Abstract
Over 100 amino acid replacements in human Cu, Zn superoxide dismutase (SOD) are known to cause amyotrophic lateral sclerosis, a gain-of-function neurodegenerative disease that destroys motor neurons. Supposing that aggregates of partially-folded states are primarily responsible for toxicity, the role of the structurally-important zinc ion in defining the folding free energy surface of dimeric SOD was determined by comparing the thermodynamic and kinetic folding properties of the zinc-free and zinc-bound forms of the protein. The presence of zinc was found to decrease the free energies of a peptide model of the unfolded monomer, a stable variant of the folded monomeric intermediate and the folded dimeric species. The unfolded state binds zinc weakly with a micromolar dissociation constant, and the folded monomeric intermediate and the native dimeric form both bind zinc tightly, with sub-nanomolar dissociation constants. Coupled with the strong driving force for the subunit association reaction, the shift in the populations towards more well-folded states in the presence of zinc decreases the steady-state populations of higher-energy states in SOD under expected in vivo zinc concentrations (∼nanomolar). The significant decrease in the population of partially-folded states is expected to diminish their potential for aggregation and account for the known protective effect of zinc. The ∼100-fold increase in the rate of folding of SOD in the presence of micromolar concentrations of zinc demonstrates a significant role for a pre-organized zinc-binding loop in the transition state ensemble for the rate-limiting monomer folding reaction in this β-barrel protein.
Keywords: ALS, beta-barrel dimer, metal binding, protein folding, thermodynamics and kinetics
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease, characterized by systematic loss of the motor neurons in the brain and spinal cord.1 Though no definitive biochemical mechanism has been discovered, a subset of familial ALS (fALS) cases has been linked to mutations at the Cu, Zn superoxide dismutase (SOD) locus.2,3 Despite constituting only 2-5% of all known cases, SOD mediated fALS is the most prominent, identified, heritable cause of the disease. Moreover, sporadic cases of ALS are clinically and pathologically similar to fALS cases,1 suggesting both forms may share a common underlying mechanism.
In its native form, SOD is a 153 amino acid, cytosolic protein that catalyzes the dismutation of superoxide into hydrogen peroxide and oxygen.4 SOD is catalytically active as a homodimer with both a copper and a zinc ion bound to each monomer. The fold of an SOD monomer is a β-sandwich composed of eight antiparallel β-strands supporting a pair of large loops that follow β4 and β7, loops IV and VII, respectively (Figure 1). Loop IV is covalently linked to β8 by a disulfide bond between Cys57 and Cys146.5 Loops IV and VII coordinate the metals and provide the catalytic and electrostatic components of the active site.6 The copper redox cycle drives the disproportionation reaction, and the zinc ion is thought to play a role in defining the structure and stability of SOD.7-9 The binding of zinc to dimeric SOD is very tight, with a Kd of <10-8 M by isothermal titration calorimetry10 and a Kd of ∼10-14 M by competition experiments.11 While the order of the metal-binding and disulfide-bonding events in vivo is not definitively known, it is thought that zinc binds first to the disulfide-reduced protein. This complex then interacts with the copper chaperone of SOD, which loads the copper and oxidizes the intramolecular disulfide bond.12
Figure 1.
(A) The crystal structure of SOD (PDB: 2C9V).34 The zinc ion is depicted in yellow, with the metal-binding region of the zinc-binding loop (loop IV), as well as the residues that bind zinc, shown in dark blue. Proximal to this region, the electrostatic loop is colored in teal. The region of loop IV involved in or near the intramolecular disulfide bond (in dark orange) between C57-C146 is shown in light yellow, and the dimer interface portion of loop IV is highlighted in orange. Shown in blue is the copper ion with the histidines to which it binds depicted in green. In red are F50 and G51 that were replaced with glutamic acid to create mAS-SOD. (B) The sequence of the zinc-binding loop with the zinc-binding residues indicated in red. The non-native residues are shown in teal for the C-terminal tryptophan and yellow for the N-terminal cysteine, which was further modified with an AEDANS molecule.
Although several hypotheses have been proposed to explain the toxic gain of function of SOD variants,13-16 a prevailing view holds that amino acid replacements increase populations of partially-folded states and induce subsequent aggregation.13,17,18 The rationale for the unfolding/aggregation hypothesis is based on the fact that well over 100 replacements and C-terminal truncations (see http://alsod.iop.kcl.ac.uk/Als/ for a complete list), covering nearly 50% of the sequence, can cause ALS.19 Many of these variants lead to the deposition of amorphous aggregates in and around the motor neurons of patients afflicted with ALS, and a debate has ensued about whether small oligomers or macroscopic aggregates are the toxic agents.20 It has been demonstrated that loss of zinc may result in aggregation, reinforcing the structural importance of zinc.21 Candidates for aggregation have focused on zinc-free partially-folded monomeric states of SOD, including those with the disulfide bond intact and those in which the disulfide bond has been reduced.22 More recently, the unfolded state, extruded from the ribosome and prior to disulfide bond formation or folding, has also been proposed as a candidate for aggregation and toxicity.23
The unfolding/aggregation hypothesis has been tested by comparative biophysical analysis of the thermodynamic folding properties of wild-type and ALS-inducing SOD variants in both their disulfide-oxidized24 and disulfide-reduced25,26 forms. Detailed analyses of the folding mechanisms of disulfide-intact SOD have shown that both apo-SOD25,27,28 and Cu, Zn-SOD29 fold by a three-state mechanism involving a folded monomeric intermediate. Left unanswered in these studies were several questions on the specific role of zinc on the folding and stability of SOD that pertain to its influence on nonnative states that might be responsible for aggregation. Pertinent to the argument that newly-synthesized chains might be the source of toxicity, at what stage of folding does zinc become bound to SOD? Once the native state is reached, what are the quantitative ramifications of zinc binding to diminish the propensity of the disulfide-bonded native dimer and the folded monomer to populate less structured, aggregation-prone states? In the event that the folded monomer is a source of aggregation, does zinc binding alter the relative populations of monomeric and dimeric forms of SOD at equilibrium to favor the fully-folded form? A comparative analysis of the thermodynamic and kinetic folding properties of disulfide-bonded AS-SOD in the presence and absence of zinc demonstrated that zinc binding modulates its entire folding free energy surface. The results provide a benchmark for future studies on metal-binding ALS-inducing variants containing disulfide bonds and on disulfide-reduced SOD.
RESULTS
Quantitative information on the effect of zinc binding on high energy states in SOD was obtained by chemical denaturation analysis of both the thermodynamic and kinetic properties of the reversible folding reaction. The C6A/C111S variant, AS-SOD, was used in place of the wild-type SOD because it has been shown to fold reversibly with wild-type like stability.30 The elimination of the free cysteines precludes intermolecular disulfide bond formation and intramolecular disulfide bond interchange. The results obtained can be compared with those from a previous study of metal-free apo-AS-SOD27 to determine the decrease in the free energy, i.e. the increase in stabilization relative to the unfolded state, of the zinc-bound states in the absence of denaturant.
Thermodynamic analysis
Chemical denaturation of dimeric AS-SOD
The effects of zinc binding on the thermodynamic properties of disulfide-intact AS-SOD were determined by chemical denaturation experiments in the presence and absence of zinc and monitored by circular dichroism spectroscopy. It has been previously shown that the ellipticity at 230 nm is a sensitive probe of global structure in AS-SOD27 and provides optimal signal to noise compared to measurements at the typical minimum for β-rich structures, 218 nm. Although urea is a sufficiently potent denaturant to unfold apo-SOD,27,31 guanidine hydrochloride (Gdn-HCl) was required to unfold zinc-bound AS-SOD. The Gdn-HCl unfolding titration curves at 230 nm, pH 7.2 and 20 °C, for apo- and stoichiometric zinc-bound Zn-AS-SOD (2 zinc ions per dimer) are shown in Figure 2. The native dimeric form of apo-AS-SOD is stable up to ∼1 M Gdn-HCl, where it experiences a cooperative unfolding transition that is complete by ∼2 M Gdn-HCl. In the presence of stoichiometric zinc, Zn-AS-SOD is stable up to ∼2 M Gdn-HCl, where it also undergoes a cooperative unfolding transition that is complete by ∼3 M Gdn-HCl. The reversibility of the Zn-AS-SOD unfolding reaction was demonstrated by the coincidence of a refolding transition curve beginning with denatured AS-SOD (data not shown). As has been observed previously with Cu, Zn SOD,29 the full equilibration of the Zn-AS-SOD samples at pH 7.2 for both unfolding and refolding curves was exceedingly slow, requiring 7 days at room temperature.
Figure 2.
Equilibrium Gdn-HCl titrations measured by the ellipticity in 20 mM HEPES, pH 7.2, at 20 °C. Representative data at 230 nm are shown with fits to a two-state model for the metal-free apo-AS-SOD (open squares) and apo-mAS-SOD (open circles) and with fits to a three-state model for stoichiometric Zn-AS-SOD (filled squares) and Zn-mAS-SOD (filled circles). Protein and zinc concentrations were 10 μM for AS-SOD and 5 μM for mAS-SOD.
Chemical denaturation of monomeric AS-SOD
Previous analyses of the chemical denaturation of apo-SOD27 and Cu, Zn SOD29 have implicated an essential role for a folded monomeric species that subsequently undergoes a diffusion-limited association reaction to produce the native dimer. Insight into the effect of zinc binding on the stability of the monomeric folding intermediate was obtained by examining the enhancement of stability for a stable monomeric version of AS-SOD, mAS-SOD, upon zinc binding. The mAS-SOD variant was created by replacing two subunit interface residues, F50 and G51, with glutamic acid.32 In the absence of zinc, the native mAS-SOD is stable up to ∼0.7 M Gdn-HCl where it undergoes a cooperative unfolding transition that is complete by ∼1 M Gdn-HCl. Similar to AS-SOD, the addition of stoichiometric zinc to mAS-SOD (1 zinc per chain) significantly stabilizes the protein. The Zn-mAS-SOD is stable up to ∼1.5 M Gdn-HCl, where it undergoes a cooperative unfolding transition that is complete by ∼2.8 M Gdn-HCl.
Stoichiometry and location of zinc binding to AS-SOD
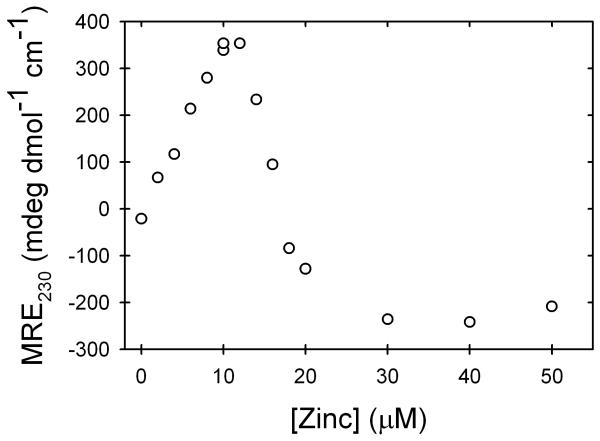
Inspection of the titration curves in Figure 2 reveal a distinctly more positive ellipticity at 230 nm under native conditions for the stoichiometric Zn-AS-SOD, and possibly Zn-mAS-SOD, compared to their metal-free counterparts. Realizing that this signal could be used to monitor zinc binding to the folded dimeric form of AS-SOD, the ellipticity at 230 nm was monitored as a function of zinc concentration. The ellipticity increased linearly with increasing concentration of zinc up to the level at which the stoichiometric zinc concentration was reached (Figure 3). Further addition of zinc lead to a decrease in the ellipticity that is complete when an equivalent of 6 zinc ions per dimer has been added. The data imply that zinc binds to at least two different sites on apo-AS-SOD, possibly the zinc and copper sites nested in loops IV and VII (Figure 1).
Figure 3.
Titration of AS-SOD in buffer monitoring the change in ellipticity at 230 nm as a function of the zinc concentration. The titration was performed in 20 mM HEPES, pH 7.2, at 20 °C. Protein concentration was 10 μM.
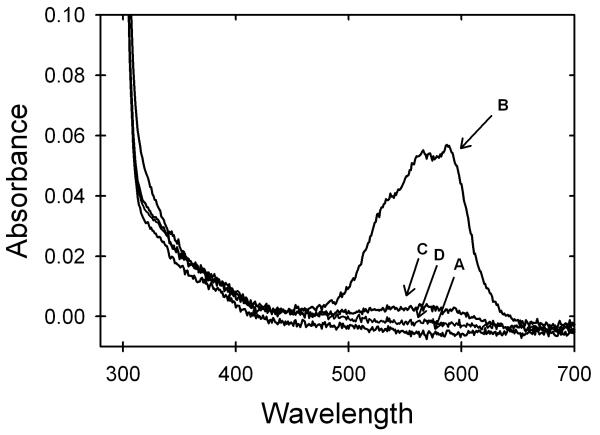
The assumption that the initial increase in ellipticity upon addition of zinc corresponds to zinc binding at the zinc site was tested using the cobalt ion as an optical probe. Cobalt is an excellent mimic for zinc because its divalent form has an ionic radius and geometric preferences that are similar to zinc. Although zinc is spectroscopically silent, cobalt gives a strong absorbance in the visible region when bound to the zinc site in SOD. Addition of stoichiometric cobalt to apo-AS-SOD yielded strong absorption bands between 500 and 650 nm, very similar to the previously determined spectra of cobalt ion bound to the zinc site (Figure 4).33 By contrast, the addition of stoichiometric cobalt after apo-AS-SOD was pre-incubated with stoichiometric zinc results in dramatically reduced absorption bands (Figure 4); further addition of zinc eliminates the small residual absorption band. These data show that zinc binds preferentially to the zinc site on AS-SOD. Excess zinc binds to one or more additional sites, possibly the copper site, on each subunit. Crystallographic studies of SOD in the presence of excess zinc show zinc binding in the copper site,34 and cobalt binding to the copper site produces absorption band in this same region.33 This conclusion is also consistent with the findings of an extensive dialysis against buffer of a sample of AS-SOD pre-incubated with 10-fold excess zinc. Atomic absorption analysis revealed an average of 2.5 zinc ions per dimer, implying a second weaker binding site for zinc in AS-SOD.
Figure 4.
Visible absorbance spectra of AS-SOD in 20mM HEPES, pH 7.2, at room temperature. Traces shown are protein with (A) no metal added, (B) after addition of 1 cobalt ion per monomer, (C) after pre-incubation of protein with 1 zinc ion per monomer followed by the addition of 1 cobalt ion per monomer, and (D) after pre-incubation with 6 zinc ions per monomer followed by the addition of 1 cobalt ion per monomer. The protein concentration for all four traces was 123 μM.
Zinc binding to unfolded AS-SOD
Although it is not surprising that the folded state of AS-SOD binds zinc in the zinc site with high affinity, it is possible that the unfolded state may also bind zinc. Previous studies on azurin35,36 have found that zinc can bind to the chemically-denatured protein, complicating estimates of stability by denaturation experiments. Binding to the unfolded state of AS-SOD was tested by synthesizing a 27-residue peptide that corresponds to the sequence containing all four zinc-binding side chains, H63, H71, H80 and D83 (Figure 1B). The potential acquisition of structure upon zinc binding to the peptide was monitored by Förster resonance energy transfer (FRET) from a tryptophan added at the C-terminus to an AEDANS acceptor covalently attached to an additional cysteine at the N-terminus.
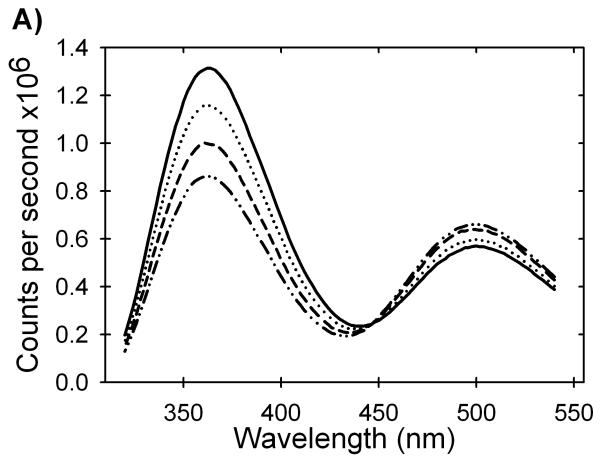
As shown in Figure 5A, the fluorescence intensity of the tryptophan emission at 360 nm decreased and the AEDANS emission at 500 nm increased with increasing zinc concentration. The quenching of the tryptophan donor by the AEDANS acceptor implies that zinc induces structure in the peptide in the micromolar concentration range. The affinity of this peptide for zinc was calculated by fitting the data in buffer to a simple binding isotherm (Figure 5B). The dissociation constant of the zinc/peptide complex is 1.1±0.1 μM, and the free energy of binding is -8.0±0.3 kcal (mol monomer)-1 at the standard state (1 M in each component). Relevant to the denaturation analysis of AS-SOD, the binding becomes progressively weaker in the presence of increasing Gdn-HCl concentrations to over 60 μM at 4 M Gdn-HCl. Significant zinc binding to the peptide is not detected above 5 M Gdn-HCl (Figure 5B). A plot of—RT ln(Kd), i.e., the free energy of the dissociation reaction at the standard state, ΔGdo, vs. the concentration of Gdn-HCl is linear (Figure S1). This behavior is typical of protein folding reactions when the free energy of folding is plotted vs. the concentration of denaturant. The free energy of the dissociation reaction in the absence of denaturant, Gdo, obtained by extrapolation, -7.6 kcal (mol monomer)-1 agrees well with the value obtained by direct measurement, -8.0 kcal (mol monomer)-1, and the denaturant dependence of ΔGdo, the m-value, is 0.42 kcal (mol monomer)-1 M-1 (Table 1). Thus, under equilibrium conditions, there is a significant perturbation of the free energy of the unfolded state of SOD from zinc binding to the zinc-binding loop peptide. This behavior influences the Gdn-HCl unfolding reaction of SOD and must be accounted for in the extraction of accurate thermodynamic parameters (see below).
Figure 5.
Zinc binding affinity of a peptide model of the unfolded state of SOD. The change in fluorescence intensity of a FRET pair at the termini of a peptide encompassing the four zinc binding residues of SOD was monitored at various zinc concentrations and in the presence of varying amounts of Gdn-HCl. (A) The addition of zinc causes a decrease in the tryptophan fluorescence (λmax= 360 nm) and a proportional increase in AEDANS fluorescence (λmax= 500 nm). Traces shown are at 1 M Gdn-HCl and represent zinc concentrations of 1 μM (solid line), 7 μM (dotted line), 20 μM (dashed line) and 60 μM (dashed and dotted line). (B) Quenching of the tryptophan emission at 360 nm by AEDANS as zinc induces structure in the peptide of the zinc-binding loop. The isotherms shown are for 0 M Gdn-HCl (filled circles), 0.5 M Gdn-HCl (open circles), 1 M Gdn-HCl (filled upside-down triangles), 2 M Gdn-HCl (open triangles), 4 M Gdn-HCl (filled squares) and 5 M Gdn-HCl (open squares). These data represent two different preparations of the peptide with different labeling efficiencies so the intensities were normalized to the zinc-free value to show relative differences. The peptide concentration for all experiments was 4 μM in 20 mM HEPES, pH 7.2, and 20 °C.
Table 1.
Thermodynamic parameters measured at 20 °C and pH 7.2 for the Gdn-HCl induced equilibrium unfolding reactions of dimeric and monomeric apo and zinc-bound AS-SOD and the zinc titration of the peptide of the zinc-binding region of SOD.
| Δ° Kinetic |
Δ° Equilibrium |
m-value Kinetic |
m-value Equilibrium |
Kd | |
|---|---|---|---|---|---|
| Apo-mAS-SODa | -3.6±0.1 | -4.3±0.1 | 4.9±0.3 | 5.1±0.5 | NAd |
| Apo-AS-SODb | -20.6±0.3 | -18.6±0.3 | NDc | 9.3±0.2 | NAd |
| Zn-mAS-SODa | -17.0±0.7 | 13.4±0.6 | 4.6±0.3 | 2.6±0.2 | 100±122 pMe |
| Zn-AS-SODb | -49.4±1.5 | -33.0±1.4 | NDc | 4.2±0.3 | 22±35 pMe |
| Peptidea | NA | -8.0±0.3 | NA | 0.42±0.08 | 1.1±0.1 μM |
Units for ΔG° are kcal (mol monomer)-1; units for m are kcal (mol monomer)-1 M-1.
Units for ΔG° are in kcal (mol dimer)-1; units for m are kcal (mol dimer)-1 M-1.
The m value could not be calculated because the protein concentration-dependent kinetic data in the transition region were inaccessible in Gdn-HCl.
Not applicable.
Calculated from kinetic parameters.
Thermodynamic parameters for AS-SOD and Zn-AS-SOD
The evident cooperativity of the Gdn-HCl unfolding reaction and the results of previous studies on apo-AS-SOD denatured in urea27 motivated the test of a simple two-state equilibrium model for the unfolding of apo-AS-SOD, 2U ⇆ N2, and a three-state equilibrium model for Zn-AS-SOD with Gdn-HCl, 2U + 2Zn ⇆ 2(U•Zn) ⇆ (N•Zn)2. The latter fits were done by fixing the change in free energy in the absence of denaturant and the m-value for the 2(U•Zn) ⇆ 2U + 2Zn step to twice the values obtained from the Zn/peptide complex described above. The accuracy of the parameters was enhanced by simultaneously fitting all of the CD data between 220 and 240 nm to the models for both forms (see Materials and Methods for a more detailed description of the analysis).
The stability of the apo-AS-SOD at standard state concentrations (1 M in each of the components), at 20 °C and pH 7.2 is -18.6±0.3 kcal (mol dimer)-1 (Figure 2 and Table 1), in good agreement with the previously determined value by Gdn-HCl denaturation monitored by CD, -17±2 kcal (mol dimer)-1,28 as well as with the value determined by urea denaturation monitored by CD, -20.4±1.0 kcal (mol dimer)-1.27 The denaturant-dependence of the free energy difference between the folded dimer and the unfolded monomer, the m-value, is 9.3±0.2 kcal (mol dimer)-1 M-1.
The binding of zinc increases the stability of AS-SOD, as evidenced by the increase in the apparent midpoint of the unfolding transition from ∼1.3 to ∼2.6 M Gdn-HCl (Figure 2). Fitting the titration data to the 3-state model, 2U + 2Zn ⇆ 2(U•Zn) ⇆ (N•Zn)2, the free energy change and the m-value for the 2(U•Zn) ⇆ (N•Zn)2 reaction were found to be -17.0±0.2 kcal (mol dimer)-1 and 3.4±0.1 kcal (mol dimer)-1 M-1. When the free energy change for this reaction is added to that for the 2U + 2Zn ⇆ 2(U•Zn) reaction, -16.0±1.2 kcal (mol dimer)-1 and 0.84 kcal (mol dimer)-1 M-1, the total free energy difference between the (N•Zn)2 and the 2U + 2Zn states is -33.0±1.4 kcal (mol dimer)-1 at the standard state.
Surprisingly, the sum of the m-values for the folding/subunit dissociation step, 3.4±0.1 kcal (mol dimer)-1 M-1, and twice the m-value for the zinc dissociation step, 0.84 kcal (mol dimer)-1 M-1, 4.2±0.2 kcal (mol dimer)-1 M-1, is significantly lower than the m-value for apo-SOD, 9.3±0.2 kcal (mol dimer)-1. The magnitude of the m-value has been shown to correlate with the exposure of buried surface area during unfolding37. Therefore, if zinc binding induces structure in native SOD, one might expect that the total m-value for the dimer unfolding and zinc dissociation steps would be at least equal to if not exceed the m-value for the unfolding of apo-SOD. As will be shown below, the aberrant m-value for the unfolding of AS-SOD in the presence of stoichiometric zinc has another explanation.
Thermodynamic parameters for mAS-SOD and Zn-mAS-SOD
The stability enhancement for zinc binding to mAS-SOD was obtained by similar two-state and three-state fits of the unfolding titration data (Figure 2) for the apo and zinc-bound stable monomeric form (see Materials and Methods). The stability of apo-mAS-SOD, -4.3±0.1 kcal (mol monomer)-1 (Table 1) is in good agreement with the value previously determined by urea denaturation, -4.27±0.48 kcal (mol monomer)-1. The m-value for the monomer unfolding reaction is 5.1±0.5 kcal (mol monomer)-1 M-1. The free energy change for the (U•Zn) ⇆ (M•Zn) reaction is -5.8±0.3 kcal (mol monomer)-1, and the m-value is 2.3±0.1 kcal (mol monomer)-1. When zinc binding to the unfolded state is considered, the total free energy change for the (M•Zn) ⇆ U + Zn reaction is -13.8±0.9 kcal (mol monomer)-1 and the total m-value is 2.6±0.2 kcal (mol monomer)-1 M-1. Similar to Zn-AS-SOD, the m-value for the unfolding of the zinc-bound stable monomeric form is significantly less than its apo-mAS-SOD counterpart.
There are two possibilities for the discrepancies in the m-values for the apo- and zinc-bound forms of AS-SOD and mAS-SOD. First, the reduced m-values in the presence of zinc could reflect the population of additional species in the transition region. Obvious candidates are the metal-free N state and zinc bound to non-zinc sites — possibly the copper site - evident at super-stoichiometric zinc concentrations (Figure 3). The possibility that zinc-bound monomers are populated is unlikely since both AS-SOD and mAS-SOD exhibit the same behavior. Kinetic analysis of the AS-SOD folding reaction resolves the discrepancies between the m-values and provides reliable estimates of the thermodynamic parameters sought in this study.
Kinetic analysis
The thermodynamic analysis of the equilibrium unfolding reactions of AS-SOD, mAS-SOD and a peptide model for the zinc-binding loop has shown that zinc can bind to all three species in the kinetic folding mechanism. Kinetic folding studies are required to determine the effects of zinc binding on the intervening transition states. Importantly, kinetic analysis also provides an alternative approach towards determining the magnitude of the stabilizing effect of zinc on the monomeric and dimeric forms of AS-SOD.
Folding kinetics of AS-SOD and Zn-AS-SOD
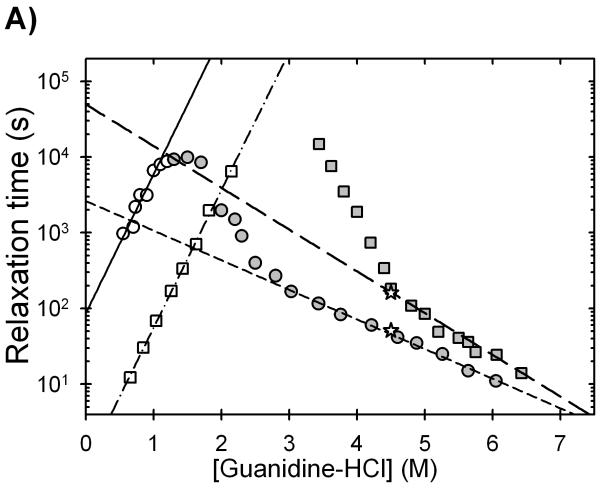
Semi-log plots of the relaxation times as a function of the final denaturant concentration, chevron plots,38 for apo-AS-SOD and Zn-AS-SOD are shown in Figure 6A. The single unfolding relaxation time for dimeric apo-AS-SOD decreases exponentially above 3 M Gdn-HCl, and the single refolding relaxation time decreases exponentially below 1 M Gdn-HCl. Neither phase depends upon the protein concentration under strongly unfolding or refolding conditions (data not shown). Previous studies of the urea denaturation reaction for apo-AS-SOD,27 have shown that the unfolding phase at high denaturant concentration corresponds to the unimolecular dissociation of the dimer. The single refolding phase observed by CD at 230 nm corresponds to the rate-limiting folding of the monomer. The non-exponential dependence in the unfolding leg of the chevron between 1.5 and 3 M Gdn-HCl reflects the coupling of the monomer refolding/unfolding and the dimer association/dissociation reactions in the transition zone.27
Figure 6.
Observed refolding (open symbols) and unfolding (filled symbols) relaxation times for (A) AS-SOD and (B) mAS-SOD as a function of final Gdn-HCl concentration in the presence (squares) and absence (circles) of zinc as monitored by CD at 230 nm. In panel A, the dashed and dotted lines represent the linear extrapolations used to determine the relaxation times for the rate-limiting folding reactions, and the dashed lines represent the extrapolations used to determine the dimer dissociation reaction in the absence of denaturant. In panel B, the lines represent the fits of the data to simple two-state chevrons. The results yield the unfolding and refolding relaxation times and their reciprocal rate constants in the absence of denaturant. The two observed relaxation times of the unfolding of 1 zinc per dimer AS-SOD is shown by the open star symbols in panel A. The filled upside-down triangle in panel B is the observed relaxation time for the unfolding double-jump experiment at stoichiometric zinc concentration. Protein concentrations were 10 μM for both AS-SOD and mAS-SOD, and the buffer contained 20 mM HEPES, pH 7.2, 20 °C.
The presence of zinc has a significant effect on both the unfolding and refolding reactions of AS-SOD (Figure 6A, Table 2). Unfolding is slowed by 5-fold at 6 M Gdn-HCl, and refolding is accelerated by a factor of ∼100-fold at 0.6 M Gdn-HCl. The denaturant dependence of the dimer dissociation relaxation time in the presence of zinc between 5.5 and 6.8 M Gdn-HCl, -0.95±0.06 kcal (mol dimer)-1 M-1, is larger than that for apo-AS-SOD between 3.8 M and 6.0 M Gdn-HCl, -0.67±0.03 kcal (mol dimer)-1 M-1. As a result, the dimer dissociation relaxation time in the absence of denaturant, estimated by linear extrapolation, is ∼20-fold longer in the presence of zinc. The small increase in the denaturant dependence of the relaxation time implies an enhancement of structure as the zinc-binding and electrostatic loops are organized by zinc binding. Given the quite similar denaturant dependence of the refolding relaxation times for apo- and Zn-AS-SOD, the kinetic refolding data yield a 100-fold acceleration of the refolding rate by zinc in the absence of Gdn-HCl. Although the linear extrapolations in Gdn-HCl may be questioned,39 the differences between the extrapolated relaxation times in the presence and absence of zinc should be minimal.
Table 2.
Microscopic rate constants, kinetic m‡ values and the effect of zinc binding on the activation free energy of the transition state ensembles of AS-SOD measured at 20 °C and pH 7.2.
| Apo-AS-SODa | Zn-AS-SODa | ΔG‡b Apo-AS-SOD |
ΔG‡b Zn-AS-SOD |
ΔΔG‡b | |
|---|---|---|---|---|---|
| kfc | 0.012±0.002 | 1.19±0.05 | 5.2±0.2 | 0.2±0.08 | -5.0±0.1 |
| c | 3.17±0.53 | 3.13±0.01 | NAh | NA | NA |
| kud | (3.1±1.5)×10-5 | (2.0±1)×10-7 | -12.0±0.6 | -18.0±0.6 | 6.0±0.7 |
| d | -1.39±0.07 | -1.45±0.11 | NA | NA | NA |
| kae | (6.9±8.3)×106 | (6.9±8.3)×106 | 18.2±1.2 | 18.2±1.2 | 0 |
| NDg | ND | NA | NA | NA | |
| kdf | (38.1±0.9)×10-5 | (2.1±0.4)×10-5 | -4.6±0.1 | -6.3±0.1 | 1.7±0.1 |
| f | -0.67±0.03 | -0.95±0.06 | NA | NA | NA |
Units for the monomer folding rate constants (kf), unfolding rate constants (ku), dimer association rate constants (ka) and dimer dissociation rate constants (kd) are s-1; units of the denaturant dependence of monomer folding () and unfolding () rate constants are kcal (mol monomer)-1 M-1, and units for the denaturant dependence of the dimer dissociation rate constant () are kcal (mol dimer)-1 M-1.
Units for the activation free energy barrier (ΔG‡) and the change in the free energy barrier upon zinc binding to the transition state ensemble(ΔΔG‡) are kcal (mol dimer)-1.
Calculated from the linear extrapolation of the refolding leg of the AS-SOD chevron (Figure 6A).
Calculated from the 2-state fit of the mAS-SOD chevron (Figure 6B).
The dimer association rate constant was taken from Svensson et al. (Manuscript in preparation)
Calculated from the linear extrapolation of the >5.5 M Gdn-HCl region of the unfolding leg of the zinc-bound AS-SOD chevron and the >3.8 M Gdn-HCl region of the unfolding leg of the apo-AS-SOD chevron (Figure 6A).
Not determined.
Not applicable.
Valentine and her colleagues,10 on the basis of isothermal titration calorimetry experiments, have proposed that the binding of the first zinc ion to the SOD dimer may contribute the majority of the stability induced by zinc binding. By implication, the 1 zinc per dimer species should comprise the entire population at half saturation. This conclusion was investigated by incubating AS-SOD with 1 zinc ion per dimer and monitoring the unfolding reaction. The observed unfolding reaction could not be fit to a single exponential expected for a homogeneous population. When fit to two exponentials, the observed relaxation times corresponded well with the unfolding relaxation times of the apo-protein and the zinc-bound protein (Figure 6). This result is better explained by a binomial distribution of zinc in the two zinc sites per dimer, reflecting the metal-free protein, the half-saturated protein and the fully-saturated protein. The absence of a third phase suggests that the unfolding relaxation time of the 1 zinc per dimer species is similar to that for the 2 zinc per dimer species. As noted above, the unfolding of AS-SOD incubated with 2 zinc per dimer is well described by a single exponential at the same concentration of Gdn-HCl (Figure 6). The discrepancies between these results and those from the Valentine laboratory may reflect the different pH values at which the two studies were performed. It is likely that the histidines that ligate zinc are protonated at pH 5.5, where the isothermal titration calorimetry studies were done. The zinc binding properties would undoubtedly be altered from those at pH 7.2, perhaps to include a preferential binding of the first zinc ion to the dimeric protein.
Attempts to visualize the protein concentration dependence in the transition region, 1.5 to 3.0 M Gdn-HCl, as previously observed for the urea denaturation reaction25,27 and required for the determination of the association rate constant, were not successful. The relaxation times in this Gdn-HCl concentration range exceed experimental limitations, >105 s, precluding accurate measurements.
Folding kinetics for mAS-SOD
In the absence of direct information on the association reaction, the effect of zinc binding on refolding/unfolding reactions of the stable monomer, mAS-SOD, was examined. Oliveberg and his colleagues24 have previously used this approach towards partitioning the global free energy change for the unfolding of AA-SOD into its monomer folding reaction and monomer/dimer association reaction components. In the absence of zinc, apo-mAS-SOD unfolds via a simple exponential reaction whose relaxation time decreases linearly with increasing denaturant concentration from ∼1 to ∼ 3.5 M Gdn-HCl (Figure 4B). The singled refolding relaxation time decreases linearly with decreasing Gdn-HCl below ∼1 M denaturant, and the two legs of the chevron intersect at ∼ 1 M Gdn-HCl. The maximum in the relaxation time corresponds closely with the midpoint in the equilibrium titration for apo-mAS-SOD (Figure 2), as expected for a simple two-state folding reaction38. Extrapolation of both relaxation times, τ, to the absence of denaturant yields rate constants, k (k = 1/τ) of ku = 3.05×10-5 s-1 for unfolding and kf = 0.015 s-1 for refolding. The denaturant dependence of the rate constants, the m‡ values, are -1.4 and 3.5 kcal (mol monomer)-1 M-1, respectively. The free energy of folding of apo-mAS-SOD, estimated from ΔG0= -RT ln(ku/kf), is -3.6±0.1 kcal (mol monomer)-1, in reasonable agreement with the stability estimated from the equilibrium titration, 4.3±0.1 kcal (mol monomer)-1 (Table 1).
The kinetic consequences of unfolding and refolding mAS-SOD in the presence of stoichiometric zinc are to increase the unfolding relaxation time by ∼110-fold at 3 M Gdn-HCl and to decrease the refolding relaxation time by ∼100-fold at 0.6 M Gdn-HCl (Figure 4B). As in the case of the dimeric AS-SOD, the refolding relaxation time is independent of the zinc concentration at and above stoichiometric zinc concentration (data not shown). Below stoichiometric zinc concentrations, a small faster phase with a relaxation time equal to that for apo-mAS-SOD appears (data not shown). The two phases reflect the independent folding of the zinc-bound unfolded and zinc-free unfolded AS-SOD. The m‡u and m‡f values in the presence of zinc are similar to those in the absence of zinc, insuring that the differences in the relaxation times observed in Gdn-HCl are maintained in the absence of denaturant.
Preferential binding of zinc to the zinc site in refolding was tested by a double-jump experiment. Metal-free mAS-SOD was unfolded in 3.9 M Gdn-HCl, and then refolded by a 10-fold dilution to 0.4 M Gdn-HCl containing stoichiometric zinc. After 30 s of refolding, approximately three times the refolding relaxation time under these conditions (Figure 4A), the protein was unfolded in 4.2 M Gdn-HCl and the unfolding reaction was monitored by CD. The observed unfolding relaxation time is within error to the observed relaxation time for a single-jump unfolding reaction to the same final conditions when the protein is incubated with stoichiometric zinc in the native state (Figure 6B). Given the preferential binding of zinc to the zinc site in native AS-SOD (Figure 4), zinc must also bind preferentially to the zinc site during the refolding reaction.
The obvious perturbations of the unfolding and refolding relaxation times for mAS-SOD at stoichiometric concentrations of monomer and zinc (Figure 6B) and the knowledge that zinc binds to both folded and unfolded protein in low concentrations of denaturant allows a simple two-state analysis of the U•Zn ⇆ M•Zn reaction from the chevron. The free energy difference between these states, ΔG = -RT ln(ku/kf), is -9.0±0.1 kcal (mol monomer)-1 in the absence of denaturant. When this value is added to the free energy of binding of zinc to the peptide model of the unfolded state, -8.0±0.3 kcal (mol monomer)-1, the free energy of the zinc-bound folded monomeric form relative to the zinc-free unfolded state is found to be -17.0±0.4 kcal (mol monomer)-1.
The kinetic chevrons for apo-mAS-SOD and Zn-mAS-SOD also provide a resolution to the anomalously low m-value for the equilibrium titration of Zn-mAS-SOD. For two-state reactions, the sum of the absolute values of the kinetic m‡-values is equal to the m-value for the equilibrium titration. The equilibrium m-value for apo-mAS-SOD, 5.1±0.5 kcal (mol monomer)-1 M-1, is in good agreement with the value determined from the rate constants for folding and unfolding, 4.9±0.1 kcal (mol monomer)-1 M-1. By contrast, the equilibrium m-value for Zn-mAS-SOD, 2.6±0.2 kcal (mol monomer)-1 M-1 is about half that of the value determined from the chevron for this system, 4.6±0.1 kcal (mol monomer)-1 M-1. These contradictory estimates for the equilibrium m-value reflect the complexity of the equilibrium Zn-mAS-SOD folding reaction. At equilibrium in the transition zone, the m-value is sensitive to the presence of all of the states that bind zinc and are significantly populated under these conditions. The kinetic experiment, by contrast, reveals the rate constants for specific steps in the mechanism, U•Zn → M•Zn, and M•Zn → U•Zn, that pass through a common transition state ensemble. The sum of the absolute values of the denaturant dependences of these rate constants provides an accurate measure of the equilibrium m-value and, with the extrapolated intercepts of the rate constants, an accurate estimate of the stability of the Zn-mAS-SOD not available from the equilibrium titration experiment.
The good agreement between the m-values for apo-mAS-SOD and Zn-mAS-SOD obtained from the kinetic analysis (Table 2) implies that the buried surface induced by zinc binding is similar for the M•Zn, the U•Zn and the intervening TSE compared to their respective apo-states. If zinc were binding to non-zinc sites in unfolded AS-SOD so as to reduce the equilibrium m-value by a factor of two, one might expect to observe a significant decrease in the m‡f-value for the folding reaction. The very similar m‡f-values for apo- and Zn-mAS-SOD (Figure 6 and Table 2) argue against this possibility.
Reaction coordinate diagram for Zn-AS-SOD
The folding data for the apo- and zinc-bound forms of AS-SOD and mAS-SOD and the thermodynamic data for the binding of zinc to the peptide model of the zinc binding loop can be combined to determine the effect of zinc on the folding reaction coordinate diagram of AS-SOD under standard state conditions. The free energies are those for the dimeric systems: 2U ⇆ 2M ⇆ N2 for apo-AS-SOD and 2(U•Zn) ⇆ 2(M•Zn) ⇆ (N•Zn)2 for Zn-AS-SOD. Although the conversion of the kinetic data to transition state energies depends upon the formalism adopted,40 the perturbations in the free energies of the transition states induced by zinc binding should, to a first-approximation, be independent of the formalism because the same kinetic mechanisms are operative.
The procedure by which the thermodynamic and kinetic data on the folding of Zn-AS-SOD were converted into a reaction coordinate diagram was as follows:
The unfolded apo-monomer, 2U, was chosen as the reference state for both the apo-AS-SOD and Zn-AS-SOD diagrams (Figure 7).
The energy of the unfolded zinc-bound monomer, U•Zn, relative to the U state was determined by converting the dissociation constant for the zinc-peptide complex using ΔG = -RT ln(Kd)= -8.0 kcal (mol monomer)-1 (Table 1). For the dimeric system, the free energy of the 2(U•Zn) state is -16.0 kcal (mol dimer)-1 lower than that of the 2U state.
The chevron analysis of apo-mAS-SOD (Figure 6B) revealed that the folded monomeric state, 2M, is -7.2 kcal (mol dimer)-1 lower than the 2U state.27 The thermodynamic stability of Zn-mAS-SOD was calculated by fitting its kinetic chevron to a two-state model, and the free energy difference between the U•Zn ⇆ N•Zn states was determined from Go = -RTln(ku/kf) to be -9.0 kcal (mol monomer)-1. The binding free energy of zinc to the unfolded state, U+Zn ⇆ U•Zn, 8.0 kcal (mol monomer)-1, was added to yield a total free energy for the U+Zn ⇆ U•Zn ⇆ N•Zn reaction of -17.0 kcal (mol monomer)-1. The energy of the 2(M•Zn) state is -34.0 kcal (mol dimer)-1 lower than the 2U state and -26.8 kcal (mol dimer)-1 less than that of the 2M state. Therefore Zn-mAS-SOD is 13.4 kcal (mol monomer)-1 more stable than apo-mAS-SOD (Table 1).
The free energy of the (N•Zn)2 state was calculated by the sum of twice the free energy of the Zn-mAS-SOD and the free energy change for the dimerization reaction. The latter free energy change was calculated from the dissociation rate constant, kd = 2.1×10-5 s-1, obtained by linear extrapolation of the unfolding relaxation time at high denaturant concentration (Figure 6A) and the association rate constant, ka = 6.9×106 M-1 s-1, determined by global kinetic analysis of the urea-denaturation reaction (Svensson et al. manuscript in preparation). These rate constants yield a ΔGo2U/N2 = 2(ΔGoU/M) — RT ln(ka/kd) = -49.4 kcal (mol dimer) for the (N•Zn)2 state relative to the 2U + 2Zn state. The same calculation performed for the metal-free AS-SOD yields a free energy difference of -20.9 kcal (mol dimer)-1 for the N2 state relative to the 2U state. Therefore, the free energy of the (N•Zn)2 state is 28.5 kcal (mol dimer)-1 less than the N2 state.
The activation free energy of the transition state ensemble for the 2M ⇆ N2 reaction, TSED, was placed relative to the M state using the Eyring formalism, ΔG‡ = -RT ln[(kah)/kBT)], where ka is the association rate constant in the absence of denaturant, 6.9 × 106 M-1 s-1, h is the Planck constant, kB is the Boltzmann constant and T is the absolute temperature; ΔG‡ =18.2 kcal (mol dimer)-1. The TSED for the 2(M•Zn) ⇆ (N•Zn)2 reaction, relative to the (M•Zn) state, is identical to that for the metal-free M state because the association reaction is near diffusion-limited and, therefore, will not be affected by zinc binding.
The activation free energy for the transition state ensemble for the 2U ⇆ 2M reaction, 2TSEM, can be placed with the rate constant for folding in the absence of denaturant, 0.008 s-1, and the Erying formalism as described above. Similarly, the 2TSEM for the 2(U•Zn) ⇆ 2(M•Zn) reaction can be placed relative to the 2(U•Zn) state, recognizing that the 100-fold acceleration in folding due to zinc binding corresponds to a decrease in the barrier of 5.0 kcal (mol dimer)-1 (Table 2).
Figure 7.
Reaction coordinate diagrams of apo-AS-SOD and Zn-AS-SOD in the dimer reference frame and at the standard state, 1 M in each of the components. Apo-states are depicted in gray, while zinc-bound states are shown in black. Solid lines are thermodynamic states, where 2U is the unfolded state, 2M is the folded monomer and N2 is the native dimer. Dotted lines represent the energies of the transitions states, where 2TSEM is the transition state ensemble of monomer folding and TSED is the transition state ensemble of dimer formation. The absolute values of the TSE energies depend upon the formalism chosen to determine these energies. All changes in free energy induced by zinc binding, reported under the bar representing the zinc-bound states, are given in units of kcal (mol dimer)-1.
It is evident that zinc binding perturbs the entire folding reaction coordinate for AS-SOD. The implications of this observation for the unfolding/aggregation hypothesis for ALS are discussed below.
DISCUSSION
Modulation of the folding free energy surface
The thermodynamic and kinetic data for the effects of zinc binding on the folding and stability of AS-SOD (Tables 1 and 2) provide quantitative answers to the following questions about the role of zinc in protecting cells against the aggregation of partially-folded states of SOD.
At what stage of the folding reaction does zinc become bound to SOD and potentially offer protection against aggregation? Zinc binds rapidly to the zinc site in the unfolded state with micromolar affinity in vitro and remains bound throughout the reaction. The binding of zinc to the peptide model for unfolded SOD provides a ready explanation for its dramatic acceleration of the monomer folding reaction in vitro and offers insights into the source of the very slow folding reaction for apo-AS-SOD (see below). The nanomolar concentration of free zinc in cells,41 however, is not sufficient to saturate the zinc-binding loop segment, and the lifetime of unfolded SOD would be that of the metal-free protein following synthesis on the ribosome.
-
What are the quantitative ramifications of zinc binding on the propensity of the native dimer and the folded monomer to populate, at equilibrium, less well structured states that may be prone to aggregate? Zinc binding significantly stabilizes the native dimer and folded monomer relative to the unfolded state by comparable amounts, -28.5±1.8 and -26.8±0.8 kcal (mol dimer)-1 respectively, at the standard state of 1 M in each component. The calculated zinc dissociation constants (Table 2) are within the range, 10-8 and 10-14 M-1, which have been reported previously.10,11 The wide range of affinities are likely the result of different buffer conditions used. Correcting to a 10 μM standard state, subtracting —RT ln[protein] = +6.7 kcal (mol monomer)-1 for mAS-SOD and -3RT ln[protein] = +20.1 kcal (mol dimer)-1 for AS-SOD, the zinc-bound monomer is stabilized 6.7 kcal (mol monomer)-1 relative to the metal-free monomer state and the relative population of the metal-free monomer would decrease by 105-fold. Zinc stabilizes dimeric AS-SOD by 14.8 kcal (mol dimer)-1, corresponding to a decrease in the relative population of the metal-free dimer of 1011. Thus, the effects of zinc binding on all of the species in the reaction coordinate diagram are dramatic.
Chakrabartty and coworkers have recently demonstrated that the major unfolding pathway of SOD at high concentrations of Gdn-HCl involves the initial dissociation of the dimer coupled with zinc release. Subsequently, the monomer unfolds and copper is released.42 The results of this study may appear surprising given the very high affinity of SOD for zinc at neutral pH. However, as acknowledged by the authors, zinc dissociation could occur either during dimer dissociation or subsequently via a rapid reaction. The results of the present study support the latter possibility in that zinc dissociation from the unfolded state, U, follows the rate-limiting dimer dissociation reaction and the faster monomer unfolding reaction.
Does zinc binding alter the relative populations of monomeric and dimeric forms of SOD at equilibrium? Although the propagation of error analysis finds the stabilities of monomeric and dimeric AS-SOD to be comparable (Table 1), the 20-fold slower dissociation of Zn-AS-SOD vs. AS-SOD shows that zinc actually stabilizes dimeric AS-SOD, relative to monomeric AS-SOD, by 1.7 kcal (mol dimer)-1. The lower free energy for the (N•Zn)2 state relative to the 2(M•Zn) state means that the equilibrium between the monomeric and dimeric zinc-bound states will shift to favor the native dimer of AS-SOD in the presence of zinc. Although the major protective effect of zinc binding on SOD would be to decrease the relative populations of metal-free forms, the decrease in the population of the zinc-bound folded monomeric form might also be a factor.
Implications for aggregation
The capacity of all of the components in the folding mechanism of wild-type SOD, including the TSEs, to bind zinc has significant implications for the aggregation propensity of ALS-inducing variants. It has been shown that zinc-bound SOD is very resistant to aggregation, reinforcing the structural and protective role of zinc.43 Two distinct scenarios for aggregation can be envisioned, an equilibrium scenario and a kinetic scenario. The equilibrium scenario involves fully-folded disulfide-bonded protein occasionally sampling partially-folded or unfolded aggregation-prone states, whereas the kinetic scenario involves the aggregation of transient folding intermediates that are highly populated during the folding reaction following synthesis on the ribosome.
Equilibrium scenario
The sub-nanomolar binding of zinc to native dimeric SOD, coupled with the intrinsic affinity of the metal-free subunits for each other,27 means that the coupled equilibria will flow towards the dimeric state for the wild-type protein. As a result, the populations of all zinc-free states will decrease. Conversely, ALS variants containing mutations that lead to the loss of metal binding44 will shift their populations toward zinc-free monomeric states. For example, the population of the monomeric metal-free form of AS-SOD would increase by 20-fold relative to its metal-bound wild-type counterpart for a 10 μM concentration of the metal-binding variants. Due to the higher order nature of aggregation reactions, even small changes in the concentration of these aggregation prone species can be amplified into a large change in the rate of aggregation.
One possible scenario is partial unfolding of the monomeric intermediate state, leaving the protein susceptible to aggregation. Oliveberg and his colleagues have argued that β5 and β6, edge strands in the β-sandwich (Figure 1), serve as gate keepers for the β-sandwich by preventing the aggregation of the monomeric species.45,46 The boundary position of these two β-strands would make them logical candidates for spontaneous dissociation in a rapidly-interconverting ensemble of conformers in a manifold of states representing the folded monomer of SOD. Supporting this model are molecular dynamics simulations which have predicted that mutations and dimer destabilization have the effect of partially unfolding β5 and β6.47 The disorder induced in the zinc-binding and electrostatic loops in the absence of zinc could propagate to the adjacent β5 strand (Figure 1) and, thereby, enhance the propensity for aggregation of monomeric SOD in the absence of zinc. The increased tendency for metal-free ALS variants to bind hydrophobic dyes may be a manifestation of this phenomenon.48
Alternatively, the unfolded state could provide a platform for aggregation. The micromolar affinity of the unfolded state for zinc and the sub-nanomolar affinities (Table 1) of the folded monomeric and dimeric states of AS-SOD means that the unfolded state will not bind zinc, but the folded monomeric intermediate and the dimeric native state will be saturated at nanomolar concentrations of zinc in vivo41 for the wild-type protein. The folded states of metal-binding variants, which would not be stabilized by metal binding, will not benefit from this shift in equilibrium for the wild-type protein towards more folded states in the presence of zinc. Once again, this effect will be amplified in the rate of aggregation by the higher order kinetics that characterize multi-component reactions.
Kinetic scenario
Arguments have also been made for a kinetic pathway for aggregation, where polypeptide chains misfold and aggregate after synthesis.49 The micromolar dissociation constant for zinc binding to a peptide model for unfolded SOD means that the folding reaction cannot be accelerated and the lifetime of the unfolded state decreased by nanomolar zinc concentrations in vivo. The increased temperature in motor neurons, 37 °C vs. 20 °C in vitro, would only serve to decrease the affinity of zinc for the unfolded state because zinc binding is exothermic.10 As a result, immediately following synthesis on the ribosome, zinc would not be expected to offer protection against aggregation of unfolded, disulfide-reduced ALS-inducing variants of SOD.
Implications for the folding mechanism of SOD
The significant acceleration of the monomer folding rate by the binding of zinc to unfolded AS-SOD provides insights into the structure of the TSEM. A previous mutational analysis of monomeric SOD revealed that β1-β4 and β7, representing contributions from both sides of the β-sandwich (β1/β2/β3 and β4/β7; Figure 1), are integral components of the TSEM.45 A surprising property of the zinc-free monomer folding reaction is its very small rate constant, 0.012 s-1 (Table 2). This value is ∼104 less than the rate constant predicted by the relative contact order, a measure of the sequence separation between contacting residues in the native structure, of the fully-folded SOD monomer.50 This significant discrepancy shows an important aspect of the rate-limiting folding reaction is not captured by this topological metric. The 100-fold acceleration of this reaction by zinc suggests that disorder in the zinc-binding loop might be partially responsible. Application of an algorithm to assess the propensity for disorder, PONDR,51 to the SOD sequence reveals that the zinc-binding loop and, to a lesser extent, the electrostatic loop as well as β5 and β6 are expected to be natively disordered. The prediction for disorder in the loops and in β5 is in accord with experimental observations on monomeric SOD.7,8 Both of these loops follow β-strands that are adjacent and integral to the β-sheet on one side of the β-sandwich, β4 and β7, and these loops pack on each other in the fully-folded structure (Figure 1). Thus, the pre-organization of the zinc-binding loop in the presence of zinc and its potential recruitment of the electrostatic loop might be expected to enhance the probability of accessing the crucial β4/β7 pair of strands in the TSEM. The significance of C-terminal organization is also consistent with slower refolding kinetics observed for disulfide-reduced SOD,25 in which the crosslink between C57 and C146 near the C-terminus is absent. It would be interesting to test the proposed involvement of the zinc and electrostatic loops in the TSEM by performing a mutational analysis on side chains in these loops in the presence and absence of zinc.
Overview on ALS-inducing variants of SOD
The biophysical analyses presented previously28,45,52,53 support a general mechanism for the gain-of-function via aggregation of partially-folded states whose populations are enhanced in ALS-inducing variants of SOD. As would be expected for a highly cooperative system, the contributions of the inherent protein stability, the disulfide bond and the affinity for metal binding are strongly coupled.26,54 This coupling is evident in the present study by the enhanced binding of zinc along the reaction coordinate. It is also evident in previous studies by the loss of affinity for zinc of aggregation-prone ALS variants.44 and by the increased susceptibility of ALS variants for disulfide bond reduction.55 Although chaperones and proteosomes may mitigate the potential for aggregation, the diminished potency of these house-keeping systems in aging cells56-58 may eventually lead to the formation of toxic aggregates by either the thermodynamic or kinetic scenario. Detailed analyses on the stability and folding of ALS-inducing variants of SOD would provide quantitative metrics on the properties of the unfolded and monomeric intermediate states for comparisons with biological measures of aggregation and toxicity. These experiments are currently in progress.
MATERIALS AND METHODS
Protein purification
Human C6A/C111S SOD and the monomeric variant C6A/C111S/G50E/F51E SOD recombinant proteins were expressed in pET3d/BL21-Gold(DE3) Escherichia coli cells (Stratagene®, Inc., Cedar Creek, TX) induced with 1 mM IPTG. All centrifugation steps were done using a Beckman JA 25.50 rotor and performed for 45 min at 4 °C unless otherwise specified. The cells were pelleted by centrifugation at 9000 RPM for 15 min and resuspended in lysis buffer (100 mM HEPES, pH 7.2, 150 mM NaCl, 1 mM ZnSO4, 1 mM DTE, 1 mg mL-1 lysozyme and one protease inhibitor tablet (F. Hoffmann-La Roche Ltd, Switzerland) per 50 mL lysate. After stirring for 30 min at room temperature, the cells were sonicated (30 s on, 90 s off) and centrifuged at 20,000 RPM. The resulting pellet was then resuspended in lysis buffer lacking lysozyme, 0.6 mL per gram of pellet, and the sonication and centrifugation steps were repeated. The combined supernatants were placed in a 47 °C water bath and incubated for 45 min and then centrifuged at 20,000 RPM (this step was not performed for mAS-SOD). The supernatant from the heat incubation was allowed to cool to room temperature, mixed with an equal volume of 4 M (NH4)2SO4, stirred at room temperature for 30 min, and centrifuged at 20,000 RPM.
The resulting supernatant was dialyzed extensively against standard buffer (20 mM HEPES, pH 7.2) and loaded onto a Q Sepharose column and eluted with a linear gradient to 40% high salt buffer (300 mM KCl, 20 mM HEPES, pH 7.2) over 10 column volumes. The presence of AS-SOD in the fractions was determined by SDS-PAGE. The pooled fractions were concentrated to <10 mL using a YM-10 membrane on an Amicon® concentrator and dialyzed against standard buffer. The protein was demetalated and tested for purity as previously described.27 The concentration of protein was calculated using a molar extinction coefficient at 280 nm of 5400 M-1 cm-1 for the monomer of AS-SOD, and, unless otherwise specified, all protein concentrations are given in terms of moles of monomer of AS-SOD per liter of solution.
Peptide labeling and purification
The peptide fragment representing the metal binding region of loop IV, amino acids 61-86, was purchased from Sigma-Aldrich and modified by adding an N-terminal tryptophan and a C-terminal cysteine. Purity was assessed by a single high pressure liquid chromatography peak and by mass spectroscopy provided by the manufacturer. The covalent modification of the cysteine with the FRET acceptor N-(iodoacetaminoethyl)-1-naphthylamine-5-sulfonic acid (IAEDANS) was performed at peptide concentrations of 1 mg mL-1 in standard buffer with the addition of 1 mM TCEP. Initially, 10-fold excess free IAEDANS was incubated with the peptide at room temperature for 2 hours. Another 10-fold excess of label was then added and the reaction mixture was incubated for another 2 hours at 4 °C. The labeled peptide was separated from free dye by running the reaction mixture on a 5 mL HiTrap™ desalting column (GE Healthcare Life Sciences).
Equilibrium folding experiments
All circular dichroism (CD) spectroscopy was performed on a Jasco-810 spectropolarimeter (Jasco Inc., Easton, MD) equipped with a water-cooled Peltier temperature control system. The Gdn-HCl induced unfolding curves were monitored from 220-240 nm in a 0.5 cm path length quartz cuvette using a scan rate of 20 nm min-1 and a response time of 8 s. Gdn-HCl concentrations were determined by refractive index59 on a Leica Mark II refractometer. Unless otherwise stated, titration samples were made from concentration matched stocks of folded protein in buffer and unfolded protein at 7 M Gdn-HCl and incubated at room temperature for 1 day for apo-protein and 7 days for zinc-bound protein. Coincident unfolding and refolding curves from companion experiments in which denaturant was added to native protein and buffer was added to unfolded protein insured that the samples were fully equilibrated in this time frame. The stocks were mixed precisely using a Hamilton Microlab 500 titrator interfaced with in-house software to calculate the mixing ratios, and the index of refraction of each sample was measured after the experiment was completed. The protein concentrations, given in monomer units, were 10 μM for AS-SOD and 5 μM for mAS-SOD for the equilibrium unfolding experiments.
Kinetic experiments
The unfolding and refolding kinetics of AS-SOD, initiated by manual mixing, were monitored by CD. Data were collected at 230 nm in a 1 cm2 cuvette under continuous mixing with a solution volume of 1.8-2.0 mL. Unfolding jumps were initiated from protein in 0 M Gdn-HCl and refolding jumps were initiated from protein denatured overnight in 5-6 M Gdn-HCl to various final concentrations. For zinc-loaded samples, the protein was incubated in stoichiometric zinc prior to dilution in the buffer. The final concentration of Gdn-HCl was measured by index of refraction. Protein concentrations were 10 μM for both AS-SOD and mAS-SOD for the kinetic folding experiments.
Analysis of equilibrium folding data
To determine the folding free energy of metal-free AS-SOD and mAS-SOD from the equilibrium unfolding titrations, the data were fit to a simple two-state mechanism. While it is known that SOD folds via a monomeric intermediate,25,27,29 it is not populated significantly at these protein concentrations,28 making a 2-state folding model an adequate approximation. For all thermodynamic analyses, the free energy of folding in the absence of denaturant was calculated assuming a linear dependence of the apparent free energy on the denaturant concentration. Equilibrium data were analyzed by globally fitting the CD signal at 20 different wavelengths (220-240 nm) to the same thermodynamic parameters. All data were fit using Savuka 6.2, an in-house non-linear least squares program using the Marquardt-Levenberg algorithm.60 In the absence of zinc, equilibrium folding data were fit to a two-state model, U ⇆ N for mAS-SOD and 2U ⇆ N2 for AS-SOD as previously described.27
For equilibrium titrations in the presence of zinc, the data were modeled as a three state reaction.
The Gdn-HCl induced unfolding transition for these titrations involves the native zinc-bound monomer (N•Zn),or dimer (N•Zn)2, state, the zinc-bound unfolded state U•Zn or 2(U•Zn) and the zinc-free unfolded chains, U+Zn and 2U+2Zn, respectively. The equilibrium constants can be defined for the stable monomeric form, Zn-mAS-SOD, as:
and for the dimeric form, Zn-AS-SOD:
An equation for total protein concentration may be written as a function of KU, the equilibrium constant of unfolding, KD, the dissociation constant for zinc, total protein concentration (Pt), total zinc concentration (Zt), and the concentration of unfolded protein ([U]) (See supplemental for a more detailed derivation).Zn-mAS-SOD:
Zn-AS-SOD:
The titration data were fit by numerically solving the equations for the concentration of unfolded protein at each Gdn-HCl concentration. The value for KD and its denaturant dependence were held fixed to the values obtained from an independent assessment of the zinc-peptide complex, and the value for KU was optimized across the denaturant concentration and the wavelength range, 220 to 240 nm. The free energy differences between thermodynamic states were assumed to depend linearly on the Gdn-HCl concentration.
Analysis of kinetic folding data
The unfolding and refolding relaxation times were obtained by jumping from 0 or 6 M Gdn-HCl to various final concentrations of denaturant and fitting the traces to exponentials as described previously.27 The monomer folding and unfolding rates, for both apo- and Zn-mAS-SOD in the absence of denaturant, were calculated by fitting the chevron to a two-state folding model
Where k is the observed rate constant, kf and ku are the folding and unfolding rate constants in the absence of denaturant respectively, mf and mu are the denaturant dependence of the folding and unfolding rates respectively, [D] is the denaturant concentration, R is the gas constant and T is the absolute temperature. Knowing the folding and unfolding rate constants in the absence of denaturant, the folding free energy can be calculated:
The dimer dissociation rate constants in the absence of denaturant were calculated from linear extrapolations to 0 M Gdn-HCl of the >5.5 M Gdn-HCl region of the unfolding leg of the Zn-AS-SOD chevron and the >3.8 M Gdn-HCl region of the unfolding leg of the apo-AS-SOD chevron.
Analysis of zinc binding to the zinc-binding loop peptide
All fluorescence spectroscopy was done on a T-format Spex Fluorolog-3 (Instruments S. A. Inc, Edison, NJ) at room temperature. The donor tryptophan was excited at 280 nm and the emission spectra were monitored from 320 to 540 nm to encompass both the tryptophan and AEDANS fluorescence. The peptide was incubated at various concentrations of Gdn-HCl ranging from 0-6 M in standard buffer. Zinc was added in a stepwise manner, from a high concentration stock solution to minimize changes to the volume, to samples at each denaturant concentration. The fluorescence spectra were obtained at various zinc and denaturant concentrations and corrected for background fluorescence and the small change in volume accompanying the addition of zinc. The fluorescence was measured as a function of zinc concentration, and all wavelengths were simultaneously fit to a binding isotherm with one binding site per peptide:
where y is the fluorescence intensity, Bmax is the signal at saturating concentrations of zinc, KD is the dissociation equilibrium constant and [Zn] is the concentration of zinc.
Supplementary Material
Acknowledgements
The authors would like to thank Drs. Osman Bilsel, Anna-Karin E. Svensson, Lawrence Hayward and Ashutosh Tiwari for their stimulating discussions and input during this project. The authors are also grateful to Dr. James Evans and the personnel at the Proteomics and Mass Spectrometry Core Facility of Massachusetts Medical School for the molecular weight and protein purity determinations. This work was supported by NIH grant GM 54836 to C.R.M.
Abbreviations used
- AS-SOD
C6A/C111S SOD
- ALS
amyotrophic lateral sclerosis
- CD
circular dichroism
- fALS
familial amyotrophic lateral sclerosis
- FRET
Förster resonance energy transfer
- Gdn-HCl
guanidine hydrochloride
- IAEDANS
5-((((2-iodoacetyl)amino)ethyl)amino)naphthalene-1- sulfonic acid
- mAS-SOD
F50E/G51E/C6A/C11S monomeric variant of SOD
- SOD
human Cu, Zn superoxide dismutase
- TCEP
(tris(2-carboxyethyl)phosphine)
- TSE
transition state ensemble.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rowland LP, Shneider NA. Amyotrophic lateral sclerosis. N Engl J Med. 2001;344:1688–700. doi: 10.1056/NEJM200105313442207. [DOI] [PubMed] [Google Scholar]
- 2.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng HX, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 3.Andersen PM, Sims KB, Xin WW, Kiely R, O’Neill G, Ravits J, Pioro E, Harati Y, Brower RD, Levine JS, Heinicke HU, Seltzer W, Boss M, Brown RH., Jr. Sixteen novel mutations in the Cu/Zn superoxide dismutase gene in amyotrophic lateral sclerosis: a decade of discoveries, defects and disputes. Amyotroph Lateral Scler Other Motor Neuron Disord. 2003;4:62–73. doi: 10.1080/14660820310011700. [DOI] [PubMed] [Google Scholar]
- 4.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–55. [PubMed] [Google Scholar]
- 5.Parge HE, Hallewell RA, Tainer JA. Atomic structures of wild-type and thermostable mutant recombinant human Cu,Zn superoxide dismutase. Proc Natl Acad Sci U S A. 1992;89:6109–13. doi: 10.1073/pnas.89.13.6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts BR, Tainer JA, Getzoff ED, Malencik DA, Anderson SR, Bomben VC, Meyers KR, Karplus PA, Beckman JS. Structural characterization of zinc-deficient human superoxide dismutase and implications for ALS. J Mol Biol. 2007;373:877–90. doi: 10.1016/j.jmb.2007.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banci L, Bertini I, Cramaro F, DelConte R, Viezzoli MS. Solution Structure of Apo Cu,Zn Superoxide Dismutase: Role of Metal Ions in Protein Folding. Biochemistry. 2003;42:9543–9553. doi: 10.1021/bi034324m. [DOI] [PubMed] [Google Scholar]
- 8.Strange RW, Antonyuk S, Hough MA, Doucette PA, Rodriguez JA, Hart PJ, Hayward LJ, Valentine JS, Hasnain SS. The structure of holo and metal-deficient wild-type human Cu, Zn superoxide dismutase and its relevance to familial amyotrophic lateral sclerosis. J Mol Biol. 2003;328:877–91. doi: 10.1016/s0022-2836(03)00355-3. [DOI] [PubMed] [Google Scholar]
- 9.Banci L, Bertini I, Cantini F, D’Onofrio M, Viezzoli MS. Structure and dynamics of copper-free SOD: The protein before binding copper. Protein Sci. 2002;11:2479–2492. doi: 10.1110/ps.0210802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potter SZ, Zhu H, Shaw BF, Rodriguez JA, Doucette PA, Sohn SH, Durazo A, Faull KF, Gralla EB, Nersissian AM, Valentine JS. Binding of a single zinc ion to one subunit of copper-zinc superoxide dismutase apoprotein substantially influences the structure and stability of the entire homodimeric protein. J Am Chem Soc. 2007;129:4575–83. doi: 10.1021/ja066690+. [DOI] [PubMed] [Google Scholar]
- 11.Crow JP, Sampson JB, Zhuang Y, Thompson JA, Beckman JS. Decreased zinc affinity of amyotrophic lateral sclerosis-associated superoxide dismutase mutants leads to enhanced catalysis of tyrosine nitration by peroxynitrite. J Neurochem. 1997;69:1936–44. doi: 10.1046/j.1471-4159.1997.69051936.x. [DOI] [PubMed] [Google Scholar]
- 12.Furukawa Y, O’Halloran TV. Posttranslational modifications in Cu,Zn-superoxide dismutase and mutations associated with amyotrophic lateral sclerosis. Antioxid Redox Signal. 2006;8:847–67. doi: 10.1089/ars.2006.8.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valentine JS, Hart PJ. Misfolded CuZnSOD and amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2003;100:3617–22. doi: 10.1073/pnas.0730423100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cleveland DW, Rothstein JD. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat Rev Neurosci. 2001;2:806–19. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- 15.Wood JD, Beaujeux TP, Shaw PJ. Protein aggregation in motor neurone disorders. Neuropathol Appl Neurobiol. 2003;29:529–45. doi: 10.1046/j.0305-1846.2003.00518.x. [DOI] [PubMed] [Google Scholar]
- 16.Bruijn LI, Miller TM, Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci. 2004;27:723–49. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 17.Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–98. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 18.Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J Mol Med. 2003;81:678–99. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 19.Valentine JS, Doucette PA, Potter S. Zittin. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu Rev Biochem. 2005;74:563–93. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 20.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–7. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 21.Khare SD, Caplow M, Dokholyan NV. The rate and equilibrium constants for a multistep reaction sequence for the aggregation of superoxide dismutase in amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2004;101:15094–9. doi: 10.1073/pnas.0406650101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hart PJ. Pathogenic superoxide dismutase structure, folding, aggregation and turnover. Curr Opin Chem Biol. 2006;10:131–8. doi: 10.1016/j.cbpa.2006.02.034. [DOI] [PubMed] [Google Scholar]
- 23.Cao X, Antonyuk S, Seetharaman SV, Whitson LJ, Taylor AB, Holloway SP, Strange RW, Doucette PA, Valentine JS, Tiwari A, Hayward LJ, Padua S, Cohlberg JA, Hasnain SS, Hart PJ. Structures of the G85R variant of SOD1 in Familial ALS. J Biol Chem. 2008 doi: 10.1074/jbc.M801522200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindberg MJ, Bystrom R, Boknas N, Andersen PM, Oliveberg M. Systematically perturbed folding patterns of amyotrophic lateral sclerosis (ALS)-associated SOD1 mutants. Proc Natl Acad Sci U S A. 2005;102:9754–9. doi: 10.1073/pnas.0501957102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindberg MJ, Normark J, Holmgren A, Oliveberg M. Folding of human superoxide dismutase: disulfide reduction prevents dimerization and produces marginally stable monomers. Proc Natl Acad Sci U S A. 2004;101:15893–8. doi: 10.1073/pnas.0403979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hornberg A, Logan DT, Marklund SL, Oliveberg M. The coupling between disulphide status, metallation and dimer interface strength in Cu/Zn superoxide dismutase. J Mol Biol. 2007;365:333–42. doi: 10.1016/j.jmb.2006.09.048. [DOI] [PubMed] [Google Scholar]
- 27.Svensson AK, Bilsel O, Kondrashkina E, Zitzewitz JA, Matthews CR. Mapping the folding free energy surface for metal-free human Cu,Zn superoxide dismutase. J Mol Biol. 2006;364:1084–102. doi: 10.1016/j.jmb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Vassall KA, Stathopulos PB, Rumfeldt JA, Lepock JR, Meiering EM. Equilibrium thermodynamic analysis of amyotrophic lateral sclerosis-associated mutant apo Cu,Zn superoxide dismutases. Biochemistry. 2006;45:7366–79. doi: 10.1021/bi0600953. [DOI] [PubMed] [Google Scholar]
- 29.Rumfeldt JA, Stathopulos PB, Chakrabarrty A, Lepock JR, Meiering EM. Mechanism and thermodynamics of guanidinium chloride-induced denaturation of ALS-associated mutant Cu,Zn superoxide dismutases. J Mol Biol. 2006;355:106–23. doi: 10.1016/j.jmb.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 30.Hallewell RA, Imlay KC, Lee P, Fong NM, Gallegos C, Getzoff ED, Tainer JA, Cabelli DE, Tekamp-Olson P, Mullenbach GT, et al. Thermostabilization of recombinant human and bovine CuZn superoxide dismutases by replacement of free cysteines. Biochem Biophys Res Commun. 1991;181:474–80. doi: 10.1016/s0006-291x(05)81443-3. [DOI] [PubMed] [Google Scholar]
- 31.Lindberg MJ, Tibell L, Oliveberg M. Common denominator of Cu/Zn superoxide dismutase mutants associated with amyotrophic lateral sclerosis: decreased stability of the apo state. Proc Natl Acad Sci U S A. 2002;99:16607–12. doi: 10.1073/pnas.262527099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertini I, Piccioli M, Viezzoli MS, Chiu CY, Mullenbach GT. A spectroscopic characterization of a monomeric analog of copper, zinc superoxide dismutase. Eur Biophys J. 1994;23:167–76. doi: 10.1007/BF01007608. [DOI] [PubMed] [Google Scholar]
- 33.Lyons TJ, Nersissian A, Huang H, Yeom H, Nishida CR, Graden JA, Gralla EB, Valentine JS. The metal binding properties of the zinc site of yeast copper-zinc superoxide dismutase: implications for amyotrophic lateral sclerosis. J Biol Inorg Chem. 2000;5:189–203. doi: 10.1007/s007750050363. [DOI] [PubMed] [Google Scholar]
- 34.Strange RW, Antonyuk SV, Hough MA, Doucette PA, Valentine JS, Hasnain SS. Variable metallation of human superoxide dismutase: atomic resolution crystal structures of Cu-Zn, Zn-Zn and as-isolated wild-type enzymes. J Mol Biol. 2006;356:1152–62. doi: 10.1016/j.jmb.2005.11.081. [DOI] [PubMed] [Google Scholar]
- 35.Pozdnyakova I, Guidry J, Wittung-Stafshede P. Probing copper ligands in denatured Pseudomonas aeruginosa azurin: unfolding His117Gly and His46Gly mutants. J Biol Inorg Chem. 2001;6:182–8. doi: 10.1007/s007750000189. [DOI] [PubMed] [Google Scholar]
- 36.Wittung-Stafshede P. Role of cofactors in protein folding. Acc Chem Res. 2002;35:201–8. doi: 10.1021/ar010106e. [DOI] [PubMed] [Google Scholar]
- 37.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4:2138–48. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matthews CR. Effect of point mutations on the folding of globular proteins. Methods Enzymol. 1987;154:498–511. doi: 10.1016/0076-6879(87)54092-7. [DOI] [PubMed] [Google Scholar]
- 39.Pace CN, Shirley BA, Thompson JA. Measuring the conformational stability of a protein. IRL Press, Oxford University press; 1990. Protein Structure, a Practical Approach. [Google Scholar]
- 40.Bodenreider C, Kiefhaber T. Interpretation of protein folding psi values. J Mol Biol. 2005;351:393–401. doi: 10.1016/j.jmb.2005.05.062. [DOI] [PubMed] [Google Scholar]
- 41.Choi DW, Koh JY. Zinc and brain injury. Annu Rev Neurosci. 1998;21:347–75. doi: 10.1146/annurev.neuro.21.1.347. [DOI] [PubMed] [Google Scholar]
- 42.Mulligan VK, Kerman A, Ho S, Chakrabartty A. Denaturational Stress Induces Formation of Zinc-Deficient Monomers of Cu,Zn Superoxide Dismutase: Implications for Pathogenesis in Amyotrophic Lateral Sclerosis. J Mol Biol. 2008 doi: 10.1016/j.jmb.2008.08.024. [DOI] [PubMed] [Google Scholar]
- 43.Banci L, Bertini I, Durazo A, Girotto S, Gralla EB, Martinelli M, Valentine JS, Vieru M, Whitelegge JP. Metal-free superoxide dismutase forms soluble oligomers under physiological conditions: a possible general mechanism for familial ALS. Proc Natl Acad Sci U S A. 2007;104:11263–7. doi: 10.1073/pnas.0704307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayward LJ, Rodriguez JA, Kim JW, Tiwari A, Goto JJ, Cabelli DE, Valentine JS, Brown RH., Jr. Decreased metallation and activity in subsets of mutant superoxide dismutases associated with familial amyotrophic lateral sclerosis. J Biol Chem. 2002;277:15923–31. doi: 10.1074/jbc.M112087200. [DOI] [PubMed] [Google Scholar]
- 45.Nordlund A, Oliveberg M. Folding of Cu/Zn superoxide dismutase suggests structural hotspots for gain of neurotoxic function in ALS: parallels to precursors in amyloid disease. Proc Natl Acad Sci U S A. 2006;103:10218–23. doi: 10.1073/pnas.0601696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson JS, Richardson DC. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci U S A. 2002;99:2754–9. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khare SD, Dokholyan NV. Common dynamical signatures of familial amyotrophic lateral sclerosis-associated structurally diverse Cu, Zn superoxide dismutase mutants. Proc Natl Acad Sci U S A. 2006;103:3147–52. doi: 10.1073/pnas.0511266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tiwari A, Xu Z, Hayward LJ. Aberrantly increased hydrophobicity shared by mutants of Cu,Zn-superoxide dismutase in familial amyotrophic lateral sclerosis. J Biol Chem. 2005;280:29771–9. doi: 10.1074/jbc.M504039200. [DOI] [PubMed] [Google Scholar]
- 49.Jahn TR, Radford SE. Folding versus aggregation: polypeptide conformations on competing pathways. Arch Biochem Biophys. 2008;469:100–17. doi: 10.1016/j.abb.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J Mol Biol. 1998;277:985–94. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 51.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 52.Khare SD, Caplow M, Dokholyan NV. FALS mutations in Cu, Zn superoxide dismutase destabilize the dimer and increase dimer dissociation propensity: a large-scale thermodynamic analysis. Amyloid. 2006;13:226–35. doi: 10.1080/13506120600960486. [DOI] [PubMed] [Google Scholar]
- 53.Rakhit R, Crow JP, Lepock JR, Kondejewski LH, Cashman NR, Chakrabartty A. Monomeric Cu,Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis. J Biol Chem. 2004;279:15499–504. doi: 10.1074/jbc.M313295200. [DOI] [PubMed] [Google Scholar]
- 54.Bruns CK, Kopito RR. Impaired post-translational folding of familial ALS-linked Cu, Zn superoxide dismutase mutants. Embo J. 2007;26:855–66. doi: 10.1038/sj.emboj.7601528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tiwari A, Hayward LJ. Familial amyotrophic lateral sclerosis mutants of copper/zinc superoxide dismutase are susceptible to disulfide reduction. J Biol Chem. 2003;278:5984–92. doi: 10.1074/jbc.M210419200. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Q, Powers ET, Nieva J, Huff ME, Dendle MA, Bieschke J, Glabe CG, Eschenmoser A, Wentworth P, Jr., Lerner RA, Kelly JW. Metabolite-initiated protein misfolding may trigger Alzheimer’s disease. Proc Natl Acad Sci U S A. 2004;101:4752–7. doi: 10.1073/pnas.0400924101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Massey AC, Kiffin R, Cuervo AM. Autophagic defects in aging: looking for an “emergency exit”? Cell Cycle. 2006;5:1292–6. doi: 10.4161/cc.5.12.2865. [DOI] [PubMed] [Google Scholar]
- 58.Derham BK, Harding JJ. Effect of aging on the chaperone-like function of human alpha-crystallin assessed by three methods. Biochem J. 1997;328(Pt 3):763–8. doi: 10.1042/bj3280763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomson JA, Shirley BA, Grimsley GR, Pace CN. Conformational stability and mechanism of folding of ribonuclease T1. J Biol Chem. 1989;264:11614–20. [PubMed] [Google Scholar]
- 60.Bilsel O, Zitzewitz JA, Bowers KE, Matthews CR. Folding mechanism of the alpha-subunit of tryptophan synthase, an alpha/beta barrel protein: global analysis highlights the interconversion of multiple native, intermediate, and unfolded forms through parallel channels. Biochemistry. 1999;38:1018–29. doi: 10.1021/bi982365q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.