Abstract
Transforming growth factor beta induced protein (TGFBIp), is secreted into the extracellular space. When fragmentation of C-terminal portions is blocked, apoptosis is low, even when the protein is overexpressed. If fragmentation occurs, apoptosis is observed. Whether full-length TGFBIp or integrin-binding fragments released from its C-terminus is necessary for apoptosis remains equivocal. More importantly, the exact portion of the C-terminus that conveys the pro-apoptotic property of TGFBIp is uncertain. It is reportedly within the final 166 amino acids. We sought to determine if this property is dependent upon the final 69 amino acids containing the integrin-binding, EPDIM and RGD, sequences. With MG-63 osteosarcoma cells, transforming growth factor (TGF)-β1 treatment increased expression of TGFBIp over 72 hours (p<0.001). At this time point, apoptosis was significantly increased (p<0.001) and was prevented by an anti-TGFBIp, polyclonal antibody (p<0.05). Overexpression of TGFBIp by transient transfection produced a 2-fold increase in apoptosis (p<0.01). Exogenous purified TGFBIp at concentrations of 37 to 150 nM produced a dose dependent increase in apoptosis (p<0.001). Mass spectrometry analysis of TGFBIp isolated from conditioned medium of cells treated with TGF-β1 revealed truncated forms of TGFBIp that lacked integrin-binding sequences in the C-terminus. Recombinant TGFBIp truncated, similarly, at amino acid 614 failed to induce apoptosis. A recombinant fragment encoding the final 69 amino acids of the TGFBIp C-terminus produced significant apoptosis. This apoptosis level was comparable to that induced by TGF-β1 upregulation of endogenous TGFBIp. Mutation of the integrin-binding sequence EPDIM, but not RGD, blocked apoptosis (p<0.001). These pro-apoptotic actions are dependent on the C-terminus most likely to interact with integrins.
Keywords: extracellular matrix, tumor, BIG-H3, beta-ig, keratoepithelin, cancer
1. Introduction
Transforming growth factor-β induced protein (TGFBIp), also known in the literature as BIG-H3, keratoepithelin and RGD-CAP, was initially characterized as a novel transcript upregulated by TGF-β1 growth-arrested A495 human adenocarcinoma cells (Skonier et al., 1992). TGFBIp is a secretory protein that supports cell adhesion and spreading (Ferguson et al., 2003a; Ferguson et al., 2003b; Kim et al., 2002; Kim et al., 2000; LeBaron et al., 1995; Nam et al., 2006; Ohno et al., 1999; Park et al., 2004) and binds molecules of the ECM (Billings et al., 2002; Hanssen et al., 2003; Hashimoto et al., 1997; Reinboth et al., 2006). In cell culture TGFBIp has been implicated in apoptosis via two mechanisms, properties that have not yet been shown in vivo, but still considered of grave importance as related to tumor growth and metastasis. First Morand and coworkers detected non-significant levels of apoptosis in HeLa and immortalized human corneal epithelial (HCE) cells when overexpressing full-length, wild type (wt) TGFBIp (TGFBIpwt) constructs that somehow prevented the posttranslational processing for C-terminal fragmentation with these transient transfections. Significant abnormal apoptosis was only detected when mutations of TGFBIp constructs were placed in sites known to occur in TGFBIp-linked human corneal dystrophies (CD), and, notably, these mutations were not within the EPDIM and RGD, integrin-binding sequences located within the C-terminal region of the molecule. Overall, apoptosis was not detected if full-length TGFBIpwt was used as a fusion protein with a C-terminal-linked green fluorescent protein. Nor was apoptosis associated with these mutations affected when the construct was truncated at amino acid 541, that is, without the final 142 amino acids (Morand et al., 2003). Second and contrary to these mutation effects, if a wt construct design did not interfere with fragmentation, then DNA laddering was evident in CHO and H1229 human lung carcinoma cells, and the final 44 amino acids seemed critical to the observed effects (Kim et al., 2003). In most instances, C-terminal fragmentation of TGFBIpwt is hypothesized to be required for apoptosis, in particular when there are no mutations to the gene.
Since the RGD sequence was implicated in triggering apoptosis in tumor cells (Kim et al., 2003), and others have implicated the integrin-binding sequence EPDIM in apoptosis (Morand et al., 2003), a closer examination of the C-terminal fragment properties was indicated. Matrix assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF/MS) characterized three forms of TGFBIp. One is the longest form that is most inclusive of the C-terminus whereas the C-termini of the two other forms mapped within the fourth fasciclin-like domain (FD4), near the integrin-binding sequences RGD and EPDIM. These early observations were corroborated by a report from another group (Andersen et al., 2004), wherein analyses of TGFBIp isolated from human and porcine cornea had likewise revealed full-length TGFBIp was a minor form; whereas, TGFBIp exhibiting C-termini at or near the RGD sequence was predicted as a major form. Collectively, the evidence substantiate cellular processing to produce C-terminal fragments, at least one of which is capable of inducing apoptosis through a suspected integrin interaction. Therefore, since the EPDIM sequence begins at amino acid 615 and the RGD follows with an intervening sequence of 29 amino acids, we focused on the final 69 amino acids of the C-terminus.
This study documents that the C-terminal 69 amino acids of TGFBIp are essential for inducing apoptosis in MG-63 osteosarcoma cells. TGF-β1 did upregulate TGFBIp expression by MG-63 osteosarcoma cells. This endogenous TGFBIp upregulation was trailed by an increase in osteosarcoma-cell apoptosis, and the apoptotic effect was blocked by anti-TGFBIp antibody. When cells were made to overexpress recombinant TGFBIpwt or when exogenous TGFBIpwt was added to the growth media of osteosarcoma cell lines, the apoptotic response increased significantly. In contrast, a truncated TGFBIp, lacking integrin-binding sequences EPDIM and RGD, failed to elicit an increase in apoptosis; however, when a TGFBIp protein fragment encoding the missing C-terminal, integrin-binding sequences was applied, the apoptotic stimulus was recovered. Mutating the C-terminal EPDIM and RGD integrin-binding sequences revealed that EPDIM elicits the apoptotic response. These results emphatically implicate an integrin-arbitrated effect.
2. Results
2.1 TGF-β1 treated MG-63 cells overexpress TGFBIp
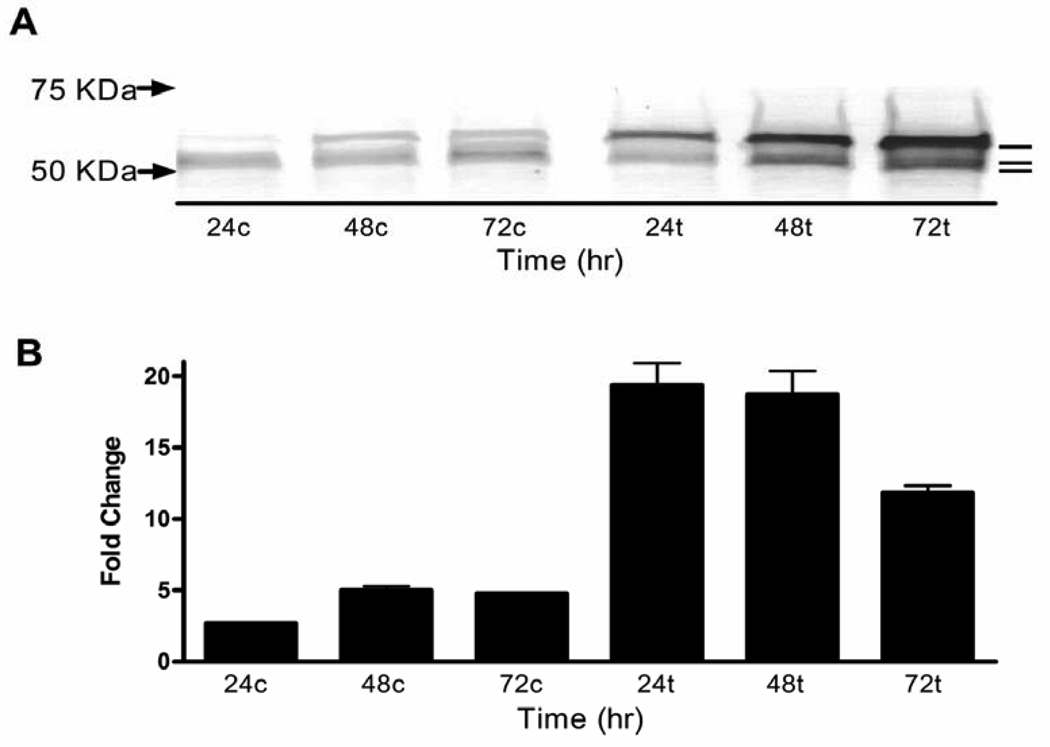
To characterize TGFBIp synthesized by unstimulated MG-63 cells, proteins in the conditioned medium resolved by SDS PAGE were stained by immunoblot using anti-TGFBIp antibody. Three protein bands were detected with relative mobility calculated at 69, 62, and 60 kDa (Figure 2A). The presence of lower molecular mass forms of TGFBIp indicates that fragmentation occurs and supports a baseline level of apoptosis. These findings raised the question of whether increased expression of the TGFBIp gene would provide sufficient TGFBIp to achieve significant lethality. To address this possibility we first determined whether the osteosarcoma TGFBIp gene was responsive to TGF-β1. Real-time quantitative reverse transcriptase PCR (qRT-PCR) analysis verified that TGF-β1 increased TGFBIp transcript levels (p< 0.001; Figure 2B). This was paralleled by increased expression of the three TGFBIp forms (Figure 2A).
Figure 2.
TGF-β1 induces TGFBIp expression in MG-63 cells. (A) Immunoblots of proteins of the conditioned medium from cells treated with 5 ng/ml TGF-β1 show TGFBIp at molecular masses estimated at 69, 62, and 60 KDa (upper, center and lower lines, respectively). Serum-free medium was used so that albumin does not interfere with the TGFBIp bands in immunoblots. Therefore, the material loaded was normalized to total protein in the conditioned media. (B) qRT-PCR performed on RNA extracted from TGF-β1 treated cells shows a 17, 14, and a 7 fold upregulation of TGFBIp expression when compared to nontreated cells at 24, 48, and 72 hours, respectively. The One Way ANOVA was significant [F(5,17)=60.05;p<0.0001]. All three time points were significantly different from the corresponding time for control p<0.001. Arrows indicate protein molecular weight standards. c - nontreated control; t - TGF-β1 treated.
2.2 TGFBIp antibody blocks TGF-β1 induced apoptosis
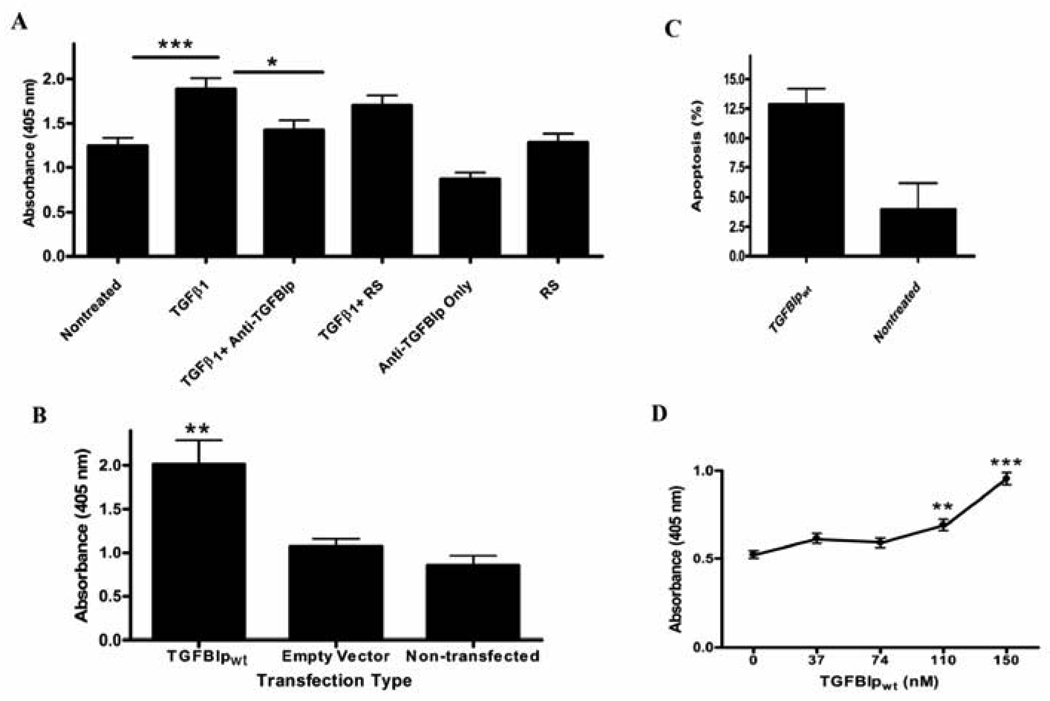
MG-63 cells treated with 5 ng/ml TGF-β1 for 72 hours exhibited a significant increase in apoptosis when compared to nontreated cells (p<0.001; Figure 3A). Moreover, anti-TGFBIp antibody diminished significantly the TGF-β1 evoked apoptosis (Figure 3A). These data suggest a relationship between upregulation of the TGFBIp gene and increased apoptosis.
Figure 3.
Endogenous and exogenous TGFBIp increases MG-63 cell apoptosis. (A) Cells were treated with recombinant TGF-β1, anti-TGFBIp antiserum or control reagents. ELISA assays as described in the methods section were used to quantify apoptosis. Apoptosis in TGF-β1 treated cells (TGF-β1) was significantly increased when compared to control (Nontreated) cells,***p<0.001. The Two Way ANOVA for TGF-β1 treatment was significant [F(1,84)=41.9;p<0.0001]. Addition of an anti-TGFBIp antibody in combination with TGF-β1 treatment (TGF-β1+Anti-TGFBIp) prevented the increase in apoptosis when compared to TGF-pi treated cells,*p<0.05. The Two Way ANOVA for the blocking antibody was significant [F(2,84)=9.7p;<0.001]. Cells treated with TGF-β1 and control preimmune rabbit serum (RS) (TGF-β1+RS) demonstrated apoptosis levels similar to TGF-β1 treated cells, p>0.05. The Two Way ANOVA was not significant for anti-TGFBIp only (p>0.05) and RS (p>0.05). N=15 (5 individual wells in each group, 3 experiments per group). (B) TGFBIp overexpression induces apoptosis of MG-63 cells. Cells transiently transfected with a TGFBIp overexpression plasmid (TGFBIpwt) show an increase in apoptosis when compared to Empty Vector and Non-transfected cells,**p<0.01, p<0.001, respectively. One Way ANOVA was significant [F(2,29)=11.64; p<0.001]. N=10 per treatment group. (C) Annexin staining increased in MG-63 cells treated with 150 nM TGFBIpwt, p<0.05. (D) TGFBIpwt-mediated apoptosis of MG-63 cells is concentration-dependent. Purified, TGFBIpwt was added at increasing concentrations, as shown on the x-axis, in MG-63 cell growth medium. One Way ANOVA [F(4,44)=32.71;p<0.0001].***p<0.001 compared to all other concentrations.**p<0.01 compared to 0 nM. The 110 nM was significantly different from 74 nM at p<0.05. N=9 per group.
2.3 Overexpression of full-length TGFBIpwt increases apoptosis
Apoptosis was quantified in cultures of transiently transfected MG-63 cells overexpressing human recombinant TGFBIpwt. Increased apoptosis was significant (p<0.01) in cells overexpressing TGFBIpwt when compared to non-transfected cells, or cells that were transfected with the plasmid only (Figure 3B), supporting the hypothesis that TGFBIp itself is sufficient to induce an apoptotic response. The results indicate that the apoptotic effect is dependent on the concentration of TGFBIp in the growth medium. Our conclusion is that amplified expression of TGFBIpwt increases the number of cells undergoing apoptosis.
2.4 Apoptosis induced by TGFBIp add back: a concentration-response effect
TGFBIpwt purified as previously described (Ferguson et al., 2003a) was mixed with fresh growth medium and apoptosis was quantified after a 3-day incubation period. In our early studies, MG-63 cells treated with 150 nM TGFBIp stained positive with anti-annexin V antibody. An evaluation of the percent apoptosis from three independent experiments revealed an increase of 13% in annexin staining in the TGFBIp-treated cells and 4% in the controls cells (Figure 3C). This finding was followed by a separate TGFBIp concentration-response trial to determine the extent of apoptosis at different TGFBIp concentrations. The results of TGFBIpwt add-back experiments demonstrated that lower concentrations of TGFBIp were sufficient to bring about a notable increase in apoptotic MG-63 cells, detected by immunological quantification of single-stranded DNA (ssDNA). Concentrations of TGFBIp at 37 and 74 nM did not increase apoptosis; however, concentrations of TGFBIp at 110 and 150 nM produced a statistically significant increase in apoptosis when compared to nontreated cells at p<0.001 (Figure 3D). The effects of these two concentrations were also significantly different from each other at p<0.001. TGFBIp in the conditioned medium of MG63 cells that were treated with 5 ng/ml TGF-β1 for 72 hours was estimated by immunoblot densitometry to be 595 ng/ml (8.7 nM), or 7.9% of the TGFBIp that induced a significant increase in apoptosis in the add-back experiments. The comparatively lower concentration of endogenous TGFBIp that achieved significant apoptosis is explained by the probability that a percentage of our purified TGFBIp was already fragmented so that the C-terminal apoptotic cue was lacking. The concentration-dependence demonstrates that there is a direct relationship between levels of TGFBIp available in the extracellular environment and the probability of apoptosis, establishing that TGFBIp itself is a key molecule of capacity sufficient to trigger apoptosis. These results are consistent with TGFBIp-dependent apoptosis as a physiological process.
2.5 Mass spectrometry analyses of secreted TGFBIp
To verify C-terminal fragmentation, each of the three forms isolated from MG-63 cell conditioned medium were characterized by MALDI-TOF mass spectrometry (Table 1). The tryptic peptides mapped to the mature amino terminus, indicating the N-terminus was essentially intact. Mapping the C-terminal tryptic peptides validated C-terminal fragmentation. Relative to the N-terminus, the most distal C-terminal peptide, derived from the high molecular mass TGFBIp, mapped to residue 676. The intermediate mass TGFBIp most-distal peptide mapped to residue 655 and the lower most-distal peptide mapped to residue 602. The C-termini of the lower mass TGFBIp molecules are sufficiently fragmented to ligand peptides that include the integrin-binding sequences, within a 69 amino acid polypeptide of the TGFBIp C-terminal region. This fragmentation pattern likely occurs from cleavages between residues 602 and 655 that could yield, within individual fragments, the EPDIM and RGD sequences. In order to test the apoptotic effects of the essential TGFBIp candidate fragments, two constructs were designed to express: 1) TGFBIp27–614 in order to exclude EPDIM and RGD; and 2) TGFBIp615–683 to include only the EPDIM and RGD sequences within the C-terminal regions.
Table 1.
Characterization of TGFBIp isolated from conditioned media. Mass spectrometry analysis mapped the N-terminal most proximal tryptic peptides (i.e., SPYQLVLQHSR) 4 residues from the mature N-terminus, shown as G24. The MG-63 cell endogenous TGFBIp C-termini mapped heterogeneously. C-terminal sequencing of recombinant TGFBIp isolated from CHO cell conditioned medium indicated heterogeneous C-termini. Shown is A614 which is immediately followed by the EPDIM cell adhesion sequence (i.e., EPVA614EPDIM).
| MG-63 cells | |
| G24PAKSPYQLVLQHSR............................................................VYQK676 | 69 KDa |
| G24PAKSPYQLVLQHSR .............................................EIFK655 | 62 KDa |
| G24PAKSPYQLVLQHSR ...............................VSLK602 | 60 KDa |
| CHO cells | |
| ..........................................................................EPVA614 | ∼60 KDa |
Additional rationale for these candidate constructs comes from C-terminal sequencing of TGFBIp isolated from medium conditioned by CHO cells overexpressing the wt form. The alanine614 residue was one of the signals albeit additional signals indicated heterogeneous C- termini in the vicinity (Table 1). This proposed fragmenting is corroborated in that CHO cells overexpressing recombinant TGFBIpwt displayed DNA laddering that was related to TGFBIp fragmentation and RGD (Kim et al., 2003). Our use of CHO cell cultures for expressing recombinant TGFBIp also corroborates the finding. First, when grown as high-density cell suspension, extensive cell demise was observed, and second, when grown as a monolayer, DNA laddering was displayed (data not shown) similar to that documented by others using CHO cells that were overexpressing TGFBIp (Kim et al., 2003). These protein characterizations strongly indicate that a C-terminal fragment is produced in TGFBIp-mediated apoptosis.
2.6 The C-terminal fragment TGFBIp615–683, but not the truncated TGFBIp27–614, induces apoptosis
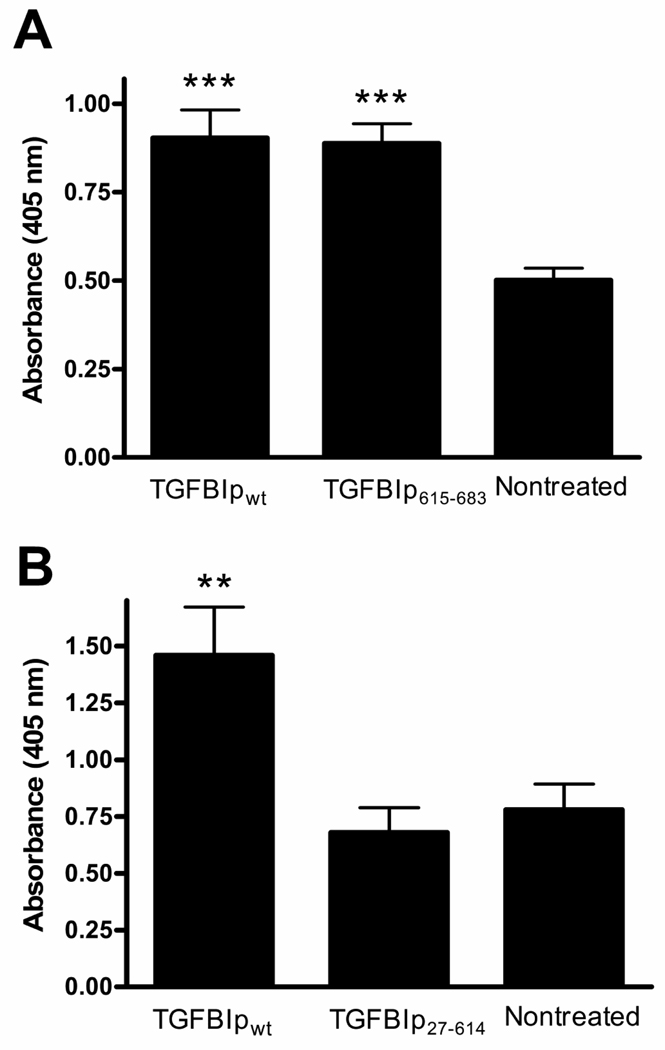
To verify whether the TGFBIp apoptotic actions might be assigned exclusively to the C-terminus, the 69 amino acid residue C-terminal fragment, TGFBIp615–683 that included the integrin-binding sequences RGD and EPDIM (Figure 1) was used in an add-back paradigm. TGFBIp615–683 induced an increase in apoptosis comparable to that quantified with TGFBIpwt (Figure 4A). The complementary TGFBIp27–614, lacking the EPDIM and RGD (Figure 1) failed to induce a significant apoptotic increase (Figure 4B). The pro-apoptotic effect on MG-63 cells was dependent upon the presence of TGFBIp’s C-terminus.
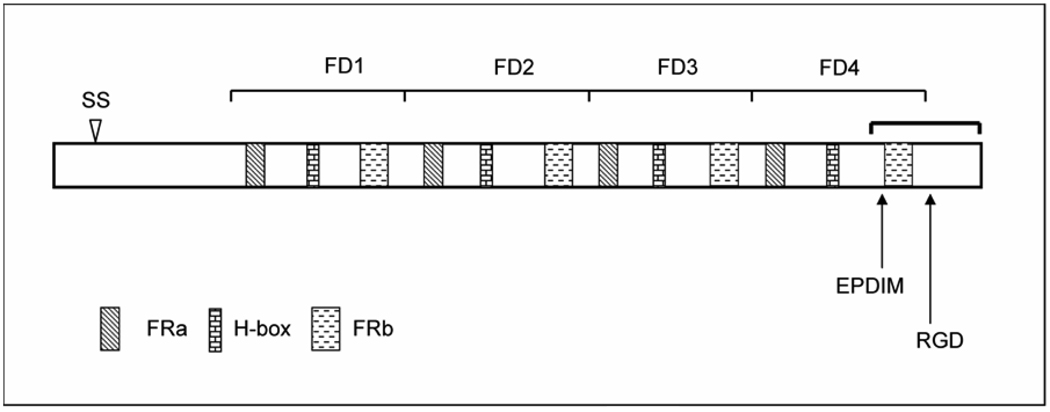
Figure 1.
Illustrated is the full-length, wt, human TGFBIp shown with known or predicted functional domains. The sequence is organized into four Fasciclin-like repeats labeled FD1–4. The arrowhead approximates the signal sequence (SS) cleavage site between amino acid residues 26 and 27. Conserved FRa and FRb regions, common to fasciclin family members, are indicated; as well, the histidine-containing region (H-box) is illustrated. Shown are the approximate locations of integrin binding sequences EPDIM and RGD (amino acid residues 615–619 and 642–644, respectively). The bracket indicates approximately the C-terminal region that is fragmented from the full-length molecule.
Figure 4.
TGFBIp’s apoptotic signal for MG-63 cells resides within the C-terminus. A) TGFBIp615–683 induced apoptosis when used at equimolar concentrations as the wt protein. One Way ANOVA was significant [F(2,26)=14.80;p<0.0001]***p<0.001 compared to nontreated. The effect of TGFBIp615–683 was not significantly different from TGFBIpwt. N=9 per group. B) TGFBIp27–614 did not elicit an apoptotic response. Its effect was not significantly different from nontreated but was significantly less than TGFBIpwt at**p<0.01. The One Way ANOVA was significant [F(2,58)=7.69;p<0.01]. N=19 for TGFBIp615–683 and N=20 for TGFBIpwt and nontreated.
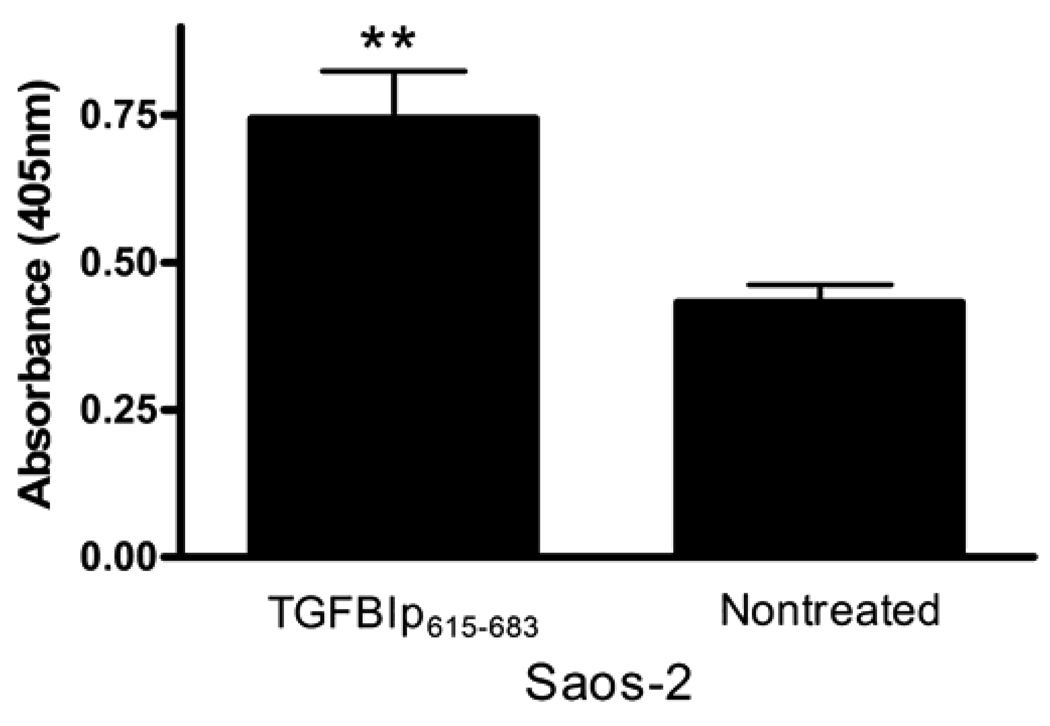
To determine whether the apoptotic effect was evident in osteosarcoma cells other than the MG-63 line, we investigated TGFBIp potential to increase apoptosis in Saos2 cells established from a human osteogenic sarcoma. When TGFBIp615–683 was added to the growth medium of Saos-2 cells a significant increase of apoptosis was the result when compared to nontreated cells (Figure 5), substantiating the determinative role of the C-terminus in cell demise in an additional cell type. Moreover, the evidence is in line with the possibility that the mechanism involves association of TGFBIp fragments with members of the integrin receptor family.
Figure 5.
TGFBIp615–683 induces apoptosis of Saos-2 osteosarcoma cells. TGFBIp615–683 induced apoptosis when added to Saos-2 osteosarcoma cell-growth medium, at the same concentration for MG-63 cells as in Figure 6A. This effect further substantiates that the apoptosis is dependent on the TGFBIp C-terminal fragment. [T(df 10)=3.63;**p<0.01].
2.7 The C-terminal EPDIM integrin-binding sequence induces apoptosis
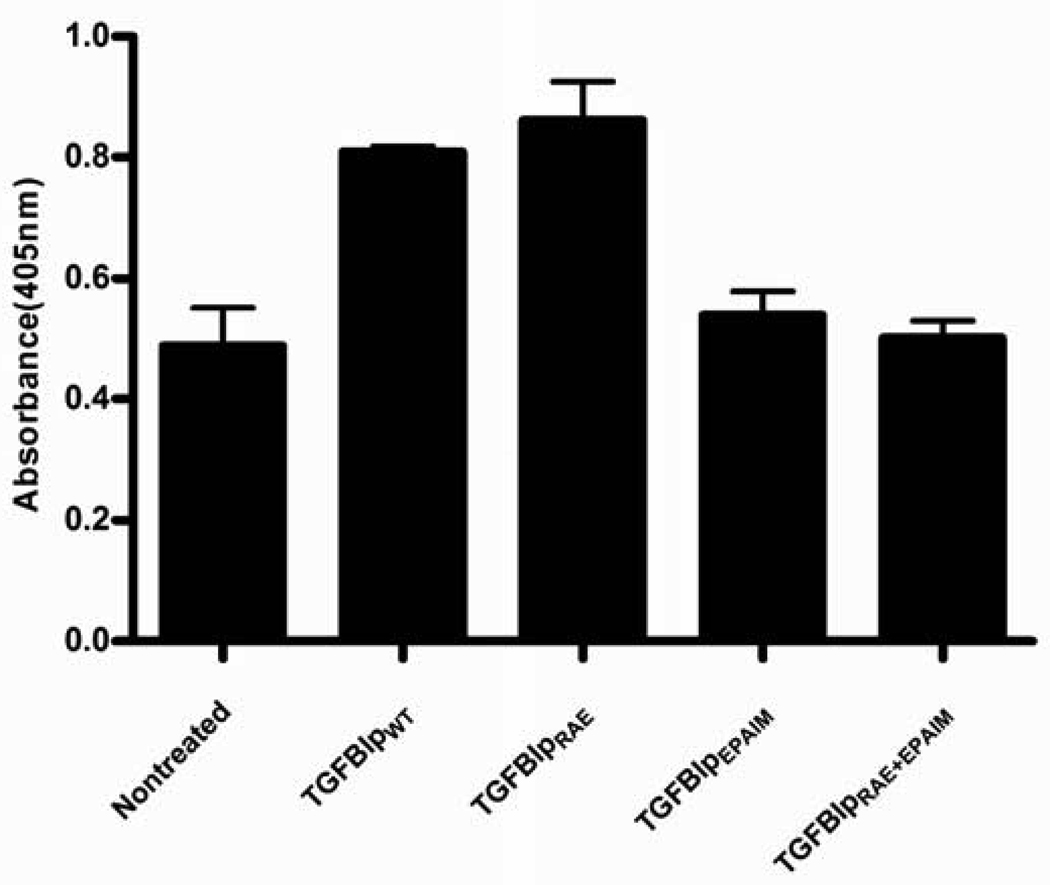
To determine whether the integrin-binding sequences that are located within the fragmented portion of TGFBIp mediate apoptosis, we introduced mutations that would alter amino acid residues critical for integrin binding. The RGD and EPDIM integrin-binding sequences were expressed as RAE (Kim et al., 2003), and EPAIM (Kim et al., 2000). In due course three mutant TGFBIp forms were constructed, TGFBIpEPAIM, TGFBIpRAE, and a double-mutant form TGFBIpEPAIM/RAE expressing an alteration in both binding sequences. These three mutant forms were used in add-back experiments essentially as described for TGFBIpwt. The results show that the mutants TGFBIpEPAIM and TGFBIpEPAIM/RAE did not induce apoptosis, implicating EPDIM in the apoptotic process. In contrast, TGFBIpRAE significantly induced apoptosis similar to that seen with the control TGFBIpwt (Figure 6). These data demonstrate that EPDIM, but not the RGD sequence, is sufficient to induce a significant apoptotic response in the MG-63 cells.
Figure 6.
TGFBIp's apoptotic signal for MG-63 cells resides within the C-terminal EPDIM integrin-binding sequence. Mutating EPDIM to EPAIM blocked TGFBIp's apoptotic action. One Way ANOVA was significant p<0.001 compared to control. TGFBIPepaim and TGFBIPepaim/rae did not induce a significant increase in apoptosis when compared to TGFBIpwt and TGFBIPrae Nontreated at p<0.01. TGFBIPraedid not significantly affect apoptosis when compared to TGFBIpwt p>0.05.
3. Discussion
Mechanisms underlying the induction and progression of apoptosis have attracted significant interest in recent years. In this study, we demonstrated conclusively that increases in MG-63 cell-apoptosis were definitive in each of three distinctive paradigms that were designed to increase the quantity of TGFBIp in the cells’ milieu. The results show that TGFBIp itself is capable of producing a graded apoptotic response. Cells treated with TGF-β1 accumulated TGFBIp in their conditioned medium, which was accompanied by a significant increase in apoptosis when compared to nontreated cells. The anti-TGFBIp antiserum blocked TGF-β1 evoked increases in apoptosis, while it did not notably block TGFBIp fragmentation (data not shown). This suggests that anti-TGFBIp antibody binds to fragments and hinders their apoptotic action. Whether blocking these fragments will definitively obstruct TGFBIp-induced apoptosis will require further investigation.
To understand the extent of TGFBIp as an element of a pathway that leads to apoptosis in MG-63 cells, MG-63 cells were made to overexpress recombinant TGFBIp in order to increase the concentration of TGFBIp in a manner independent of the use of exogenous TGF-β1. Transiently transfected MG-63 cells exhibited apoptosis that was significantly greater than that in control cells, again indicating that TGFBIp is capable of triggering events that lead to apoptosis. We conclude that when TGFBIp originates from within the cell it is secreted and the apoptotic signal is extracellular.
The next question was, if there is an exogenous source, does TGFBIp still trigger apoptosis, and does this occur at physiological concentrations? At lower concentrations, similar to TGFBIp in the cornea (35 nM) (Andersen et al., 2004) apoptosis was not significant. With 110 and 150 nM TGFBIp, a significant increase in apoptosis was achieved. These results indicate that at typical, normal, concentrations found in the cornea, apoptosis is nominal, but if tissue concentrations are close to equilibrium with plasma, then some level of apoptosis above baseline might be expected. Because they could not detect the smallest predicted C-terminal fragments in the cell culture media, Kim et al. hypothesized that the pro-apoptotic fragments are rapidly degraded within tissues keeping normal apoptotic levels under control (Kim et al., 2003). Most likely the apoptotic property of TGFBIp is removed before reaching the plasma.
Additionally, TGFBIp at concentrations that induce apoptosis could play important roles in pathologies. However, the only information available is that the TGFBIp concentrations are three or more fold greater than normal in corneas from individuals with particular types of CD (Klintworth et al., 1998), and in the urine of diabetic patients (Cha et al., 2005). In particular types of TGFBIp-linked CD, TGFBIp mutations are localized between EPDIM and RGD (Kannabiran and Klintworth, 2006), seemingly contrary to C-terminal fragmentation within this vicinity. In these instances, the C-terminal fragmentation may not have progressed through the CD mutations, or the development of TGFBIp-linked CD may proceed somehow despite fragmentation.
Recently, high levels of apoptosis were reported for the human kidney with chronic diabetes (Verzola et al., 2007) leading to the speculation that increased TGFBIp burden of the kidneys is related to the increased apoptosis. Fragments that encompass EPDIM and RGD have been implicated in TGFBIp-mediated apoptosis (Kim et al., 2003; Morand et al., 2003) through a mechanism associated with fragmentation of TGFBIp’s C-terminus (Kim et al., 2003). Supposedly, the rate of TGFBIp synthesis, secretion, and fragmentation would be increased in pathology producing the higher level of apoptosis.
Although the fragmentation is hypothesized to occur extracellularly (Kim et al., 2003), not much is known about the actual processing of the TGFBIp molecule. TGFBIp characterization demonstrated that the higher mass protein form essentially represents intact TGFBIp, whereas the two lower molecular mass forms lack a full-length C-terminus consistent with the idea that C-terminal fragmentation is a decisive event in TGFBIp’s role in apoptosis. Mass spectrometric analysis mapped heterogeneous C-termini within and downstream of FD4, and in that light, the C-termini are comparable to those C-termini of TGFBIp derived from human, porcine, and bovine tissues (Andersen et al., 2004; Gibson et al., 1996). These results collectively indicate the predominant TGFBIp forms in tissues and in vitro are C-terminally fragmented.
TGFBIp fragmentation might be progressive, yielding TGFBIp exhibiting a relative mobility of 60 kDa as the lowest mass form detected in this study. We report that the minimal TGFBIp that includes both integrin-binding sites causes apoptosis. The work of others suggests that further fragmentation of TGFBIp614–683 to at least an RGD hexapeptide can also lead to apoptosis (Kim et al., 2003). The ultimate fate of TGFBIp fragments is unknown, but the process exists in vivo.
The pathway activated by de novo and recombinant TGFBIp expression reliably and predictably resulted in osteosarcoma cell death, and may evoke a similar outcome when upregulated by TGF-β1 in vivo. Substantiating TGFBIp roles in tumor growth and metastasis in vivo, CHO cells and human transformed bronchial epithelial cells overexpressing recombinant TGFBIp exhibited a diminished capacity to maintain tumor growth in subcutaneous loci of nude mice (Skonier et al., 1994; Zhao et al., 2006; Zhao et al., 2004). More recently, TGFBIp−/− mice were shown to be prone to develop spontaneous tumors (Zhang et al., 2009), and TGFBIp was shown to sensitize ovarian cancer cells to paclitaxel (Ahmed et al., 2007), demonstrating respectively a TGFBIp-mediated suppressive effect on tumor progression in vivo and on various transformed cell types. Conversely, a mechanism that blocks TGFBIp’s influence on the progression of tumor growth has been linked to hypermethylation of the TGFBIp promoter, effectively inactivating the gene (Shah et al., 2008; Shao et al., 2006). The RGD and EPDIM motifs implicate integrin actions in apoptosis. In addition to TGFBIp fragmentation, TGFBIp-mediated apoptosis involves executioner caspase-3 activation (Kim et al., 2003; Morand et al., 2003; Nam et al., 2005) and is likely regulated by the expression of particular integrin types, suggesting characteristically anoikis (Frisch and Screaton, 2001). Ours is the first report to demonstrate that the apoptotic mechanism in osteosarcoma cells involves TGFBIp, particularly EPDIM, and in addition, corroborates actions of C-terminal integrin-binding sequences in apoptosis (Kim et al., 2003; Morand et al., 2003).
Though activation of various signaling pathways TGF-β1 affects a considerable complement of cellular processes during embryogenesis and in adult tissues including an increase in apoptosis (Rahimi and Leof, 2007). These data implicate TGFBIp expression and C-terminal fragmentation as an apoptotic switch affecting tumor biology, embryonic development, tissue remodeling and morphogenesis. Natural steps in our studies are to determine the particular integrin receptor that mediates TGFBIp apoptosis and further elucidate the apoptotic pathways activated by TGFBIp, in comparison to pathways activated by TGF-β1.
4. Experimental Procedures
4.1 Materials
A549 lung adenocarcinoma cells (CCL-185), MG-63 (CCL-1427) and Saos-2 osteosarcoma cells (HTB-85) were purchased from the American Type Culture Collection (Rockville, MD). Dr. Gary Sunter (UTSA) provided Spodoptera frugiperda (Sf9) insect cells. NovaBlue GigaSingles™ competent cells and Origami (DE3) pLacI cells were from Novagen (San Diego, CA). Ni-NTA agarose and RNeasy were purchased from Qiagen (Valencia, CA). TaqMan was purchased from Applied Biosystems (Foster City, CA), and reagents to detect ssDNA were from Chemicon International (Temecula, CA). Bicinchoninic acid protein reagents were from Pierce Biochemicals. Dulbecco’s Modified Eagle Medium (DMEM) was from Gibco BRL. Vectors pTriEX-4 Ek/LIC and pIEX-3 Ek/LIC, and Insect GeneJuice® Transfection Reagent were from Novagen (Novagen, WI). CHO-VGS cell growth media was purchased from Irvine Scientific (Santa Ana, CA), and SF900 II medium and annexin staining reagents were from Invitrogen (Carlsbad, CA).
4.2 cDNA Constructs
The cDNA that encodes full-length TGFBIp was obtained by reverse transcription-PCR from RNA isolated from human A549 cells. Coding sequences of TGFBIpwt were amplified using the forward primer 5’-GACGACGACAAGATGTCGCCCTACCAGCTG-3’ and reverse primer 5’-GAGGAGAAGCCCGGTCTAATGCTTCATCC-3’. Coding sequences for TGFBIp truncated at amino acid 614 (TGFBIp27–614) were obtained using the wt forward primer above and the reverse primer 5’-GAGGAGAAGCCCGGTCTAGGCAACAGGCTCC-3’. Amplicons cloned into the expression vector pIEX-3 Ek/LIC were expressed with GST and 6His-tag sequences at the N-terminus. The coding sequence for a TGFBIp C-terminal protein encoding amino acid residues 615 to 683 (TGFBIp615–683) was amplified using the forward primer (5’-GACGACGACAAGATGAGCCTGACATCATGGCC-3’). The reverse primer was the same as wt. The amplified product was cloned into pTriEx-4 EK/LIC. Non-tagged TGFBIp constructs for CHO expression have been previously described (Skonier et al., 1994).
Oligonucleotide-directed mutagenesis (QuikChange, Stratagene) introduced the mutations D617A, and G643A and D644E, into pIEX-5 encoding 6His-tagged TGFBIpwt, thereby mutating EPDIM and RGD to EPAIM and RAE (amino acid residues 615–619 and 642–644, respectively). Mutation-specific primers for EPAIM were sense 5' CTGTTGCCGAGCCTGCCATCATGGCCACAAA- 3' and anti-sense 5- TTTGTGGCCATGATGGCAGGCTCGGCAACAG-3', and for RAE, sense 5-ACAGACCTCAGGAAAGAGCTGAAGAACTTGCAGACTCTGCG-3' and anti-sense 5 CGCAGAGTCTGCAAGTTCTTCAGCTCTTTCCTGAGGTCTGT- 3'. Sequencing both strands of the mutated TGFBIps confirmed that the mutations were integrated into the constructs. Mutated TGFBIps were expressed in Sf9 cells and purified as described below.
4.3 Quantitative real-time PCR
MG-63 RNA was extracted using the RNeasy protocol. cDNA was synthesized with TaqMan polymerase. For TGFBIp qRT-PCR analysis, the TGFBIp primer pair (forward 5’-CACGATGCTTGAAGGTAACG-3’ and reverse 5’-GGCTTCAGCACACATAGCTG-3’) was used. Measurements of cyclophilin A were used to normalize measurements of the TGFBIp expressions. Quantitative measurements were performed using the relative quantification method recommended for ABI model 7900. Relative measurements were quantified using the ΔΔCT method.
4.4 Antibodies
New Zealand White rabbits immunized with a TGFBIp bacterial fusion protein produced TGFBIp antiserum that has been previously characterized (Ferguson et al., 2003b). Preimmune serum of the same animal was used as negative control.
4.5 Cell lines, transfections and TGFBIp purification
CHO cells expressing non-tagged human recombinant TGFBIpwt were conditioned for propagation in CHO-VGS growth medium. The conditioned medium was used for TGFBIp purification as previously described (Ferguson et al., 2003a). Sf9 cells were maintained in SF900 II medium at 28°C in orbital suspension at 140 rpm. Using GeneJuice reagent, cells were transfected with the pIEX-3 Ek/LIC plasmid encoding a 6His-tagged TGFBIpwt, a TGFBIp27–614, and the RGD and EPDIM mutated forms. NovaBlue and Origami cells were transfected with pTriEx-4 Ek/LIC encoding a 6His-tagged TGFBIp615–683 sequence using standard heat shock methods. TGFBIp proteins were isolated using Ni-NTA resin.
MG-63 cells were propagated in DMEM containing 0.1 mM non-essential amino acids, 2 mM L-glutamine and 10% FBS (DMEM+). Standard calcium phosphate co-precipitation was used to transiently transfect MG-63 cells with pTriEX-4 encoding full-length TGFBIp. TGFBIp expression by Sf9 and MG-63 cells was verified by TGFBIp immunoblot analysis.
4.6 Apoptosis Assay
Apoptosis was quantified by immunological detection of ssDNA and annexin V staining. To quantify ssDNA, wells of a microtiter plate were seeded with an average of 8 × 103 osteosarcoma cells in 0.1 ml DMEM+ containing 10 µg/ml TGFBIp. After 3 days, the cells were detached, and 5 × 103 were seeded for TGF-β1 experiments, and 3 × 103 were seeded for add-back apoptosis experiments and apoptosis was quantified by following the manufacturer’s recommendations. Briefly, the plates were centrifuged for 5 minutes at 200 × g. The cells were fixed for 30 minutes in 80% methanol in PBS, dried, and sequentially immersed in 22°C, 75°C and 4°C formamide for 10, 20 and 5 minutes, respectively. After incubation in 3% nonfat dry milk, wells were incubated for 30 minutes with a mixture of primary monoclonal antibody to ssDNA and horseradish peroxidase (HRP)-labeled anti-mouse IgM. Wells were rinsed and incubated for 1 hour with the substrate 2,2’-AZINO-bis [3-ethylbenziazoline-6-sulfonic acid]. Absorbance at 405 nm was recorded using a standard microplate reader.
To quantify annexin V staining, cells were treated with TGFBIp (10 µg/ml). After three days the cells were treated with annexin V antibody (Vybrant Apoptosis Assay reagents, Invitrogen) as recommended by the manufacturer. Stained cells from three different experiments were quantified for apoptosis using a fluorescence microscope. The average number of cells of the combined experiments is reported.
4.7 Protein immunoblots
Proteins in conditioned media were quantified and 1 µg of total protein was resolved per lane on 4–15% polyacrylamide gels. Proteins were transferred to Immobilon-P membrane and probed with anti-TGFBIp antibody and a second antibody conjugated to HRP. Antibody was detected with the substrate diaminobenzidine.
4.8 Mass Spectrometry
Bands of interest were excised from the gel and digested in situ with trypsin (Promega, modified). MALDI-TOF mass spectra were obtained on an Applied Biosystems Voyager-DE STR with positive ion detection in reflectron mode and α-cyano-4-hydroxycinnamic acid as matrix. The instrument was calibrated with Calibration Mixture I (des-Arg1-bradykinin, angiotensin I, glu1-fibrinopeptide B, neurotensin; Applied Biosystems) prior to each set of analyses; each spectrum was internally calibrated based on the trypsin autolysis peaks at m/z 842.51 and m/z 2211.11. Spectra represent an average of 100 laser shots. Spectral processing included the following: advanced baseline correction (peak width, 32; flexibility, 0.5; degree, 0.1), noise filter (correlation factor, 0.7), noise removal (std. dev. to remove, 2), Gaussian smooth (filter width, 5 points). Monoisotopic masses were assigned by the instrument software (Data Explorer) and were verified by visual inspection. Peak lists were searched against the Swiss-Prot database by means of Mascot (Matrix Science; in-house license).
4.9 Statistical Analyses
All data are reported as mean +/− SEM. Statistical comparisons of TGF-β1 treated and nontreated cells using apoptosis assays were performed using a two-way analysis of variance (ANOVA) followed by a Bonferroni post-test. Statistical comparison between TGFBIp-treated and nontreated Saos-2 cells was assessed by t-test. All other statistical comparisons were assessed by One-way ANOVA followed by Newman-Keuls multiple comparison test. Statistical significance was accepted when p<0.05.
Acknowledgements
This study was supported by an MBRS/SCORE grant (GM-08194, RGL) a Faculty Research Award from the University of Texas at San Antonio (RGL), an MBRS/RISE (GM-6065, RZ) and a Sloan Fellowship to RZ. The research described here is part of a dissertation by R. Zamilpa submitted to The University of Texas at San Antonio in partial fulfillment of the requirements of the Ph.D. degree. Mass spectrometric analyses were performed at the UTSA Proteomics Laboratory under the direction of Dr. William Haskins, and at the University of Texas Health Science Center San Antonio Institutional Mass Spectrometry Laboratory under the direction of Dr. Susan T. Weintraub. The assistance of Christopher A. Carroll for performing the MS analyses is greatly appreciated.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Office of the Dean, School of Health Professions, University of Texas Health Science Center, San Antonio, TX, 78229.
References
- Ahmed AA, Mills AD, Ibrahim AE, Temple J, Blenkiron C, Vias M, Massie CE, Iyer NG, McGeoch A, Crawford R, Nicke B, Downward J, Swanton C, Bell SD, Earl HM, Laskey RA, Caldas C, Brenton JD. The extracellular matrix protein tgfbi induces microtubule stabilization and sensitizes ovarian cancers to paclitaxel. Cancer Cell. 2007;12:514–527. doi: 10.1016/j.ccr.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen RB, Karring H, Moller-Pedersen T, Valnickova Z, Thogersen IB, Hedegaard CJ, Kristensen T, Klintworth GK, Enghild JJ. Purification and structural characterization of transforming growth factor beta induced protein (TGFBIp) from porcine and human corneas. Biochemistry. 2004;43:16374–16384. doi: 10.1021/bi048589s. [DOI] [PubMed] [Google Scholar]
- Billings PC, Whitbeck JC, Adams CS, Abrams WR, Cohen AJ, Engelsberg BN, Howard PS, Rosenbloom J. The transforming growth factor-beta-inducible matrix protein (beta)ig-h3 interacts with fibronectin. J Biol Chem. 2002;277:28003–28009. doi: 10.1074/jbc.M106837200. [DOI] [PubMed] [Google Scholar]
- Cha DR, Kim IS, Kang YS, Han SY, Han KH, Shin C, Ji YH, Kim NH. Urinary concentration of transforming growth factor-beta-inducible gene-h3 (beta ig-h3) in patients with type 2 diabetes mellitus. Diabet Med. 2005;22:14–20. doi: 10.1111/j.1464-5491.2004.01295.x. [DOI] [PubMed] [Google Scholar]
- Ferguson JW, Mikesh MF, Wheeler EF, LeBaron RG. Developmental expression patterns of beta-ig (betaig-h3) and its function as a cell adhesion protein. Mech Dev. 2003a;120:851–864. doi: 10.1016/s0925-4773(03)00165-5. [DOI] [PubMed] [Google Scholar]
- Ferguson JW, Thoma BS, Mikesh MF, Kramer RH, Bennett KL, Purchio A, Bellard BJ, LeBaron RG. The extracellular matrix protein big-h3 is expressed at myotendinous junctions and supports muscle cell adhesion. Cell Tiss Res. 2003b;313:93–105. doi: 10.1007/s00441-003-0743-z. [DOI] [PubMed] [Google Scholar]
- Frisch SM, Screaton RA. Anoikis mechanisms. Curr Opin Cell Biol. 2001;13:555–562. doi: 10.1016/s0955-0674(00)00251-9. [DOI] [PubMed] [Google Scholar]
- Gibson MA, Hatzinikolas G, Kumaratilake JS, Sandberg LB, Nicholl JK, Sutherland GR, Cleary EG. Further characterization of proteins associated with elastic fiber microfibrils including the molecular cloning of MAGP-2 (MP25) J Biol Chem. 1996;271:1096–1103. doi: 10.1074/jbc.271.2.1096. [DOI] [PubMed] [Google Scholar]
- Hanssen E, Reinboth B, Gibson MA. Covalent and non-covalent interactions of betaig-h3 with collagen VI. Beta ig-h3 is covalently attached to the amino-terminal region of collagen VI in tissue microfibrils. J Biol Chem. 2003;278:24334–24341. doi: 10.1074/jbc.M303455200. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Noshiro M, Ohno S, Kawamoto T, Satakeda H, Akagawa Y, Nakashima K, Okimura A, Ishida H, Okamoto T, Pan H, Shen M, Yan W, Kato Y. Characterization of a cartilage-derived 66-kda protein (RGD-CAP/beta ig-h3) that binds to collagen. Biochimica et Biophysica Acta. 1997;1355:303–314. doi: 10.1016/s0167-4889(96)00147-4. [DOI] [PubMed] [Google Scholar]
- Kannabiran C, Klintworth GK. TGFBI gene mutations in corneal dystrophies. Hum Mutat. 2006;27:615–625. doi: 10.1002/humu.20334. [DOI] [PubMed] [Google Scholar]
- Kim JE, Jeong HW, Nam JO, Lee BH, Choi JY, Park RW, Park JY, Kim IS. Identification of motifs in the fasciclin domains of the transforming growth factor-beta-induced matrix protein betaig-h3 that interact with the alphavbeta5 integrin. J Biol Chem. 2002;277:46159–46165. doi: 10.1074/jbc.M207055200. [DOI] [PubMed] [Google Scholar]
- Kim JE, Kim SJ, Jeong HW, Lee BH, Choi JY, Park RW, Park JY, Kim IS. Rgd peptides released from beta ig-h3, a TGF-beta-induced cell-adhesive molecule, mediate apoptosis. Oncogene. 2003;22:2045–2053. doi: 10.1038/sj.onc.1206269. [DOI] [PubMed] [Google Scholar]
- Kim JE, Kim SJ, Lee BH, Park RW, Kim KS, Kim IS. Identification of motifs for cell adhesion within the repeated domains of transforming growth factor-beta-induced gene, betaig-h3. J Biol Chem. 2000;275:30907–30915. doi: 10.1074/jbc.M002752200. [DOI] [PubMed] [Google Scholar]
- Klintworth GK, Valnickova Z, Enghild JJ. Accumulation of beta ig-h3 gene product in corneas with granular dystrophy. American Journal of Pathology. 1998;152:743–748. [PMC free article] [PubMed] [Google Scholar]
- LeBaron RG, Bezverkov KI, Zimber MP, Pavelec R, Skonier J, Purchio AF. Beta ig-h3, a novel secretory protein inducible by transforming growth factor-beta, is present in normal skin and promotes the adhesion and spreading of dermal fibroblasts in vitro. Journal of Investigative Dermatology. 1995;104:844–849. doi: 10.1111/1523-1747.ep12607024. [DOI] [PubMed] [Google Scholar]
- Morand S, Buchillier V, Maurer F, Bonny C, Arsenijevic Y, Munier FL, Schorderet DF. Induction of apoptosis in human corneal and HeLa cells by mutated bigh3. Invest Ophthalmol Vis Sci. 2003;44:2973–2979. doi: 10.1167/iovs.02-0661. [DOI] [PubMed] [Google Scholar]
- Nam EJ, Sa KH, You DW, Cho JH, Seo JS, Han SW, Park JY, Kim SI, Kyung HS, Kim IS, Kang YM. Up-regulated transforming growth factor beta-inducible gene H3 in rheumatoid arthritis mediates adhesion and migration of synoviocytes through alpha v beta3 integrin: Regulation by cytokines. Arthritis Rheum. 2006;54:2734–2744. doi: 10.1002/art.22076. [DOI] [PubMed] [Google Scholar]
- Nam JO, Jeong HW, Lee BH, Park RW, Kim IS. Regulation of tumor angiogenesis by fastatin, the fourth fas1 domain of betaig-h3, via alphavbeta3 integrin. Cancer Res. 2005;65:4153–4161. doi: 10.1158/0008-5472.CAN-04-2705. [DOI] [PubMed] [Google Scholar]
- Ohno S, Noshiro M, Makihira S, Kawamoto T, Shen M, Yan W, Kawashima-Ohya Y, Fujimoto K, Tanne K, Kato Y. RGD-CAP ((beta)ig-h3) enhances the spreading of chondrocytes and fibroblasts via integrin alpha(l)beta(l) Biochimica et Biophysica Acta. 1999;1451:196–205. doi: 10.1016/s0167-4889(99)00093-2. [DOI] [PubMed] [Google Scholar]
- Park SW, Bae JS, Kim KS, Park SH, Lee BH, Choi JY, Park JY, Ha SW, Kim YL, Kwon TH, Kim IS, Park RW. Beta ig-h3 promotes renal proximal tubular epithelial cell adhesion, migration and proliferation through the interaction with alpha3beta1 integrin. Exp Mol Med. 2004;36:211–219. doi: 10.1038/emm.2004.29. [DOI] [PubMed] [Google Scholar]
- Rahimi RA, Leof EB. TGF-beta signaling: A tale of two responses. J Cell Biochem. 2007;102:593–608. doi: 10.1002/jcb.21501. [DOI] [PubMed] [Google Scholar]
- Reinboth B, Thomas J, Hanssen E, Gibson MA. Beta ig-h3 interacts directly with biglycan and decorin, promotes collagen VI aggregation, and participates in ternary complexing with these macromolecules. J Biol Chem. 2006;281:7816–7824. doi: 10.1074/jbc.M511316200. [DOI] [PubMed] [Google Scholar]
- Shah JN, Shao G, Hei TK, Zhao Y. Methylation screening of the TGFBI promoter in human lung and prostate cancer by methylation-specific PCR. BMC Cancer. 2008;8:284. doi: 10.1186/1471-2407-8-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao G, Berenguer J, Borczuk AC, Powell CA, Hei TK, Zhao Y. Epigenetic inactivation of betaig-h3 gene in human cancer cells. Cancer Res. 2006;66:4566–4573. doi: 10.1158/0008-5472.CAN-05-2130. [DOI] [PubMed] [Google Scholar]
- Skonier J, Bennett K, Rothwell V, Kosowski S, Plowman G, Wallace P, Edelhoff S, Disteche C, Neubauer M, Marquardt H, et al. Beta ig-h3: A transforming growth factor-beta-responsive gene encoding a secreted protein that inhibits cell attachment in vitro and suppresses the growth of CHO cells in nude mice. DNA & Cell Biology. 1994;13:571–584. doi: 10.1089/dna.1994.13.571. [DOI] [PubMed] [Google Scholar]
- Skonier J, Neubauer M, Madisen L, Bennett K, Plowman GD, Purchio AF. cDNA cloning and sequence analysis of beta ig-h3, a novel gene induced in a human adenocarcinoma cell line after treatment with transforming growth factor-beta. DNA & Cell Biology. 1992;11:511–522. doi: 10.1089/dna.1992.11.511. [DOI] [PubMed] [Google Scholar]
- Verzola D, Gandolfo MT, Ferrario F, Rastaldi MP, Villaggio B, Gianiorio F, Giannoni M, Rimoldi L, Lauria F, Miji M, Deferrari G, Garibotto G. Apoptosis in the kidneys of patients with type II diabetic nephropathy. Kidney Int. 2007;72:1262–1272. doi: 10.1038/sj.ki.5002531. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wen G, Shao G, Wang C, Lin C, Fang H, Balajee AS, Bhagat G, Hei TK, Zhao Y. TGFBI deficiency predisposes mice to spontaneous tumor development. Cancer Res. 2009;69:37–44. doi: 10.1158/0008-5472.CAN-08-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, El-Gabry M, Hei TK. Loss of betaig-h3 protein is frequent in primary lung carcinoma and related to tumorigenic phenotype in lung cancer cells. Mol Carcinog. 2006;45:84–92. doi: 10.1002/mc.20167. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Shao G, Piao CQ, Berenguer J, Hei TK. Down-regulation of betaig-h3 gene is involved in the tumorigenesis in human bronchial epithelial cells induced by heavy-ion radiation. Radiat Res. 2004;162:655–659. doi: 10.1667/rr3270. [DOI] [PubMed] [Google Scholar]