Abstract
Recent studies have indicated that nuclear protein of 95 kDa (Np95) is essential for maintaining genomic methylation by recruiting DNA methyltransferase (Dnmt) 1 to hemi-methylated sites. Here, we show that Np95 interacts more strongly with regulatory domains of the de novo methyltransferases Dnmt3a and Dnmt3b. To investigate possible functions, we developed an epigenetic silencing assay using fluorescent reporters in embryonic stem cells (ESCs). Interestingly, silencing of the cytomegalovirus promoter in ESCs preceded DNA methylation and was strictly dependent on the presence of either Np95, histone H3 methyltransferase G9a or Dnmt3a and Dnmt3b. Our results indicate a regulatory role for Np95, Dnmt3a and Dnmt3b in mediating epigenetic silencing through histone modification followed by DNA methylation.
Keywords: DNA methylation, histone modification, epigenetics, silencing, Uhrf1
Introduction
In mammals, DNA methylation contributes to the establishment and maintenance of cell-type-specific gene expression programmes, imprinting, X-chromosome inactivation and genome stability (Bird, 2002). The majority of genomic methylation occurs at cytosine residues within CpG dinucleotides and is catalysed by the DNA methyltransferases (Dnmt) 1, 3a and 3b. Dnmt1 is responsible for maintaining genomic methylation, whereas Dnmt3a and Dnmt3b are mainly involved in de novo establishment of methylation patterns during cellular differentiation (Leonhardt et al, 1992; Li et al, 1992; Lei et al, 1996; Okano et al, 1999; Spada et al, 2007). Nuclear protein of 95 kDa (Np95; also known as Uhrf1) has recently been identified as an essential co-factor for maintaining genomic methylation (Bostick et al, 2007; Sharif et al, 2007; Achour et al, 2008). dnmt1−/− and np95−/− embryonic stem cells (ESCs) and embryos have similar reduced levels of DNA methylation. In addition, Np95 interacts with Dnmt1, binds hemi-methylated CpG sites through its Set and Ring associated (SRA) domain and both Np95 and Dnmt1 accumulate at replication sites (Uemura et al, 2000; Bostick et al, 2007; Papait et al, 2007; Arita et al, 2008; Avvakumov et al, 2008; Hashimoto et al, 2008). Thus, it has been proposed that Np95 mediates maintenance of genomic methylation by recruiting Dnmt1 to hemi-methylated CpG sites generated during replication.
Here, we investigated a possible involvement of Np95 in epigenetic regulation beyond its role in Dnmt1-mediated maintenance of DNA methylation. We found that Np95 interacts with the de novo methyltransferases, Dnmt3a and Dnmt3b, and mediates promoter silencing before DNA methylation is detected.
Results And Discussion
Np95 interacts with Dnmt3a and Dnmt3b
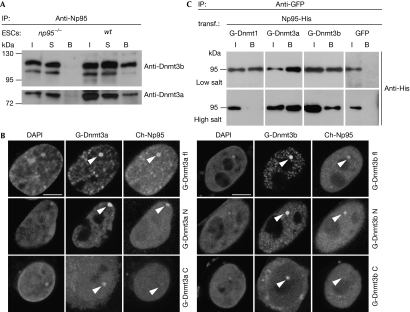
Immunoprecipitation experiments showed that different isoforms of both de novo methyltransferases Dnmt3a and Dnmt3b interact with Np95 in wild-type (wt) ESCs, including the more abundant Dnmt3a2 and Dnmt3b1 (Fig 1A). Furthermore, using a green fluorescent protein (GFP) trap (Rothbauer et al, 2008), we co-immunoprecipitated endogenous Dnmt1 and isoforms of Dnmt3a and Dnmt3b with a GFP–Np95 fusion construct transiently expressed in np95−/− ESCs and, vice versa, endogenous Np95 co-immunoprecipitated with GFP–Dnmt3a or GFP–Dnmt3b1 fusions in dnmt3a and 3b double knockout (DKO) ESCs (supplementary Fig S1A,B online). In addition, we observed co-immunoprecipitation of endogenous DNMT3b and inverted CCAAT box binding protein of 90 kDa—the human homologue of Np95—from human embryonic kidney 293T (HEK293T) cell extracts (supplementary Fig S1C online). We confirmed the interaction of Np95 with Dnmt3a/b by using a recently developed fluorescent two hybrid assay (F2H; Zolghadr et al, 2008). GFP–Dnmt3 fusion constructs were used as bait by tethering them to a lac operator array present in baby hamster kidney (BHK) cells, so that the array was visible as a distinct nuclear spot of enriched GFP fluorescence (Fig 1B). A Cherry–Np95 fusion (prey) accumulated at this spot only when GFP fusions of full-length Dnmt3a and Dnmt3b1 or their amino-terminal regions were used as bait and not when their isolated Carboxy-terminal catalytic domains were used. We further mapped the interaction of Np95 with Dnmt3a/b through co-immunoprecipitation of deletion constructs and isolated domains transiently expressed in HEK293T cells (supplementary Fig S2 online). The results were consistent with those produced by F2H: the N-terminal regions of Dnmt3a and Dnmt3b1, but not their C-terminal catalytic domains, interacted with Np95. Deletion of the PHD or PWWP domains of Dnmt3a and Dnmt3b did not eliminate the interaction with Np95. We then determined the domains of Np95 involved in this interaction. We found that the SRA domain and the N-terminal 298 amino acids of Np95, which include the ubiquitin-like domain, interacted with Dnmt3a and Dnmt3b1, whereas the PHD domain and the C-terminal 132 amino acids, including the Ring domain, did not. Furthermore, we observed co-immunoprecipitation of endogenous Np95 with GFP–Dnmt3a and GFP–Dnmt3b transiently expressed in dnmt1−/− ESCs, indicating that Dnmt3a and Dnmt3b interact with Np95 independently of Dnmt1 (supplementary Fig S1D online). To compare the relative association between endogenous Np95 and Dnmts, we re-probed the blot in Fig 1A with a Dnmt1 antibody and observed a substantially weaker signal for the co-immunoprecipitated Dnmt1 relative to the input than in the case of Dnmt3a2 and Dnmt3b1 (supplementary Fig S3A online). To compare further the stability of Np95 interactions with the Dnmts, we transiently co-expressed Np95-His with GFP–Dnmt1, GFP–Dnmt3a or GFP–Dnmt3b1 in HEK293T cells and immunoprecipitated with the GFP trap in the presence of different salt concentrations (Fig 1C; supplementary Fig S3B online). Interestingly, under high salt conditions, the interaction between Np95-His and GFP–Dnmt1 was lost, whereas co-immunoprecipitation of GFP–Dnmt3a and GFP–Dnmt3b1 remained relatively unaffected. These data clearly indicate that Np95 interacts more strongly with the de novo methyltransferases, Dnmt3a and Dnmt3b, than with Dnmt1.
Figure 1.
Np95 interacts with de novo methyltransferases Dnmt3a and Dnmt3b. (A) Co-immunoprecipitation of Dnmt3a and Dnmt3b with Np95 in wt and np95−/− E14 ESCs. The Dnmt3a2 isoform is shown in the lower panel. (B) F2H shows recruitment of Cherry–Np95 (prey) at the lac operator array (indicated by arrowheads) when GFP fusions of full-length Dnmt3a and Dnmt3b1 (G-Dnmt3a/b fl) or their amino-terminal regions (G-Dnmt3a/b N) are used as bait and not with their isolated C-terminal catalytic domains (G-Dnmt3a/b C). Scale bars, 5 μm. (C) Co-immunoprecipitation of Np95-His with GFP-tagged Dnmt1, Dnmt3a and Dnmt3b1 (G-Dnmt) transiently co-expressed in HEK293T cells. Co-expression of GFP was used as the control. In the upper row, immunoprecipitations carried out in the presence of 150 mM NaCl throughout the procedure are shown, whereas in the lower row, immunoprecipitation and wash buffers were carried out using 300 and 500 mM NaCl, respectively. Two per cent of input and supernatant relative to bound fractions were loaded in (A) and (C). B, bound; Dnmt, DNA methyltransferase; ESCs, embryonic stem cells; F2H, fluorescent two-hybrid assay; GFP, green fluorescent protein; HEK293T, human embryonic kidney 293T; I, input; Np95, nuclear protein of 95 kDa; S, supernatant; wt, wild type.
Np95, Dnmt3a/3b and G9a mediate epigenetic silencing
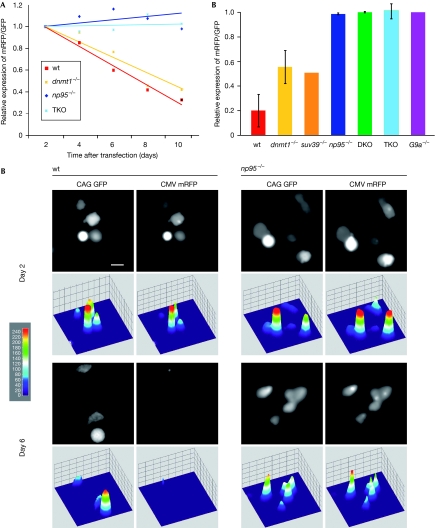
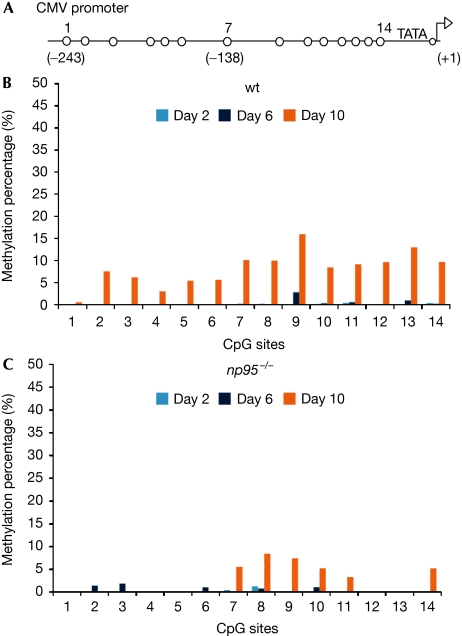
As DNA methylation has a central role in epigenetic silencing, we investigated the requirement of DNA methyltransferases and Np95 for promoter silencing in ESCs. We found that, on transient transfection of wt ESCs, constructs driven by the cytomegalovirus (CMV) promoter were rapidly silenced, as opposed to longer-lasting expression of constructs driven by the chimeric CMV early enhancer/chicken β actin (CAG) promoter (Fig 2), which is consistent with the popularity of the CAG promoter for stable transgene expression in ESCs and mice. We then established an epigenetic silencing assay based on this observation. ESCs were co-transfected with two distinct plasmids, one expressing monomeric red fluorescent protein (mRFP) under the CMV promoter, the other expressing GFP driven by the CAG promoter. mRFP and GFP expression was monitored after transfection for up to ten days by using automated image acquisition and quantification of fluorescent signals (supplementary Fig S4A online). The ratio between mRFP and GFP expression declined steadily in wt ESCs, reflecting preferential silencing of the CMV promoter (Fig 2). By contrast, DKO ESCs and ESCs lacking all three major DNA methyltransferases (dnmt1, 3a and 3b triple knockout) showed no preferential silencing of the CMV promoter. Surprisingly, np95−/− ESCs were also unable to silence the CMV promoter, whereas dnmt1−/− ESCs showed only partly reduced silencing under these conditions. Similar results were obtained on swapping GFP and mRFP reporter sequences, ruling out potential artefacts due to differences in their coding sequences or stability of the reporter proteins (supplementary Fig S4B online). Thus, despite expressing a full complement of DNA methyltransferases, ESCs lacking Np95 are as deficient in promoter silencing activity as ESCs lacking all three major Dnmts. We next investigated whether silencing of the CMV promoter correlates with CpG methylation. Interestingly, promoter methylation was detected only ten days after transfection and was lower in np95−/− than in wt ESCs, whereas none of the dnmt mutant ESCs showed appreciable DNA methylation (Fig 3; supplementary Fig S5A online). At the same time no obvious methylation was detected in any of the ESC lines within the CpG island of the CAG promoter construct (supplementary Fig S5B online). Thus, CMV promoter silencing depends on the presence of both Np95 and de novo Dnmts, but ensues well before de novo methylation of the promoter is detected. This prompted us to investigate the involvement of repressive histone methylation as a possible mechanism for the observed silencing. We found that in the absence of histone H3 lysine 9 methyltransferases (H3K9MTs), G9a or Suv39h1/2, silencing of the CMV promoter was completely abolished or reduced, respectively, indicating that G9a and, in part, Suv39h1/2 are also required for silencing (Fig 2B).
Figure 2.
Promoter silencing activity in wild-type and mutant ESCs. ESCs with the indicated phenotypes were transiently co-transfected with CMV promoter-driven mRFP and CAG promoter-driven GFP reporter constructs. Between 90 and 150 images per sample were acquired either (A) every second day after transfection from a single experiment or (B) only at days 2 and 7–10 after transfection from 3–5 independent experiments. Relative levels of red over green fluorescence are shown with values for day 2 (first day of imaging) set to 1 (A). Wild-type J1 and E14 cells gave similar results (data not shown). suv39−/− stands for Suv39h1/2 double null ESCs. (C) Representative images of wt and np95−/− E14 ESCs co-transfected as in (A) and (B) (upper panels) and respective heat map intensity plots (lower panels). Scale bar, 15 μm. CAG, CMV early enhancer/chicken β actin promoter; CMV, cytomegalovirus promoter; DKO, double knockout; Dnmt, DNA methyltransferase; ESCs, embryonic stem cells; GFP, green fluorescent protein; mRFP, monomeric red fluorescent protein; Np95, nuclear protein of 95 kDa; TKO, triple knockout; wt, wild type.
Figure 3.
Methylation of the CMV promoter 2, 6 and 10 days after transfection. Wild-type and np95−/− ESCs were transfected as in Fig 2 and GFP-positive cells were sequentially sorted at the indicated days after transfection. Total DNA was isolated from sorted cells and bisulphite-treated. A proximal part of the CMV promoter was amplified and subjected to pyrosequencing. (A) Schematic drawing of the 14 proximal CpG sites analysed (shown as open circles). The numbers above correspond to the CpG sites shown in (B) and (C) and numbers in brackets refer to the position of CpG sites with respect to the transcription start site. (B,C) Methylation percentage at individual CpG sites for (B) wt and (C) np95−/− ESCs as measured by pyrosequencing. CMV, cytomegalovirus; ESCs, embryonic stem cells; GFP, green fluorescent protein; wt, wild type.
The results shown here indicate that Np95 interacts with Dnmt3a and Dnmt3b and mediates silencing of the CMV promoter by mechanisms that are, at least initially, independent of de novo DNA methylation. Importantly, our data also show the involvement of H3K9MTs, G9a and Suv39h1/2 in CMV promoter silencing. H3K9MTs were reported to associate with de novo Dnmts, and major satellite repeats were found to be hypomethylated in ESCs lacking either Suv39h1/2 or Dnmt3 enzymes. However, major satellite transcript levels were altered only in Suv39h1/2-deficient cells and not in Dnmt3-deficient cells (Fuks et al, 2003; Lehnertz et al, 2003; Martens et al, 2005). A recent study showed that G9a, Dnmt1, Dnmt3a and Dnmt3b are required for normal methylation at long terminal repeats of endogenous retrotransposable elements, although transcription of these elements was increased in Dnmt-deficient ESCs, but not G9a-deficient ESCs (Dong et al, 2008). Furthermore, recent studies have shown that G9a interacts with Dnmt3a and Dnmt3b and mediates de novo methylation of the oct4, nanog and dnmt3l promoters on retinoic-acid-induced differentiation of ESCs (Feldman et al, 2006; Li et al, 2007; Epsztejn-Litman et al, 2008). However, two of these studies showed that neither G9a nor de novo Dnmts are required to silence the oct4 promoter, and G9a was also found to be dispensable for silencing the nanog and dnmt3l promoters (Feldman et al, 2006; Epsztejn-Litman et al, 2008). In the third study, nanog, but not oct4, was shown to be silenced in differentiating ESCs lacking both Dnmt3a and Dnmt3b (Li et al, 2007). We found that silencing of oct4 during embryoid body differentiation is largely independent from the presence of Np95 as well as all three major Dnmts, and occurs in the absence of DNA methylation (D.M., F.S., S.B., and H.L., unpublished data). These data, together with our findings on silencing of the CMV promoter in ESCs, indicate that Dnmts, Np95 and H3K9MTs mediate silencing through many mechanisms that do not necessarily involve DNA methylation and might depend on the presence of different cis elements and an intricate interplay with other epigenetic and transcription factors. Interestingly, Np95 was recently shown to interact with G9a (Kim et al, 2009) and here we show that silencing of the CMV promoter in ESCs strictly depends on Np95 and on de novo Dnmts as well as G9a. Taken together these observations suggest that Np95, de novo Dnmts and G9a might be involved in a common silencing pathway.
In summary, our data clearly support a crucial role of Np95 in epigenetic silencing mediated by de novo DNA and histone methyltransferases, and make Np95 an attractive target for epigenetic reprogramming strategies.
Methods
Cell culture and transfection. HEK293T cells, BHK cells and ESCs were cultured and transfected as described by Schermelleh et al (2007), except FuGENE HD (Roche, Mannheim, Germany) was used for transfection of ESCs. The dnmt1−/− J1 ESCs used in this study were homozygous for the c allele (Lei et al, 1996). BHK cells were co-transfected on glass coverslips with GFP–Dnmt3 and Cherry–Np95 constructs using Transfectin (Bio-Rad, Munich, Germany) according to the manufacturer's instructions. Cell fixation and microscopy were carried out as described by Zolghadr et al (2008).
Co-immunoprecipitation. ESCs and HEK293T cell extracts were prepared in lysis buffer (20 mM Tris–HCl (pH 7.5), 0.5 mM EDTA, 2 mM phenylmethyl sulphonyl fluoride and 0.5% NP40) containing 150 or 300 mM NaCl (high-salt condition) and diluted with lysis buffer without NP40. GFP trap (Rothbauer et al, 2008) and a specific rabbit antiserum (Citterio et al, 2004) were used for immunoprecipitation of GFP fusions and endogenous Np95, respectively. GFP trap and protein G beads (Sigma, Taufkirchen, Germany) were washed with dilution buffer containing increasing salt concentrations (150 and 300 mM, or 300 and 500 mM NaCl for the high-salt condition) and re-suspended in SDS–PAGE sample buffer. The following mouse monoclonal antibodies were used for immunoblotting: anti-His (C-terminal, Invitrogen, Karlsruhe, Germany), anti-Dnmt3a (clone 64B1446, Imgenex, San Diego, CA, USA) and anti-Dnmt3b (clone 52A1018, Abcam, Cambridge, UK). Np95 was detected with the same antiserum used for immunoprecipitation and a rabbit antiserum was used for detection of Dnmt1 (Grohmann et al, 2005). Horseradish peroxidase-conjugated rabbit anti-mouse or goat anti-rabbit secondary antibodies (Sigma) and ECL Plus reagent (GE Healthcare, Munich, Germany) were used for detection.
Silencing assay. ESCs were co-transfected with pCAG-eGFP-IB and pCMV-mRFP as described above and images from live cells were acquired at the indicated time points with an InCell Analyser 1000 (GE Healthcare) using a × 20 air objective (NA=0.45) and standard filter settings for GFP and RFP. A total of 90–150 images were acquired for each channel, using the same exposure time throughout the time course. Cells were passaged every second day and images were taken 4–5 h after seeding. Images were analysed using ImageJ v1.42a software. To calculate fluorescent reporter expression, pictures were processed using a Gaussian blur algorithm (radius (sigma)=2), and a threshold for maximal signal and minimal background coverage was adjusted and applied to each channel (supplementary Fig S4A online). The threshold was converted into area selection and the total size of the selected area was measured.
DNA methylation analysis. ESCs were transfected as in the silencing assay with pCAG-eGFP-IB and pCMV-mRFP, and GFP-positive cells were sequentially sorted with a FACSVantage or FACSAria II (Becton Dickinson, Heidelberg, Germany) at days 2, 6 and 10 after transfection. After each sorting, total DNA was isolated using the QIAmp DNA Mini kit (Qiagen, Hilden, Germany) and bisulphite treated with the EZ DNA Methylation-Gold kit (Zymo research, Orange, CA, USA). The following primers were used for PCR amplification: CMV-forward TGGGATTTTTTTATTTGGTAGT; CMV-reverse ATGGGAGTTTGTTTTGGTATTA; CAG-forward GGAGAGGTGAGGAGGTAGTTAATTAGA and CAG-reverse CCCCAAACCCCTCAAAACTT. Pyrosequencing was carried out by Varionostic GmbH (Ulm, Germany).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We are grateful to the following colleagues for providing ESCs: M. Muto and H. Koseki for wt and np95−/− E14; E. Li and T. Chen for wt, dnmt1−/− and DKO J1; Masaki Okano for TKO J1; Gunnar Schotta for Suv39h1/2 dn; Y. Shinkai and T. Jenuwein for wt and G9a−/− TT2. We thank L. Schermelleh for help with image processing and K. Zolghadr for assistance with the InCell Analyzer. This study was supported by the Nanosystems Initiative Munich and BioImaging Network Munich, and by grants from the Deutsche Forschungsgemeinschaft to H.L.
Footnotes
The authors declare that they have no conflict of interest.
References
- Achour M et al. (2008) The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene 27: 2187–2197 [DOI] [PubMed] [Google Scholar]
- Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M (2008) Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455: 818–821 [DOI] [PubMed] [Google Scholar]
- Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S (2008) Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 455: 822–825 [DOI] [PubMed] [Google Scholar]
- Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21 [DOI] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve P-O, Clark A, Pradhan S, Jacobsen SE (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317: 1760–1764 [DOI] [PubMed] [Google Scholar]
- Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, Bonapace IM (2004) Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol Cell Biol 24: 2526–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong KB et al. (2008) DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J 27: 2691–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epsztejn-Litman S et al. (2008) De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol 15: 1176–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y (2006) G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol 8: 188–194 [DOI] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Deplus R, Kouzarides T (2003) The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res 31: 2305–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann M, Spada F, Schermelleh L, Alenina N, Bader M, Cardoso MC, Leonhardt H (2005) Restricted mobility of Dnmt1 in preimplantation embryos: implications for epigenetic reprogramming. BMC Dev Biol 5: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X (2008) The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 455: 826–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Esteve PO, Jacobsen SE, Pradhan S (2009) UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res 37: 493–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH (2003) Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 13: 1192–1200 [DOI] [PubMed] [Google Scholar]
- Lei H, Oh S, Okano M, Juttermann R, Goss K, Jaenisch R, Li E (1996) De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122: 3195–3205 [DOI] [PubMed] [Google Scholar]
- Leonhardt H, Page AW, Weier HU, Bestor TH (1992) A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71: 865–873 [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926 [DOI] [PubMed] [Google Scholar]
- Li JY et al. (2007) Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol Cell Biol 27: 8748–8759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JH, O'Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T (2005) The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J 24: 800–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257 [DOI] [PubMed] [Google Scholar]
- Papait R, Pistore C, Negri D, Pecoraro D, Cantarini L, Bonapace IM (2007) Np95 is implicated in pericentromeric heterochromatin replication and in major satellite silencing. Mol Biol Cell 18: 1098–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H (2008) A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics 7: 282–289 [DOI] [PubMed] [Google Scholar]
- Schermelleh L, Haemmer A, Spada F, Rosing N, Meilinger D, Rothbauer U, Cristina Cardoso M, Leonhardt H (2007) Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res 35: 4301–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J et al. (2007) The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450: 908–912 [DOI] [PubMed] [Google Scholar]
- Spada F, Haemmer A, Kuch D, Rothbauer U, Schermelleh L, Kremmer E, Carell T, Langst G, Leonhardt H (2007) DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J Cell Biol 176: 565–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Kubo E, Kanari Y, Ikemura T, Tatsumi K, Muto M (2000) Temporal and spatial localization of novel nuclear protein NP95 in mitotic and meiotic cells. Cell Struct Funct 25: 149–159 [DOI] [PubMed] [Google Scholar]
- Zolghadr K, Mortusewicz O, Rothbauer U, Kleinhans R, Goehler H, Wanker EE, Cardoso MC, Leonhardt H (2008) A fluorescent two-hybrid (F2H) assay for direct visualization of protein interactions in living cells. Mol Cell Proteomics 7: 2279–2287 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information