M, Wassarman KM, Martin GR. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr. Biol. 1997;7:148–151. doi: 10.1016/s0960-9822(06)00059-5. [DOI] [PubMed] [Google Scholar]
- 30.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]