Abstract
Background:
Given the increasing number of indications for liver surgery and the growing complexity of operations, many trainees in surgical, imaging and related subspecialties require a good working knowledge of the complex intrahepatic anatomy. Computed tomography (CT), the most commonly used liver imaging modality, enhances our understanding of liver anatomy, but comprises a two-dimensional (2D) representation of a complex 3D organ. It is challenging for trainees to acquire the necessary skills for converting these 2D images into 3D mental reconstructions because learning opportunities are limited and internal hepatic anatomy is complicated, asymmetrical and variable. We have created a website that uses interactive 3D models of the liver to assist trainees in understanding the complex spatial anatomy of the liver and to help them create a 3D mental interpretation of this anatomy when viewing CT scans.
Methods:
Computed tomography scans were imported into DICOM imaging software (OsiriX™) to obtain 3D surface renderings of the liver and its internal structures. Using these 3D renderings as a reference, 3D models of the liver surface and the intrahepatic structures, portal veins, hepatic veins, hepatic arteries and the biliary system were created using 3D modelling software (Cinema 4D™).
Results:
Using current best practices for creating multimedia tools, a unique, freely available, online learning resource has been developed, entitled Visual Interactive Resource for Teaching, Understanding And Learning Liver Anatomy (VIRTUAL Liver) (http://pie.med.utoronto.ca/VLiver). This website uses interactive 3D models to provide trainees with a constructive resource for learning common liver anatomy and liver segmentation, and facilitates the development of the skills required to mentally reconstruct a 3D version of this anatomy from 2D CT scans.
Discussion:
Although the intended audience for VIRTUAL Liver consists of residents in various medical and surgical specialties, the website will also be useful for other health care professionals (i.e. radiologists, nurses, hepatologists, radiation oncologists, family doctors) and educators because it provides a comprehensive resource for teaching liver anatomy.
Keywords: 3D liver model, web-based learning, hepatic anatomy, surgical anatomy
Introduction
Young surgical trainees experience a steep learning curve in the process of acquiring even a rudimentary understanding of hepatic anatomy, as formal learning opportunities are limited and only basic liver anatomy is taught to medical students.1 Hepatic surgical procedures require the surgeon to be sufficiently comfortable with anatomy to be able to rely on his or her own three-dimensional (3D) mental reconstruction of the 2D images provided by computed tomography (CT) scans. Interpreting CT scans is difficult and students find it a frustrating task that hinders their ability to optimize their educational experiences on hepatobiliary surgical rotations.2,3
It is difficult to demonstrate the superiority of online education resources compared with traditional methods of teaching,4 However, there is considerable evidence that web-based learning is a preferred method for students.5–7 This modality allows users to control content, learning pace and learning sequence,8 and can also be accessed by large numbers of students, on demand.9 Because learning increases with repeated exposure that is distributed over time,10 the utility of a teaching tool will increase if it can be accessed as needed in the clinical setting. Provided that online tutorials are well designed, from both an instructional and user-centred perspective,11–13 they are effective teaching tools and have been shown to improve subsequent face-to-face teaching sessions.14 Such a tool would have particular value for surgical residents engaged in interpreting CT scans during clinical sessions with surgeons.
We describe the creation of a website referred to as the Visual Interactive Resource for Teaching, Understanding And Learning Liver Anatomy (VIRTUAL Liver), which uses interactive 3D models to aid understanding of complex spatial internal hepatic anatomy and to assist trainees in creating a 3D mental interpretation of this anatomy from CT scans.
Materials and methods
Although there are no specific guidelines for designing and constructing web-based 3D interactive teaching tools, there are a number of recommendations11–13 describing current best practices for producing high-quality, cognitively efficient, multimedia learning tools. These practices were used to guide the development of the VIRTUAL Liver website.
Website development
The development of VIRTUAL Liver included the following two simultaneous steps:
building an accurate 3D model of the liver, and
constructing the website.
3D models
Although there are a number of methods for generating spatially accurate 3D models of anatomy, we required a model of common liver anatomy that could be directly related to a specific CT dataset. These CT scans were acquired from a library of scans of healthy individuals with no underlying liver disease. The scans were reviewed to select the most representative example of common hepatic anatomy. These CT images were imported into an open-source DICOM imaging software (OsiriX™; Geneva, Switzerland) to produce 3D surface renderings of the liver surface, hepatic veins, portal veins, hepatic arteries and biliary system. The surface renderings were then imported into 3D modelling software (MAXON Cinema 4D™ R10; Maxon Computer, Newbury Park, CA, USA) and used as a template to build accurate and informative models (Fig. 1). Second- and third-order divisions were modelled of each internal structure. Surface anatomy features that cannot be visualized using CT scans (e.g. the falciform ligament) were modelled according to anatomical atlases and by viewing livers from a medical anatomy museum at the University of Toronto.
Figure 1.
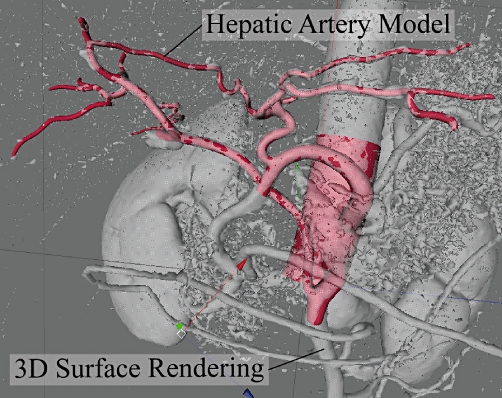
3D surface renderings developed in OsiriX™ are imported into Cinema4D™ and used as a template for building spatially accurate models of liver structures, such as the hepatic artery shown here
Website construction
Adobe Flash CS3™ (Adobe Systems, Inc., San Jose, CA, USA) was chosen to create the user interface and interactivity because of its capabilities for producing rich Internet content, interactive elements and web-based animations15 and because the Flash Player™ is available for most computer systems and is the most prevalent web browser software plug-in worldwide.16
The user interface of VIRTUAL Liver was designed to reduce cognitive load17 by providing a simple, refined user interface, presenting the content in discrete digestible chunks, presenting visuals simultaneously with text for increased comprehension and ease of reference, and integrating interactive elements to engage the user.
Results
The design, development and implementation of a web-based 3D interactive teaching model of surgical liver anatomy entitled VIRTUAL Liver has been completed (http://pie.med.utoronto.ca/VLiver).
When users first enter the application they are presented with a simple, easily navigable interface (Fig. 2). To the right is a welcome screen stating the purpose of the site; this is replaced by the 3D model once users begin working with the application. In the top right-hand corner is a glossary and help button. On the left-hand side of the screen is a menu bar with which users can navigate through the content of the VIRTUAL Liver application. The content has been broken down into two main sections designated Common Anatomy and Liver Segmentation, respectively.
Figure 2.
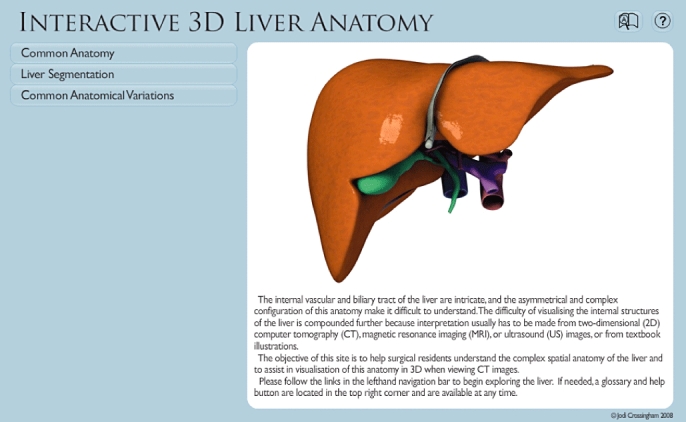
The VIRTUAL Liver website homepage (http://pie.med.utoronto.ca/VLiver/), showing the navigational toolbar located on the left and help tools located in the top right-hand corner
The Common Anatomy section (Fig. 3) is designed to assist learners in developing their understanding of basic hepatic anatomy. Its material is divided into five main categories: liver surface; portal veins; hepatic veins; hepatic arteries, and biliary system. The anatomy can be explored in the following ways:
Figure 3.
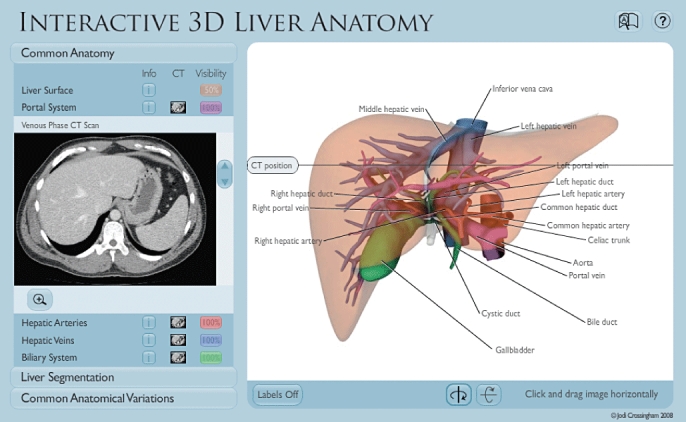
The Common Anatomy section allows users to explore common liver anatomy, and to view text or computed tomography images while interacting with the 3D liver model
A 3D liver model is available to review liver anatomy. Users are able to interact with this model to gain a deeper understanding of the anatomy. The model includes the following features: it can be rotated 360 degrees in both the vertical and horizontal axes to enable understanding of spatial relationships; the liver surface can be made transparent to show the intrahepatic structures in relation to the liver surface; each of the intrahepatic structures can be isolated for better analysis of its configuration, and labels can be applied to the model to aid in the identification of anatomy.
Users can view text that describes anatomical and surgically relevant information. The text is opened in the left-hand menu bar under the relevant subheadings, which allows for simultaneous interaction with the 3D liver model and reading of the text.
CT scans are available for each phase, including venous, arterial and biliary phases. These are located in the left-hand menu bar and can be viewed alongside the liver model. These are the same CTs that were used to create the 3D model and thus are directly relatable. As users scroll through the CT images, a moving line shows the position of the CT on the 3D liver model. This helps enhance 3D understanding of the structures shown in the CT scans.
Once an understanding of common hepatic anatomy has been obtained, users of the VIRTUAL Liver website can next explore Liver Segmentation. This section contains two modules: the first describes the eight Couinaud liver segments (Fig. 4), and the second discusses the nine common liver resections (Fig. 5). The Liver Segmentation section includes the following features:
Figure 4.
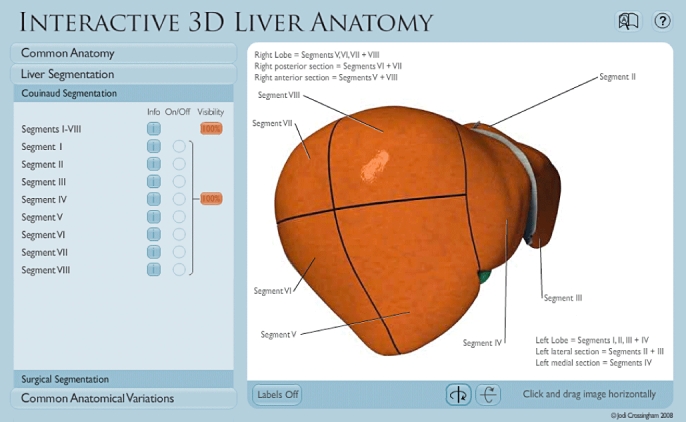
The Couinaud Segmentation module in the Liver Segmentation section allows users to examine the eight liver segments through text and interaction with the liver model
Figure 5.
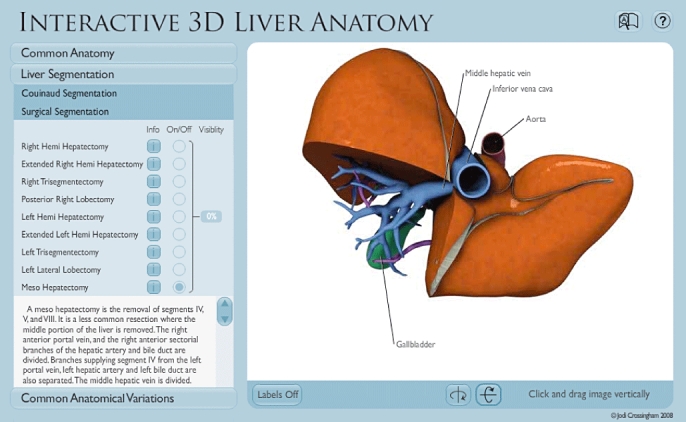
The Surgical Segmentation module in the Liver Segmentation section discusses nine different common resections, showing users the portion of liver that is removed and vessels that need to be divided during the procedure
Depending on which module is being viewed, the 3D model can be divided to show the eight Couinaud segments or the nine common liver resections. The model includes the following features: it can be rotated 360 degrees in both the vertical and horizontal axes; each segment can be isolated and made transparent to show the vessels and ducts that supply each segment, and component structures can be labelled.
Descriptive text can be viewed in the left-hand menu bar. The content is pertinent to each of the two liver segmentation modules.
The VIRTUAL Liver website was posted online in September 2008 (http://pie.med.utoronto.ca/VLiver). Analysis of web logs was used to track online use and showed that, at the end of February 2009, 5044 visitors from 51 countries had entered the site, including approximately 44 visitors per day during the month of February.
Discussion
Interpretation of 3D liver anatomy from 2D CT scans is a difficult and frustrating task for residents. Using current best practices for designing and developing multimedia learning tools, we have created a sophisticated, web-based, 3D interactive educational tool to facilitate the learning of common hepatic anatomy and the interpretation of this anatomy from CT scans.
The use of OsiriX™ was instrumental in producing spatially accurate 3D models, but using this method has some limitations. The quality of surface rendering obtained is directly dependent on the quality of the CT scans used. For example, poor surface renderings are obtained if there is not enough contrast between a desired vessel structure and the surrounding tissue. Even if high-quality CTs are acquired, details visible in the CT are often lost. This occurs if structures are too close together (i.e. the portal vein and hepatic vein in a venous phase CT), which sometimes causes them to join together in the 3D surface rendering. When OsiriX™ tries to translate small structures, such as the delicate sectorial branches of the hepatic artery, the surface renderings will break apart in these areas, making it difficult to interpret their flow.
The necessity of the CT to accurately match the 3D model required a CT dataset with arterial and venous phases as well as a CT cholangiogram from the same patient. Because CT cholangiograms are rarely performed, the pool of potential CT datasets was limited. The CT most representative of normal anatomy was chosen, with the recognition that it included anatomical variants, as noted in the website text.
By building an educational website using current best practices, we have constructed a potentially valuable and engaging learning resource. Future development of the site will include a section covering common anatomical variations and a step-by-step teaching tutorial that will take the learner through specific modules and offer a quiz after each one. Studies are underway to assess the face and content validity and usability of the VIRTUAL Liver website, all of which are important to refining the content of the site. A final website evaluation using an experimental study design will determine the effectiveness of VIRTUAL Liver as an educational resource.
VIRTUAL Liver was created as a learning aid primarily for surgical residents and is intended to assist their understanding of 3D hepatic anatomy and to aid their ability to interpret this anatomy from CT images. We believe it will be a valuable tool for all health care professionals who require knowledge of intrahepatic anatomy. It is also potentially useful for educators teaching liver anatomy and for health professionals educating patients prior to liver surgery.
Conflicts of interest
None declared.
References
- 1.Jurgaitis J, Paškonis M, Pivoriūnas J, Martinaitytè I, Juška A, Jurgitienè R, et al. The comparison of 2-dimensional with 3-dimensional hepatic visualization in the clinical hepatic anatomy education. Medicina. 2008;44:428–438. [PubMed] [Google Scholar]
- 2.Heylings DJ. Anatomy 1999–2000: the curriculum, who teaches it and how? Med Educ. 2002;36:702–710. doi: 10.1046/j.1365-2923.2002.01272.x. [DOI] [PubMed] [Google Scholar]
- 3.Marks SC., Jr Recovering the significance of 3-dimensional data in medical education and clinical practice. Clin Anat. 2001;14:90–91. doi: 10.1002/1098-2353(200101)14:1<90::AID-CA1014>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 4.Cook DA. Where are we with web-based learning in medical education? Med Teach. 2007;28:594–598. doi: 10.1080/01421590601028854. [DOI] [PubMed] [Google Scholar]
- 5.Granger NA, Calleson DC, Henson OW, Juliano E, Wineski L, McDaniel MD, et al. Use of web-based materials to enhance anatomy instruction in the health sciences. Anat Rec. 2006;2898:121–127. doi: 10.1002/ar.b.20104. [DOI] [PubMed] [Google Scholar]
- 6.Hopkins MA, Nachbar M, Kalet A. Educational imperatives drive technological advancement in surgery clerkship. Med Educ. 2004;38:1186–1187. doi: 10.1111/j.1365-2929.2004.01993.x. [DOI] [PubMed] [Google Scholar]
- 7.Yeazel MW, Center BA. Demonstration of the effectiveness and acceptability of self-study module use in residency education. Med Teach. 2004;26:57–62. doi: 10.1080/0142159032000150476. [DOI] [PubMed] [Google Scholar]
- 8.Ruiz JG, Mintzer MJ, Leipzig RM. The impact of e-learning in medical education. Acad Med. 2006;81:207–212. doi: 10.1097/00001888-200603000-00002. [DOI] [PubMed] [Google Scholar]
- 9.Vernon T, Peckham D. The benefits of 3D modelling and animation in medical teaching. J Audiov Media Med. 2002;25:142–148. doi: 10.1080/0140511021000051117. [DOI] [PubMed] [Google Scholar]
- 10.Moulton CA, Dubrowski A, Macrae H, Graham B, Grober E, Reznick R. Teaching surgical skills: what kind of practice makes perfect? A randomized, controlled trial. Ann Surg. 2006;244:400–409. doi: 10.1097/01.sla.0000234808.85789.6a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grunwald T, Corsbie-Massay C. Guidelines for cognitively efficient multimedia learning tools: educational strategies, cognitive load, and interface design. Acad Med. 2006;81:213–223. doi: 10.1097/00001888-200603000-00003. [DOI] [PubMed] [Google Scholar]
- 12.Huang C. Designing high-quality interactive multimedia learning modules. Comput Med Imaging Graph. 2005;29:223–233. doi: 10.1016/j.compmedimag.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 13.Khalil MK, Paas F, Johnson TE, Payer AF. Design of interactive and dynamic anatomical visualizations: the implication of cognitive load theory. Anat Rec. 2005;286B:15–20. doi: 10.1002/ar.b.20078. [DOI] [PubMed] [Google Scholar]
- 14.Goldberg HR, Haase E, Shoukas A, Schramm L. Redefining classroom instruction. Adv Physiol Educ. 2006;30:124–127. doi: 10.1152/advan.00017.2006. [DOI] [PubMed] [Google Scholar]
- 15.Guttmann GD. Animating functional anatomy for the web. Anat Rec. 2000;261:57–63. doi: 10.1002/(SICI)1097-0185(20000415)261:2<57::AID-AR5>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 16.Adobe.com [Flash Player Penetration. San Jose: Adobe Systems, Inc http://www.adobe.com/products/player_census/flashplayer/. [Accessed 10 March 2009.
- 17.Sweller J. Cognitive load during problem solving: effects on learning. Cogn Sci. 1988;12:257–285. [Google Scholar]


