Abstract
Background
Age-related macular degeneration (AMD) is a prevalent cause of blindness in Western societies. Variants in the genes encoding complement factor H (CFH), complement component 3 (C3) and age-related maculopathy susceptibility 2 (ARMS2) have repeatedly been shown to confer significant risks for AMD; however, their role in disease progression and thus their potential relevance for interventional therapeutic approaches remains unknown.
Methodology/Principal Findings
Here, we analyzed association between variants in CFH, C3 and ARMS2 and disease progression of geographic atrophy (GA) due to AMD. A quantitative phenotype of disease progression was computed based on longitudinal observations by fundus autofluorescence imaging. In a subset of 99 cases with pure bilateral GA, variants in CFH (Y402H), C3 (R102G), and ARMS2 (A69S) are associated with disease (P = 1.6×10−9, 3.2×10−3, and P = 2.6×10−12, respectively) when compared to 612 unrelated healthy control individuals. In cases, median progression rate of GA over a mean follow-up period of 3.0 years was 1.61 mm2/year with high concordance between fellow eyes. No association between the progression rate and any of the genetic risk variants at the three loci was observed (P>0.13).
Conclusions/Significance
This study confirms that variants at CFH, C3, and ARMS2 confer significant risks for GA due to AMD. In contrast, our data indicate no association of these variants with disease progression which may have important implications for future treatment strategies. Other, as yet unknown susceptibilities may influence disease progression.
Introduction
Age-related macular degeneration (AMD [MIM 603075]) is the leading cause of blindness in Western countries [1]. Two major forms of advanced AMD are responsible for vision loss. Geographic atrophy (GA) or “dry” AMD is characterized by an extensive loss of the choriocapillaris and the overlying retinal pigment epithelium, while the neovascular form or “wet” AMD develops due to invasion of neovascular complexes, a process known as choroidal neovascularization (CNV) [2]–[4]. Although constituting only 10–15% of all AMD cases, CNV accounts for almost 80% of AMD-related blindness. Nevertheless, with an increasingly aging population, blindness due to GA is predicted to become a significant socioeconomic burden in the near future [5]–[7]. To date, successful therapeutic intervention is available only for active CNV, while GA still remains untreatable [8], [9].
AMD is a complex condition with both genetic and environmental factors contributing to disease [2], [3], [10], [11]. Genetic variants at two chromosomal loci, 1q31 [12]–[15] and 10q26 [16], [17] confer major disease risks, together likely accounting for over 50% of cases. Chromosome 1q31 association is tightly linked to the complement factor H (CFH) gene suggesting an important role of inflammation and the alternative pathway of complement in AMD pathogenesis. This is further supported by strong association of AMD with variants in the complement component 3 (C3 [MIM 120700]) gene [18], [19]. At 10q26, variants at two genes in strong linkage disequilibrium, namely age-related maculopathy susceptibility 2 (ARMS2 [MIM 611313]) [17], [20] and HtrA serine peptidase 1 (HTRA1 [MIM 602194]) [21], [22], were strongly linked to AMD susceptibility. Further studies will be needed however to clarify which of the two candidates plays the causal role in AMD pathology.
Going beyond cross-sectional association between AMD and genetic variants, Seddon et al. reported a significant association between the polymorphisms CFH-Y402H and ARMS2-A69S and progression from early or intermediate stages of AMD to the advanced forms [23]. It should be noted, however, that this study has not addressed the question of whether genetic variants are associated with disease progression once late AMD has developed. Nevertheless, it is the progression of the two late forms, GA and CNV, which is the most important variable with regard to future therapeutic intervention.
A quantitative measure of geographic atrophy progression represents the detection and quantification of atrophic areas on the fundus at different time points in order to calculate atrophy enlargement rates over time. The gold standard to define such areas has been fundus photography; however, the reproducibility for defining and measuring GA by fundus photography has been reported by several groups to be only moderate, particularly for smaller lesions [24]–[27]. An attractive alternative for non-invasive imaging of atrophy and its progression over time is fundus autofluorescence (FAF) imaging with the confocal scanning laser ophthalmoscope (cSLO). This method potentially allows direct visualization of loss of key cells in the disease process by tracing metabolic RPE changes. Therefore, areas of GA can now reliably be mapped facilitating absolute quantification of atrophic areas and their progression in the disease process [28]. Here, we used FAF imaging for phenotyping patients with GA in a longitudinal study, analyzed GA progression and correlated this biologically-based quantitative phenotype with genetic variants in the CFH, C3, and ARMS2 genes.
Results
Among the 619 participants of the FAM study cohort, 136 exhibited GA with no sign of CNV throughout subsequent follow-up examinations. Of these, 99 patients exhibited pure GA in both eyes throughout the study period and were included in the further analysis (for a comprehensive compilation of the phenotypic and genotypic data, see Table S1). Mean follow-up was 3.0 years (SD, 2.1; range, 6 months to 10 years). Mean number of measurements of GA area was 3.3 per eye (range, 2 to 12) and 3.5 per patient (range, 2 to 12). Mean age of patients was 71.8±7.4 years (range, 53–89 years) and 76.2±5.3 years (range, 65–97 years) for the controls (p<0.0001; t-test). Overall, 61% of the AMD patients and 62% of the controls were female (p = 0.837). Considering the most general measurement of smoking history as a binary “ever or never” variable, AMD patients were significantly more likely to smoke than controls (49% versus 13%; p<0.0001).
In a case-control association analysis (N = 99 cases of pure bilateral GA; N = 612 controls), polymorphisms Y402H (rs1061170) in CFH, A69S (rs10490924) in ARMS2 and R102G (rs2230199) in the C3 gene were strongly associated with GA due to AMD ( Table 1 ). In a case-only analysis (N = 99 GA cases), the median growth rate of GA was 1.61 mm2/year (interquartile range [IQR], 1.19 to 2.12; range, 0.26 to 3.45), which is very similar to our previous report [28]. A typical example of serial FAF images over a study period of 109 months is shown in Figure 1 . Median GA area at baseline was 6.50 mm2 (IQR, 3.26 to 11.03; range, 0.05 to 34.6). As a measure of concordance of the GA progression rate between the right and left eye, the concordance correlation coefficient [29] was calculated at 0.726, 95% CI [0.587; 0.8297] indicating a high intra-individual symmetry of the GA progression rate in our cohort.
Table 1. SNP association in 99 AMD patients with bilateral pure GA versus 612 matched controls.
| Gene (Marker) | Group | Genotypes (Frequency) | MAF | ATT P-value | ||
| CFH (rs1061170) | T/T | T/C | C/C | |||
| Cases | 18 (0.184) | 42 (0.429) | 38 (0.388) | 0.602 | ||
| Controls | 214 (0.350) | 327 (0.535) | 70 (0.115) | 0·382 | ||
| Odds Ratio [95% CI] | 1 [Ref.] | 1.53 [0.86,2.72] | 6.45 [3.46,12.03] | 1.63×10−9 | ||
| ARMS2 (rs10490924) | G/G | G/T | T/T | |||
| Cases | 36 (0.364) | 42 (0.424) | 21 (0.212) | 0.424 | ||
| Controls | 402 (0.658) | 184 (0.301) | 25 (0.041) | 0.191 | ||
| Odds Ratio [95% CI] | 1 [Ref.] | 2.55 [1.58,4.11] | 9.38 [4.79,18.39] | 2.58×10−12 | ||
| C3 (rs2230199) | G/G | G/C | C/C | |||
| Cases | 54 (0.557) | 35 (0.361) | 8 (0.082) | 0.263 | ||
| Controls | 394 (0.675) | 176 (0.301) | 14 (0.024) | 0.175 | ||
| Odds Ratio [95% CI] | 1 [Ref.] | 1.45 [0.92,2.30] | 4.17 [1.67,10.40] | 0.0032 | ||
Abbreviations: MAF, minor allele frequency; ATT, Armitage's trend test; CI, confidence interval.
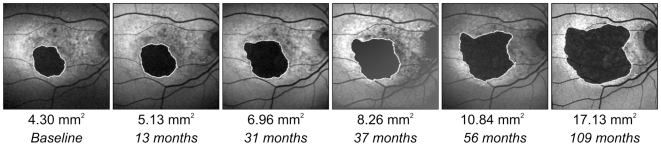
Figure 1. Progression of GA in a patient with age-related macular degeneration over a period of nine years.
Atrophic areas are outlined in each image. Baseline GA area was 4.3 mm2; GA progression rate was 1.50 mm2/year based on 12 observations within nine years of clinical follow-up. GA growth rate of the fellow eye was 1.59 mm2/year. The patient's genotype was C/T for CFH-rs1061170, G/G for ARMS2-rs10490924, and C/G for C3-rs2230199.
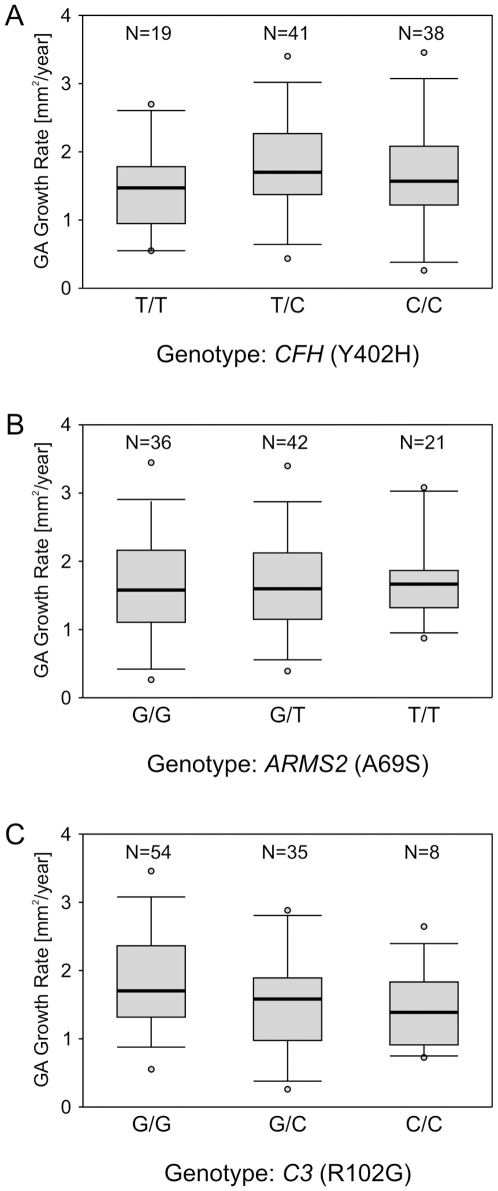
To visualize the distribution of GA growth rate for individual genotypes of CFH (Y402H), ARMS2 (A69S), and C3 (R102G), patients were grouped for genotypes of these three major risk genes ( Figure 2 ). The association between the quantitative trait variable “GA progression” and SNP alleles in the CFH, ARMS2 and C3 genes was then analyzed ( Table 2 ). Adjusting for age, smoking history and body mass index (BMI), and after a Sidak correction for multiple testing, none of the single-marker P values remained significant at alpha = 0.05. Similarly, there was no significant association between GA progression and the Y402H (rs1061170) and I62V (rs800292) haplotype of CFH ( Table 2 ). Power calculations revealed a power of 80% (at alpha = 0.05) for detecting an additive allele effect of 0.278 mm2/year (Y402H, CFH), 0.275 mm2/year (A69V, ARMS2) and 0.292 mm2/year (R102G, C3), respectively. There was 95% power to detect an effect of less than 0.4 mm2/year for any of the three genetic risk factors. A detailed power calculation is given in Table S2.
Figure 2. Growth rates of geographic atrophy for the subgroups of AMD patients grouped by genotypes for the three major risk alleles, CFH (A), ARMS2 (B), and C3 (C).
Shown are the median (central bar), the 25th and 75th percentiles (grey box), the 5th and 95th percentiles (bars) and the minimum and maximum (small circles).
Table 2. Association between the quantitative trait variable “geographic atrophy progression” and SNP alleles and haplotypes.
| Gene (Marker) | Allele/Haplotype | Frequency (N = 99) | Additive Allele Effect in mm2/year (95% CI)a | P valueb (unadjusted) | P valueb (adjusted for age, smoking and BMI) | P valueb (corrected for multiple testingc) |
| CFH (rs800292) | G | 172 (0.89) | Reference | |||
| A | 22 (0.11) | 0.0963 (−0.50, 0.70) | ||||
| 0.377 | 0.425 | 0.937 | ||||
| CFH (rs1061170) | T | 78 (0.40) | Reference | |||
| C | 118 (0.60) | 0.2055 (−0.19, 0.60) | ||||
| 0.156 | 0.097 | 0.4 | ||||
| CFH (rs800292–rs1061170) | G–T | 55 (0.29) | Reference | |||
| G–C | 115 (0.60) | 0.3048 (−0.15, 0.75) | ||||
| A–T | 22 (0.11) | 0.3301 (−0.36, 1.02) | ||||
| 0.376d | 0.204d | 0.68 | ||||
| ARMS2 (rs10490924) | G | 114 (0.58) | Reference | |||
| T | 84 (0.42) | 0.0776 (−0.31;0.47) | ||||
| 0.233 | 0.376 | 0.905 | ||||
| C3 (rs2230199) | G | 143 (0.73) | Reference | |||
| C | 51 (0.26) | −0.5141 (−0.98, −0.05) | ||||
| 1 | 1 | 1 |
Progression rates relative to the reference allele/haplotype (non risk) which was set by definition to 0 mm2/year.
Testing for a positive trend of progression (one-sided test).
Sidak correction for five tests (4 SNPs and haplotype test).
Two-sided test.
Discussion
Here, we demonstrate a significant association between our study group with late AMD manifestations of pure GA and known variants at CFH, C3, and ARMS2. The odds ratios for heterozygotes and homozygotes for any of the three genes are highly consistent with previous reports [18], [19], [30]. Those reports typically included mixed study populations including both forms of late AMD, CNV and GA. This was also true for recent reports of R102G in C3 as a significant risk variant for AMD where similar results were found for CNV and GA [18], [19]. DeWan and co-workers reported HTRA1 as a risk gene for CNV due to AMD, implicating that polymorphisms in the ARMS2-HTRA1 region are responsible for this specific phenotype of late AMD. This is a surprising notion since others, including our own study, did not find any difference in the association data between GA and CNV due to AMD [17]. Our current data now unambiguously demonstrate that the risk for GA is similarly associated with ARMS2-HTRA1 variants and suggests that the association between the CFH and ARMS2 polymorphisms is independent of disease evolution towards the atrophic or the neovascular form of AMD.
A significant original finding of this study is that none of the AMD-associated polymorphisms in CFH, C3 and ARMS2 contribute to GA progression once GA has developed. It is of note that our analysis had good power to detect an additive allele effect of less than 0.3 mm2 of GA progression per year. A GA progression rate of 0.3 mm2 or less represents a relatively small effect with less than 1/5 of the mean effect in our study population. Moreover, it is clearly below the threshold of recent phase II studies with therapeutic intervention in GA (e.g. fenretinide trial; ClinicalTrials.gov identifier: NCT00429936) which aim to slow GA progression by at least 0.6 mm2/year.
Although the number of patients (N = 99; baseline, N = 619) may appear limited, the significance of this study mainly originates from its dependency on the major outcome variable which is a biologically based continuous variable. To put the size of our patient study group into perspective, the major population-based studies such as the Beaver Dam Eye Study as well as the major clinical trial sponsored by the National Eye Institute (e.g. Age-Related Eye Disease Study, AREDS) all include similarly limited numbers of longitudinally phenotyped patients with pure bilateral GA. Specifically, AREDS has 70 such patients (baseline, N = 3640) [31], while the Beaver Dam Eye Study has only 27 patients of at least 5-year follow-up (baseline, N = 4926; Ronald Klein, personal communication).
The strength of the current data is based on the phenotypic assessment of GA by cSLO-based FA imaging, semi-automated image analysis and complex statistics condensing longitudinal data into one continuous phenotypic variable. Although its accuracy may improve with the number of measurements over time, we have previously shown that GA progression is linear, which makes two consecutive measurements sufficient to estimate GA growth rate, although the number of longitudinal observations improves the accuracy of the estimated GA progression rate. [28], [32] An additional strong point in this study is the substantial number of patients with bilateral pure GA recruited through the multicenter FAM study.
Strong linkage disequilibrium (LD) across the ARMS2-HTRA1 region makes it difficult to distinguish between ARMS2 [16], [17] and HTRA1 [21], [22] as causal factors in AMD pathogenesis. We recently identified a deletion-insertion polymorphism in ARMS2 (*372_815del443ins54) that is in almost perfect LD with the A69S variant and directly affects the stability of the transcript by removing the polyadenylation signal and inserts a 54-bp element known to mediate rapid mRNA turnover. [20] Immunohistochemistry has associated ARMS2 with mitochondria [33], specifically within the photoreceptor inner segments [20], suggesting that mechanisms of mitochondrial dysfunction may be involved in AMD pathogenesis.
Experimental [34] and clinical [35] data propose a key role of the major lipofuscin fluorophore A2-E in GA progression. Interestingly, A2-E biogenesis was recently linked to complement activation [36]. There is now compelling evidence that the complement system is involved in the pathogenesis of AMD and that the Y402H variant of CFH is associated with AMD susceptibility. Our data confirm this finding specifically for GA. Recently, we have detected systemic complement activation in AMD and found that markers of chronic complement activation were associated with the CFH risk haplotype (including the Y402H polymorphism). [37] However, since the Y402H variant of CFH confers similar risk of soft drusen and both forms of advanced AMD, it may contribute to the increased risk of advanced AMD largely or entirely through its impact on precursors of visually disabling AMD such as drusen. [38] Since polymorphisms in CFH, C3, and ARMS2 are not associated with progression of GA and since our previous analysis did not detect a significant contribution of other characteristics (including smoking and BMI) [28] other modifying genetic factors may be involved.
Our findings may have strong implications for future efforts to design therapeutic interventions for AMD, although replication in an independent cohort and empirical treatment data are needed for further substantiation. Since variants within CFH, C3, and ARMS2 confer risk for susceptibility of GA but appear unrelated to its progression, addressing the complement cascade or ARMS2-, and possibly HTRA1-associated pathways per se may not be promising strategies for alleviating disease progression once GA has developed. Instead, defining the exogenic and possibly the genetic factors responsible for GA progression could be crucial in helping those patients already suffering from advanced stages of the disease.
Methods
Ethics Statement
The study followed the tenets of the Declaration of Helsinki and was approved by the local Ethics Review Board at the University of Bonn. Informed written consent was obtained from each patient after explanation of the nature and possible consequences of the study.
Cases and Controls
Patients with GA secondary to AMD were included from the longitudinal natural history arm of the multicenter FAM–Study (Fundus Autofluorescence in Age-Related Macular Degeneration; registration www.clinicaltrials.gov: NCT00393692). The study procedures have been previously reported. [28], [39] A total of 619 patients with dry (early or late) AMD in the study eye were enrolled (mean age, 73.9 years; mean follow-up, 35 months) and were recruited at six centres throughout Germany. Because of differences in genotype frequencies due to race and geographical origin, only white probands originating exclusively from Germany were included in the study.
Patients with uni- or multifocal GA and with clear vitreous to allow FAF imaging were included in the analysis. Exclusion criteria were a history of retinal surgery, laser photocoagulation and radiation therapy or other retinal diseases in the study eye such as diabetic retinopathy or hereditary retinal dystrophies. Fluorescein angiography was only performed if funduscopic signs were present indicative of neovascular AMD in addition to patches of GA. Such eyes were excluded from further analysis.
612 unrelated control individuals of German origin served as controls for case-control analysis to investigate the role of variants in CFH, C3 and ARMS2 for AMD susceptibility. [17] Each control subject underwent a single ophthalmic examination including visual acuity, slit lamp biomicroscopy and fundus ophthalmoscopy. Patients and controls underwent a general health interview protocol including a standardized smoking history.
Phenotyping by FAF Imaging
FAF was measured using a confocal laser scanning ophthalmoscope (cSLO; Heidelberg Retina Angiograph, HRA classic and HRA 2, Heidelberg Engineering, Dossenheim, Germany), the optical and technical principles of which have been described previously. [40] An argon blue laser (HRA classic) or optically pumped solid state laser (HRA 2) were used for excitation (both 488 nm). The emitted light above 500 nm was detected with a barrier filter. FAF images were recorded in accordance with standard operating procedures (SOP) including focussing in the blue reflection mode, acquisition of a series of 30°×30° images (488 nm) and calculation of mean images after automated alignment in order to amplify signal to noise ratio with image analysis software (Heidelberg Eye Explorer, Heidelberg Engineering, Dossenheim, Germany). [41]
Only patient eyes with at least two follow-ups in a 6 month interval and with sufficient image quality to accurately determine the size of atrophy were included. To further improve on our previous analytical strategy [28], [32], serial FAF images of the same eye were aligned by the “4-point-alignment” function using Picture Window Pro 4.0.1.2 analysis software (Digital Light & Color, Cambridge, MA, USA). The total size of GA was measured in the processed FAF images by automated imaging analysis software that uses region-growing techniques to segment the areas of GA. [42] This procedure significantly reduced the estimated within-group error variance from 0.761 mm2/year, 95% CI [0.645; 0.898] previously [32] to 0.212 mm2/year, 95% CI [0.182; 0.246].
Genotyping
Genomic DNA was extracted from peripheral blood leukocytes according to established protocols. Genotyping was done by TaqMan SNP Genotyping or by the matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry method (Sequenom, San Diego, CA, USA). TaqMan Pre-Designed SNP Genotyping Assays (Applied Biosystems, Foster City, CA, USA) were performed according to the manufacturer's instructions and were analyzed with a 7900HT Fast Real-Time PCR System (Applied Biosystems). All SNPs showed high genotyping quality with an average call rate of 98.5%.
Statistical Analysis
Data management and statistical analysis were conducted using SAS Version 9.1.3 (Cary, NC, USA). The statistical analyses were carried out using software package R, Version 2.7.0 (http://www.r-project.org/), and the R-library “non-linear mixed effects” (NLME) was applied for hierarchical regression modelling. A linear mixed effects model was used to quantify GA growth. A detailed description of the longitudinal modelling process is provided elsewhere. [32], [43] The two-level-random effects model separates ocular-specific as well as patient-specific effects and helps to handle related observations. The mixed model methodology allows the study of variance components and fixed effects simultaneously. Using this two-level-random effects model, a single variable of GA progression rate was computed based on longitudinal observations of either one or both eyes for each patient studied. [32] This variable represents a continuous, biology-based quantitative phenotype and was used for association analysis with genetic data.
We used the case-control tool box from FAMHAP to perform quality control of the genetic data. [44] SNP genotypes in cases and controls showed no deviation from Hardy-Weinberg-Equilibrium at a level of alpha = 0.001. UNPHASED [45] was used to test for association between the quantitative trait variable “GA progression” and SNP alleles and haplotypes. We adopted the standard practice of genetic epidemiological studies to test in an allele-based manner when the underlying disease model is unknown. This has the advantage of reducing the number of degrees of freedom to 1. Although the resulting test is additive by nature, it is the most powerful test under a much broader range of true disease scenarios. The UNPHASED program provides P values as well as additive effect estimators for the effects of alleles/haplotypes on the quantitative trait. The reference alleles represent the respective major SNP alleles within Caucasian populations. The respective confidence intervals are provided as well. We also adjusted for the covariates age, BMI, and smoking, using the “modifiers” option of the UNPHASED software package. Posthoc power values for a t-test as an approximation of power for quantitative trait analysis for single SNPs were computed with the R software package, with the two groups of the t-test constituting the two alleles and the group sizes the allele counts. Covariates were not accounted for in the power calculation. Since the disease-associated allele is known, one-sided power values were computed.
Supporting Information
Clinical and molecular genetic findings in 99 patients with late-stage AMD
(0.27 MB DOC)
Power values for alpha = 0.05 and additive allele effects of growth of geographic atrophy (n = 99)
(0.04 MB DOC)
Acknowledgments
We are grateful to the patients and control subjects for their participation in this study.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported in part by grants from the European Commission (EU FP6) (HPNS, FGH), the Integrated Project “EVIGENORET” (LSHG-CT-2005-512036) (HPNS, FGH); the German Research Foundation (DFG) Heisenberg Fellowship SCHO 734/2-1 (HPNS); the DFG Priority Research Programme Age-related Macular Degeneration (SPP1088) (FGH); DFG grants HO1926/1-3 (FGH); WE1259/18-1 (BHFW), and WE1259/19-1 (BHFW), a Research Grant from the German Society of Ophthalmology (DOG) 2006/2007 (MF); the Kroener Foundation (Germering, Germany); The Ruth and Milton Steinbach Foundation New York (BHFW) and the Alcon Research Institute (BHFW). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Congdon N, O'Colmain B, Klaver CC, Klein R, Munoz B, et al. Causes and prevalence of visual impairment among adults in the United States. Arch Ophthalmol. 2004;122:477–485. doi: 10.1001/archopht.122.4.477. [DOI] [PubMed] [Google Scholar]
- 2.de Jong PT. Age-related macular degeneration. N Engl J Med. 2006;355:1474–1485. doi: 10.1056/NEJMra062326. [DOI] [PubMed] [Google Scholar]
- 3.Holz FG, Pauleikhoff D, Klein R, Bird AC. Pathogenesis of lesions in late age-related macular disease. Am J Ophthalmol. 2004;137:504–510. doi: 10.1016/j.ajo.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 4.Jager RD, Mieler WF, Miller JW. Age-related macular degeneration. N Engl J Med. 2008;358:2606–2617. doi: 10.1056/NEJMra0801537. [DOI] [PubMed] [Google Scholar]
- 5.Friedman DS, O'Colmain BJ, Munoz B, Tomany SC, McCarty C, et al. Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol. 2004;122:564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 6.Klein R, Klein BE, Knudtson MD, Meuer SM, Swift M, et al. Fifteen-year cumulative incidence of age-related macular degeneration: the Beaver Dam Eye Study. Ophthalmology. 2007;114:253–262. doi: 10.1016/j.ophtha.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 7.Klein R, Peto T, Bird A, Vannewkirk MR. The epidemiology of age-related macular degeneration. Am J Ophthalmol. 2004;137:486–495. doi: 10.1016/j.ajo.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 8.Brown DM, Kaiser PK, Michels M, Soubrane G, Heier JS, et al. Ranibizumab versus verteporfin for neovascular age-related macular degeneration. N Engl J Med. 2006;355:1432–1444. doi: 10.1056/NEJMoa062655. [DOI] [PubMed] [Google Scholar]
- 9.Rosenfeld PJ, Brown DM, Heier JS, Boyer DS, Kaiser PK, et al. Ranibizumab for neovascular age-related macular degeneration. N Engl J Med. 2006;355:1419–1431. doi: 10.1056/NEJMoa054481. [DOI] [PubMed] [Google Scholar]
- 10.Haddad S, Chen CA, Santangelo SL, Seddon JM. The genetics of age-related macular degeneration: a review of progress to date. Surv Ophthalmol. 2006;51:316–363. doi: 10.1016/j.survophthal.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Scholl HP, Fleckenstein M, Charbel Issa P, Keilhauer C, Holz FG, et al. An update on the genetics of age-related macular degeneration. Mol Vis. 2007;13:196–205. [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards AO, Ritter R, 3rd, Abel KJ, Manning A, Panhuysen C, et al. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 13.Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 15.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jakobsdottir J, Conley YP, Weeks DE, Mah TS, Ferrell RE, et al. Susceptibility genes for age-related maculopathy on chromosome 10q26. Am J Hum Genet. 2005;77:389–407. doi: 10.1086/444437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivera A, Fisher SA, Fritsche LG, Keilhauer CN, Lichtner P, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14:3227–3236. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 18.Maller JB, Fagerness JA, Reynolds RC, Neale BM, Daly MJ, et al. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat Genet. 2007;39:1200–1201. doi: 10.1038/ng2131. [DOI] [PubMed] [Google Scholar]
- 19.Yates JR, Sepp T, Matharu BK, Khan JC, Thurlby DA, et al. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357:553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- 20.Fritsche LG, Loenhardt T, Janssen A, Fisher SA, Rivera A, et al. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat Genet. 2008;40:892–896. doi: 10.1038/ng.170. [DOI] [PubMed] [Google Scholar]
- 21.Dewan A, Liu M, Hartman S, Zhang SS, Liu DT, et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314:989–992. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- 22.Yang Z, Camp NJ, Sun H, Tong Z, Gibbs D, et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314:992–993. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- 23.Seddon JM, Francis PJ, George S, Schultz DW, Rosner B, et al. Association of CFH Y402H and LOC387715 A69S with progression of age-related macular degeneration. JAMA. 2007;297:1793–1800. doi: 10.1001/jama.297.16.1793. [DOI] [PubMed] [Google Scholar]
- 24.Davis MD, Gangnon RE, Lee LY, Hubbard LD, Klein BE, et al. The Age-Related Eye Disease Study severity scale for age-related macular degeneration: AREDS Report No. 17. Arch Ophthalmol. 2005;123:1484–1498. doi: 10.1001/archopht.123.11.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sunness JS, Bressler NM, Tian Y, Alexander J, Applegate CA. Measuring geographic atrophy in advanced age-related macular degeneration. Invest Ophthalmol Vis Sci. 1999;40:1761–1769. [PubMed] [Google Scholar]
- 26.Pirbhai A, Sheidow T, Hooper P. Prospective evaluation of digital non-stereo color fundus photography as a screening tool in age-related macular degeneration. Am J Ophthalmol. 2005;139:455–461. doi: 10.1016/j.ajo.2004.09.077. [DOI] [PubMed] [Google Scholar]
- 27.Scholl HP, Peto T, Dandekar S, Bunce C, Xing W, et al. Inter- and intra-observer variability in grading lesions of age-related maculopathy and macular degeneration. Graefes Arch Clin Exp Ophthalmol. 2003;241:39–47. doi: 10.1007/s00417-002-0602-8. [DOI] [PubMed] [Google Scholar]
- 28.Holz FG, Bindewald-Wittich A, Fleckenstein M, Dreyhaupt J, Scholl HP, et al. Progression of geographic atrophy and impact of fundus autofluorescence patterns in age-related macular degeneration. Am J Ophthalmol. 2007;143:463–472. doi: 10.1016/j.ajo.2006.11.041. [DOI] [PubMed] [Google Scholar]
- 29.Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989;45:255–268. [PubMed] [Google Scholar]
- 30.Schaumberg DA, Hankinson SE, Guo Q, Rimm E, Hunter DJ. A prospective study of 2 major age-related macular degeneration susceptibility alleles and interactions with modifiable risk factors. Arch Ophthalmol. 2007;125:55–62. doi: 10.1001/archopht.125.1.55. [DOI] [PubMed] [Google Scholar]
- 31.The AREDS Research Group. Change in Area of Geographic Atrophy in the Age-Related Eye Disease Study: AREDS Report Number 26. Arch Ophthalmol. 2009;127:1168–1174. doi: 10.1001/archophthalmol.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dreyhaupt J, Mansmann U, Pritsch M, Dolar-Szczasny J, Bindewald A, et al. Modelling the natural history of geographic atrophy in patients with age-related macular degeneration. Ophthalmic Epidemiol. 2005;12:353–362. doi: 10.1080/09286580591005723. [DOI] [PubMed] [Google Scholar]
- 33.Kanda A, Chen W, Othman M, Branham KE, Brooks M, et al. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci U S A. 2007;104:16227–16232. doi: 10.1073/pnas.0703933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergmann M, Schutt F, Holz FG, Kopitz J. Inhibition of the ATP-driven proton pump in RPE lysosomes by the major lipofuscin fluorophore A2-E may contribute to the pathogenesis of age-related macular degeneration. FASEB J. 2004;18:562–564. doi: 10.1096/fj.03-0289fje. [DOI] [PubMed] [Google Scholar]
- 35.Holz FG, Bellman C, Staudt S, Schutt F, Volcker HE. Fundus autofluorescence and development of geographic atrophy in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2001;42:1051–1056. [PubMed] [Google Scholar]
- 36.Zhou J, Jang YP, Kim SR, Sparrow JR. Complement activation by photooxidation products of A2E, a lipofuscin constituent of the retinal pigment epithelium. Proc Natl Acad Sci U S A. 2006;103:16182–16187. doi: 10.1073/pnas.0604255103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scholl HP, Charbel Issa P, Walier M, Janzer S, Pollok-Kopp B, et al. Systemic complement activation in age-related macular degeneration. PLoS ONE. 2008;3:e2593. doi: 10.1371/journal.pone.0002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magnusson KP, Duan S, Sigurdsson H, Petursson H, Yang Z, et al. CFH Y402H confers similar risk of soft drusen and both forms of advanced AMD. PLoS Med. 2006;3:e5. doi: 10.1371/journal.pmed.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bindewald A, Schmitz-Valckenberg S, Jorzik JJ, Dolar-Szczasny J, Sieber H, et al. Classification of abnormal fundus autofluorescence patterns in the junctional zone of geographic atrophy in patients with age related macular degeneration. Br J Ophthalmol. 2005;89:874–878. doi: 10.1136/bjo.2004.057794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holz FG, Bellmann C, Rohrschneider K, Burk RO, Volcker HE. Simultaneous confocal scanning laser fluorescein and indocyanine green angiography. Am J Ophthalmol. 1998;125:227–236. doi: 10.1016/s0002-9394(99)80095-6. [DOI] [PubMed] [Google Scholar]
- 41.Schmitz-Valckenberg S, Bultmann S, Dreyhaupt J, Bindewald A, Holz FG, et al. Fundus autofluorescence and fundus perimetry in the junctional zone of geographic atrophy in patients with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2004;45:4470–4476. doi: 10.1167/iovs.03-1311. [DOI] [PubMed] [Google Scholar]
- 42.Deckert A, Schmitz-Valckenberg S, Jorzik J, Bindewald A, Holz FG, et al. Automated analysis of digital fundus autofluorescence images of geographic atrophy in advanced age-related macular degeneration using confocal scanning laser ophthalmoscopy (cSLO). BMC Ophthalmol. 2005;5:8. doi: 10.1186/1471-2415-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dreyhaupt J, Dolar-Szczasny J, Bindewald A, Holz FG, Mansmann U. Discovery of factors influencing the growth of geographic atrophy in patients with age-related macular degeneration. Methods Inf Med. 2007;46:432–439. doi: 10.1160/me0328. [DOI] [PubMed] [Google Scholar]
- 44.Becker T, Cichon S, Jonson E, Knapp M. Multiple testing in the context of haplotype analysis revisited: application to case-control data. Ann Hum Genet. 2005;69:747–756. doi: 10.1111/j.1529-8817.2005.00198.x. [DOI] [PubMed] [Google Scholar]
- 45.Dudbridge F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered. 2008;66:87–98. doi: 10.1159/000119108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical and molecular genetic findings in 99 patients with late-stage AMD
(0.27 MB DOC)
Power values for alpha = 0.05 and additive allele effects of growth of geographic atrophy (n = 99)
(0.04 MB DOC)