Abstract
Myosin VI challenges the prevailing theory of how myosin motors move on actin: the lever arm hypothesis. While the reverse directionality and large powerstroke of myosin VI can be attributed to unusual properties of a subdomain of the motor (converter with a unique insert), these adaptations cannot account for the large step size on actin. Either the lever arm hypothesis needs modification, or myosin VI has some unique form of extension of its lever arm. We determined the structure of the region immediately distal to the lever arm of the motor and show that it is a 3-helix bundle. Based on C-terminal truncations that display the normal range of step sizes on actin, CD, fluorescence studies, and a partial deletion of the bundle, we demonstrate that this bundle unfolds upon dimerization of two myosin VI monomers. This unprecedented mechanism generates an extension of the lever arm of myosin VI.
Keywords: unconventional myosin, motility, converter, lever arm, single stable α-helix
INTRODUCTION
Within the myosin superfamily there are at least 35 classes of molecular motors that move along actin filaments (Odronitz and Kollmar, 2007). Myosin motor activity is initiated by actin binding, which drives conformational changes allowing sequential release of ATP hydrolysis products coupled to movement of the myosin “lever arm”. This is referred to as the swinging lever arm hypothesis (Holmes and Geeves, 2000; Holmes et al., 2004). The lever arm is a variable length, extended α-helix containing calmodulin (CaM) and/or CaM-like light chain binding sites (IQ motifs).
Myosin VI is the only class of myosin that has been shown to move toward the minus-end of actin filaments (Wells et al., 1999). The myosin VI dimer is capable of processive movement (i.e. can move as a single molecule) along an actin filament (Park et al., 2006) as well as load-dependent anchoring (Altman et al., 2004), and thus can fulfill a number of specialized cell biological functions (Buss et al., 2004; Frank et al., 2004; Sweeney and Houdusse, 2007). Our two previously solved myosin VI structures, representing the beginning (pre-powerstroke state; Ménétrey et al., 2007) and the end (rigor state; Ménétrey et al., 2005) of the lever arm swing on actin, reveal the magnitude of the myosin VI powerstroke. This is often referred to as the stroke size, which is determined by the net displacement of the lever arm from its pre-powerstroke position to its rigor position.
The stepping behavior of myosin VI is also unusual compared to other processive myosins. Instead of a narrow range of step sizes, as seen for myosin V (Mehta et al., 1999), myosin VI has a broad distribution of step sizes centered around 30–36nm (Rock et al., 2001 ; Nishikawa et al., 2002). Since the apparent lever arm of myosin VI contains only two CaM binding sites, this stepping behavior is not easily explained. We previously demonstrated that it requires some additional structure, distal to the apparent lever arm (Rock et al., 2005). This structure functions as a lever arm extension and perhaps contains greater flexibility than the standard myosin lever arm.
Intriguingly, a number of myosin classes, including VI, VIIa and X contain a domain that is predicted to form an extended, stable single α-helix (SAH) following their lever arms (Knight et al., 2005) ; see Figure 1. It has been suggested that these domains are sufficiently rigid to provide functional extensions of the lever arms of myosins such as VI, VIIa and X (Knight et al., 2005; Spink et al., 2008). SAH domains have also been described in caldesmon (Wang et al., 1991), and recently were reported to exist in more than 123 different proteins (Sivaramakrishnan et al., 2008).
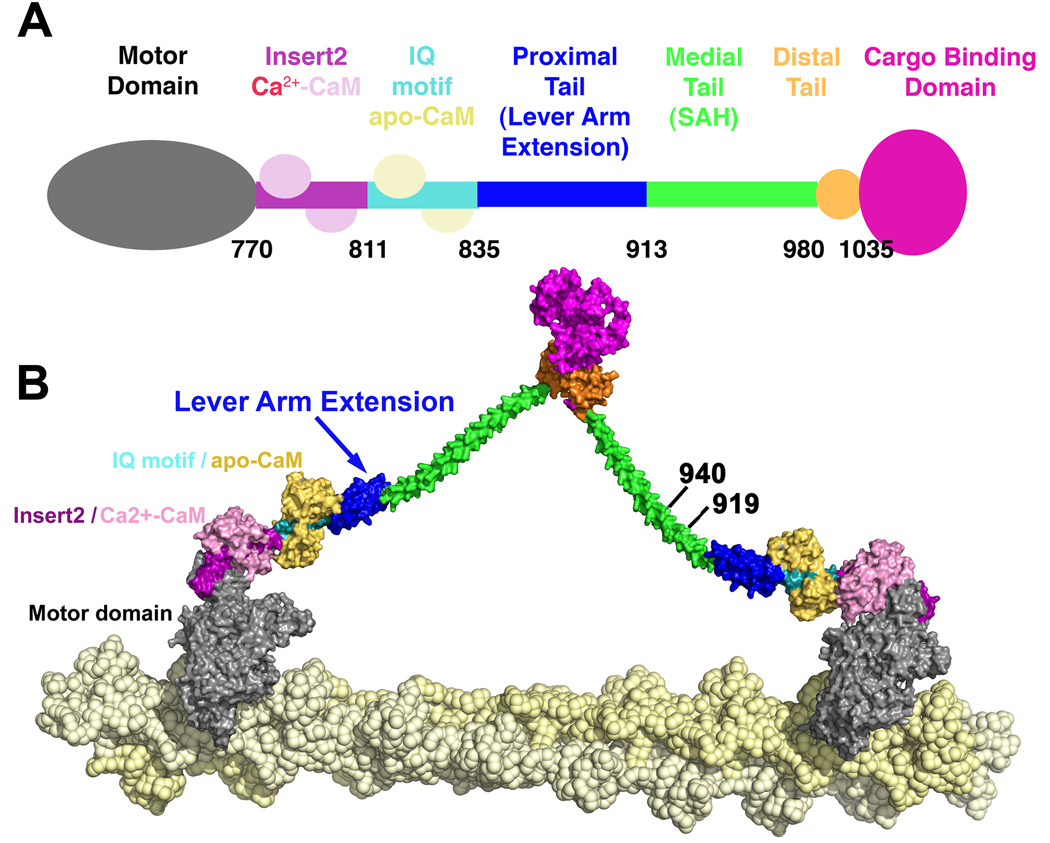
Figure 1. Schematic of full-length myosin VI.
A. The N-terminal motor domain is followed by a unique insert (Insert 2) that binds the structural calmodulin (CaM), followed by an IQ motif that is bound to an exchangeable CaM. This is adjacent to the proximal tail domain that may form an extension of the lever arm formed by the bound CaMs. The proximal tail is followed by the medial tail, a region that can weakly dimerize, as we have previously shown (Park et al., 2006). This same region has been predicted to contain a stable single alpha helix (SAH) by Spink et al. (2008). In this study we provide evidence that the medial tail likely contains a short dimerization domain (likely coiled coil) that may be followed by a SAH. The rest of the molecule is composed of a small distal tail region and a cargo-binding domain. B. Model of myosin VI dimer proposed by Spink et al. (2008), using stable single alpha helices (SAH) as lever arm extensions. The sites of truncations used for this study are noted by the C-terminal amino acid number.
In the case of myosin VI, there is an additional domain, which has been referred to as the proximal tail, between the conventional CaM-containing lever arm and the predicted SAH. In a recent study Spink et al., (2008) demonstrated that the region of myosin VI immediately following the lever arm is largely α-helical and forms a highly compacted domain, which the authors postulated was most consistent with a 3-helix bundle. They further suggested that the region in between the 3-helix bundle and the cargo-binding domain is a stable single α-helix that forms the bulk of the lever arm extension necessary for the large step sizes of myosin VI. Lastly, they postulated that the cargo-binding domain of full-length myosin VI is solely responsible for dimerization (see Figure 1). However, this postulate is inconsistent with our earlier observations (Park et al., 2006) that somewhere proximal to the cargo-binding domain is a region that can allow dimerization of myosin VI, resulting in processive step sizes identical to the full-length dimer. The data of Park et al. (2006) might be compatible with a model in which a SAH domain provides the lever arm extension of myosin VI if there is a residual component of weak dimerization at the end of the constructs examined by Park et al. (2006).
To clarify the mechanism that allows myosin VI to achieve its large step size, we made additional C-terminal truncations in order to further delineate the location of the lever arm extension and the minimal length structure that is capable of dimerizing with a normal step size (see Figure 1). We also crystallized and solved the structure of a truncated construct that included the region of the approximately 80 amino acids immediately following the lever arm (proximal tail). What we observe is that the region following the lever arm is indeed a 3-helix bundle in the monomeric crystal structure. Surprisingly, we also observe that a construct extending only 28 amino acids beyond this structure is capable of forming a dimer that moves processively, with step sizes identical to that of the full-length dimer. The only way to explain all of the data is to propose that upon dimerization, the 3-helix bundle must unfold and form the extension of the myosin VI lever arm. Finally, we performed experiments involving fluorescence quenching and deletion of two helices of the bundle to provide definitive evidence that this is the case.
RESULTS
The proximal tail is a 3-helix bundle
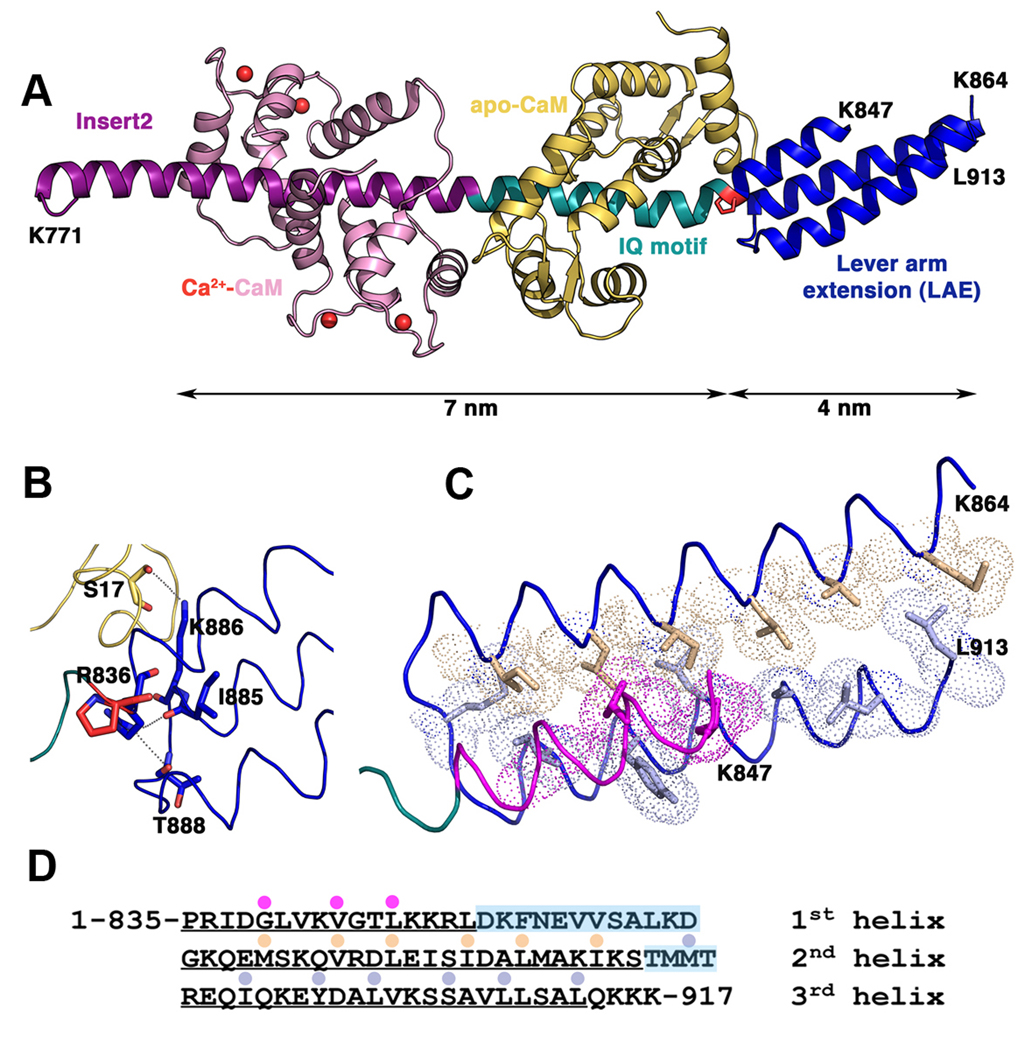
Crystallization of a myosin VI construct (residues 1–917) co-expressed with calmodulin (CaM) gave rise to a structure that surprisingly contained only residues 770–913 with two bound CaMs. (Statistics on data collection and refinement can be found in Table 1.) It is evident that we crystallized a proteolytic fragment of the originally expressed construct. At 2.7 Å resolution, the crystals allowed the determination of the structure of the full lever arm (FLA) containing two CaMs bound to Insert 2 (Ins2) and IQ motif followed by the 77 residues of the heavy chain (residues 835–913) which form an antiparallel 3-helix bundle. As shown in Figure 2A, the length of the FLA is 11 nm, with the CaM-bound Ins2-IQ helix segment accounting for 7nm and 4 nm for the 3-helix bundle.
Table 1.
Statistics on data collection and refinement
| Data Collection | |
|---|---|
| Space group | P212121 |
| Cell dimensions | 51.79, 118.39, 182.06 |
| a, b, c (Å) | |
| Resolution (Å) | 50-2.7 (2.85-2.7) |
| Rmeas | 8.3 (48.1) |
| I/σ | 7.7 (1.7) |
| Completeness (%) | 100 |
| Redundancy | 7.2 |
| Refinement | |
| Resolution (Å) | 45-2.7 |
| No. reflections | 30044 |
| Rwork/Rfree | 22.35/27.22 |
| No. atoms | |
| Protein | 6592 |
| Hetero atoms | 14 |
| Water | 87 |
| Mean B-factors | 48.97 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.068 |
Figure 2. The Myosin VI full-length lever arm.
A. Structure of the myosin VI lever arm with the Insert 2 (purple, Ca2+-CaM pink) and the IQ motif (cyan, apo-CaM yellow) helix followed by the three helix bundle (blue) that forms the lever arm extension (LAE). Note the 30º kink at the junction between the IQ motif and the LAE at P835 (red). B. Details of the interactions between the apo-CaM bound to the IQ motif and the triple helix bundle. Two hydrogen bonds mediated by the helix 1 R836 also stabilize the bundle by interacting with two carbonyls of the loop between helices 2 and 3 (I885 and T888). C. Apolar interactions that stabilize the bundle in the proximal part. Note that helix 1 is shorter than the other two helices and the loop that follows is not visible in the electron density. Fewer interactions are found between the helices in the distal part of the bundle, which is not well stabilized. D. Sequence of the LAE with the residues found within the bundle marked by a colored dot. The loops between the helices are highlighted in blue.
From Ins2 to the end of the IQ motif, the MVI heavy chain (HC) adopts a conformation of an elongated and straight α-helix embraced by the CaMs. Note that the two CaMs do not interact with one another. As previously described (Ménétrey et al., 2005), Ins2 contains an unusual 1–6–14 motif which binds to Ca2+-CaM through strong interactions with three key anchoring residues (W793, W798 and L806). As predicted from the sequence of the IQ motif, “IQxxxRGxxxR”, the C-lobe of an apo-CaM binds to the N-terminus of the myosin VI IQ motif in a conventional manner (Houdusse et al., 1996). In contrast, the C-terminus of the IQ motif sequence (MQktiRMwlcK) where the N-lobe of apo-CaM binds, is unusual in myosin VI. Instead of the conventional glycine, a residue with a large side-chain (M826) is found and the last arginine of the motif is replaced by a lysine (K830). Modelling studies proposed that this motif could recruit CaM by interaction with the C-lobe only, leaving the N-lobe free in solution (Terrak et al., 2005). However, the structure of the FLA shows that the N-lobe of CaM does interact with the HC helix and its position relative to the HC is analogous to that found for classic IQ motif/CaM complexes. (See details in Supplementary Data.)
Structure of the three-helix bundle
The three-helix bundle or proximal tail domain (PTD) starts at P835. The proline, in part, imposes a 30° kink relative to the HC helix that corresponds to the first part of the lever arm: the Ins2 and IQ helical motif (see Figure 2A). The PTD consists of a helical bundle formed by three antiparallel amphipathic α-helices. The bundle, which is 3.8 nm long and ∼1.7 nm in diameter, is organized in such a way that several residues at the proximal part of the bundle participate in a compact hydrophobic core. In contrast, the distal part of the bundle is very loose and is composed of two helices with little packing of the hydrophobic side chains. Helix 1 is smaller and a long loop (residues K848-K864 for which no electron density could be observed) connects helices 1 and 2.
There are several indications that the bundle may not be very stable. Although the core of the bundle is mostly hydrophobic, the side chains are short and thus the bundle is loosely packed (Figure 2C). While the proximal part has interactions that give some stability, the distal part of the bundle is particularly loose. In agreement with this, the electron density is not very well defined for most of the side chains of the bundle. This is in contrast to other stable triple helix bundles for which the hydrophobic patch between the helices tightly associate the helices along their whole length (see Supplementary Figure 2).
In addition to the hydrophobic interactions within the bundle, two hydrogen bonds between the beginning of helix 1 (R836) and the carbonyls at the end of helix 2 of residues I885 and T888 stabilize the proximal part of the bundle (Figure 2B). The kink at the beginning of the bundle (P835) is stabilized by a number of van der Waals interactions between the N-lobe of the CaM bound to the IQ motif (residues L18, F19 and K21) and the first helix of the bundle (R836, I837, V841). Note that these interactions with the first helix of the bundle would be preserved if the bundle melted and would thus help define the orientation of the elongated proximal tail domain. In contrast, no interaction occurs with helix 3 and the CaM bound to the IQ motif and only a few interactions occur with the end of helix 2, although one hydrogen bond is seen between K886 and the carbonyl of S17 of CaM. Thus the main interactions that stabilize the CaM bound to the IQ occur with the first helix of the triple helix bundle which would be retained even if the triple helix melted.
Truncated constructs can dimerize and step processively
We previously reported that a myosin VI construct truncated at Arg 991 is capable of forming dimers and taking processive steps that are identical in size (∼30nm) to the full-length dimer and to a zippered dimer in which a leucine zipper (GCN4) is appended following amino acid 991 (Park et al., 2006). Based on this, we concluded that both the lever arm extension and dimerization domain of myosin VI are located between residues 834 (end of the IQ motif) and 991. (See Figure 1.)
We probed this further by first truncating at Arg940 (creating MVI-940) and performed previously described (Park et al., 2006) dimerization assays (ATPase assays, rotary shadowing and single molecule analyses). As shown in Table 2, the ATPase activity of MVI-940 reveals gating (decreased ATPase activity per head) following actin clustering of the monomers or binding and release of the monomers to an antibody (via a C-terminal FLAG tag), indicative of a dimer in which the lead head cannot complete its ATPase cycle until the rear head releases from actin. Note that the fact that this clustering of monomers via actin or antibodies to increase the effective concentration is necessary indicates that the dimerization is weak, requiring concentrations greater than the low micro-molar concentrations of our ATPase assays, in order for dimers to form.
Table 2.
Actin-Activated ATPase Activity (Vmax) of Myosin VI Constructs
| CONSTRUCT | Vmax (head−1second−1) | After Antibody Binding Vmax (head−1second−1) | After Actin Saturation Vmax (head−1second−1) |
|---|---|---|---|
| MVI-917 | 6.0±1.3* | 6.4±1.5* | 5.6±1.3* |
| MVI-940 | 7.5±1.0 | 3.1±0.7 | 2.9±0.8 |
| MVI-991 | 7.1±1.6* | 2.5±1.7* | 2.9±0.8* |
| Zippered Dimer | 2.4±0.8* | 2.5±1.1* | 2.4±1.3* |
Mean values (±S.D.) of 3–5 independent protein preparations are shown for each construct and condition
Values reported in Park et al., 2006.
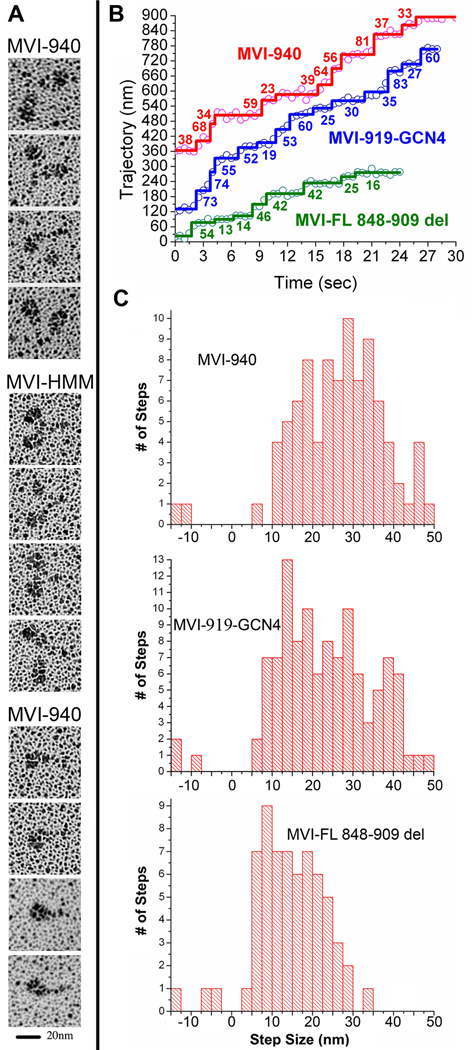
As shown in Figure 3A, dimers of 940 could be visualized via rotary shadowing. These dimers were only a small fraction of the total population (10±3%) as compared to the zippered-HMM included for comparison (98±1%). In part, this appears due to the instability of the dimers once released from actin, as increasing the time between release from actin and spraying onto the EM grid decreases the number of dimers detected. We also noted that the glycerol concentrations needed for rotary shadowing destabilize the dimerization (demonstrated by the loss of gating of the MVI-940 at glycerol concentrations of >10%). Nonetheless, this was a smaller percentage of dimers than we observed (Park et al., 2006) for constructs truncated at either amino acid 991 or 1049 (50–70%), indicating that additional interactions distal to amino acid 940 further stabilize dimerization.
Figure 3.
A. Rotary shadowing EM of myosin VI constructs. Representative images for the MVI-940 construct following dimerization using actin clustering are shown in the first row and images of the zippered HMM (MVI-991-GCN4) are shown in the second row. These dimers show two closely spaced motor domains with an intervening link. The third row contains panel with representative images of MVI-940 monomers. B. Observed movements of myosin VI constructs. Representative stepping traces, based on FIONA values as a function of time, are shown for three truncated myosin VI constructs. Stepping traces for the labeled head (Cy3 on IQ-bound CaM) are shown for MVI-940 (red), MVI-919-GCN4 (blue), and for the full-length construct with the last two helices of the bundle deleted, MVI-FL 848–909 del (green). Positions of the dye were tracked and steps were calculated using a custom Student’s t-test algorithm coded for by IDL (ITT Visual Information Solutions, Boulder, CO). In all cases, the observed steps are actually two steps, since the probe is only on one head. C. Distribution of step sizes of Cy3-labeled truncated myosin VI constructs. For MVI-940, following dimerization by binding to actin, the average forward step (1/2 observed movement) was 27.2 ± 9.7 nm (n =87). The average backward step (1/2 observed movement) of MVI-940 was –12.3 ± 1.7 nm (n = 2). The distribution of the MVI-940 was similar to what we previously observed for both the full-length myosin VI dimer and the zippered HMM. The average forward step size of MVI-919-GCN4 was 23.7 ± 10.6 nm (n = 108) whereas the average backward step size was –12.5 ± 2.7 nm (n = 3). For the full-length construct with the second and third helices of the bundle deleted, the average forward step size was 16.1 ± 7.0 nm (n=61), and the average backward step size was −7.7 ± 4.3 nm (n=3),
Further evidence that these dimers were sufficiently stable to function as processive motors was provided by single molecule FIONA (Fluorescence Imaging with One Nanometer Accuracy) assays. In these assays a small percentage of monomers were labeled with Cy3-CaM, as previously described (Park et al., 2006). The population of sparsely labeled monomers was bound to an actin filament in the absence of ATP to induce dimerization. Following addition of ATP, a percentage of the labeled molecules were observed to initiate processive movement on actin, indicating dimerization with an unlabeled monomer. These actin-induced dimers were capable of processive movement along actin filaments with step sizes that were indistinguishable from the full-length dimer, the zippered HMM (991 + zipper) or the dimerized construct truncated at 991 without an added zipper (Table 3 and Figure 3B&C). Additionally, the MVI-940 dimers had average run lengths of 0.9 µm, longer than any construct other than the full-length dimer (Table 3).
Table 3.
Single Molecule Motility of Myosin VI Constructs as Assessed by FIONA (following actin saturation in rigor)
| CONSTRUCT | % of Molecules that were Processive (Dimers) | Average Step Size (nm ± S.D.) (1/2 observed movement) | Average Run Length (µm) |
|---|---|---|---|
| MVI-917 | 0 | 0 | 0 |
| MVI-940 | 12 | 27.2±9.7 | 0.9 |
| MVI-991 | 10* | 27.1±8.7* | 0.6* |
| Zippered Dimer (MVI-991 + GCN4) | >98* | 27.6±9.8* | 0.3* |
| Full-Length | 15–30* | 27.6±8.6* | 1.1* |
Values reported in Park et al., 2006.
The 3-helix bundle must unfold to form the lever arm extension (LAE)
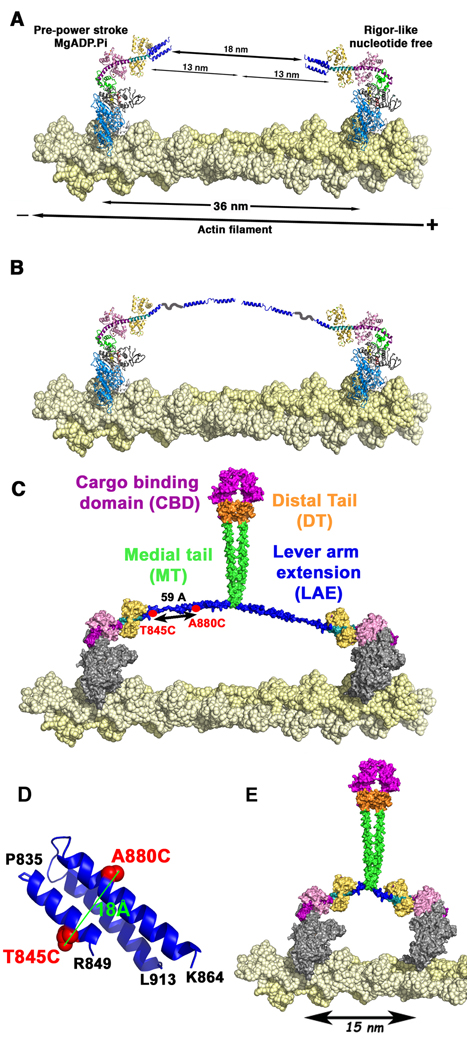
In Figure 4A we illustrate that if the 3-helix bundle remains folded, then there must be additional lever arm extensions of 9nm per head (18nm/2). Even if the region between the end of the 3-helix bundle and 940 was a SAH with none of the structure participating in dimerization, this would only provide another 4nm and would not allow for dimer formation. A possible solution is for the 3-helix bundle to unfold and form three separate α-helices, perhaps connected by hinges, providing the bulk of the lever arm extension. This would require that the bundle would be stable in the monomeric form of full-length myosin VI, but not in the dimer.
Figure 4. Dimerization-mediated unfolding of the 3-helix bundle of myosin VI.
Our working model for myosin VI in a cell is that the full-length protein exists as a monomer if not bound to cargo. Binding of myosin VI monomers to cargo alters the conformation of the molecule, possibly exposing a region capable of dimerization. The dimerization in turn triggers an unfolding of the 3-helix bundle that follows the CaM, generating a lever arm extension. A. The lead head (left) and the rear head (right) are bound to the actin filament (yellow spheres) 36 nm apart. If the triple helix is not unfolded, the bundle covers only 4 nm out of the 13 nm necessary. 18nm are thus unaccounted for. B. When the triple helix unfolds, three helices separated by segments of undefined structure account for the 13 nm necessary to extend the lever arm. C. Model of the dimerized molecule showing that the region immediately following the lever arm extension (LAE), the most proximal part of the medial tail, participates in dimerization, as do segments of the distal tail (orange) and the cargo-binding domain (magenta). Note that most of the medial tail (green) is shown as a SAH and acting as a spacer between motor and cargo. Thus the SAH is not contributing to the lever arm extension or to dimerization. The distance between the two cysteines introduced for fluorescence measurements at 845 and 880 are shown in the unfolded bundle with maximal extension of the helices. D. The positions of the cysteines that were introduced into the 3-helix bundle of myosin VI for fluorescence measurements (T845C and A880C) are shown in red. The distance between the two residues (Cα), to which two tetramethylrhodamine 5-iodoacetamide fluorophores were attached, is 18Å as depicted. E. Model of the dimerized full-length molecule in which the last two helices of the 3-helix bundle have been removed. If dimerization occurs immediately following this remaining helix, then the step size will be greatly decreased, as illustrated.
Circular dichroism (CD) data on melting of the 3-helix bundle provide further evidence for the existence of the 3-helix bundle and also suggest a mechanism in which dimerization per se could drive unfolding of the bundle. As shown in Supplementary Figure 3, a bacterially expressed proximal tail construct that should contain the 3-helix bundle (834–917) shows high α-helical content (deep trough at 222 nm in upper panel) and a cooperative (steep transition) melting at 55° (lower panel), consistent with a 3-helix bundle. This is in good agreement with the results of Spink et al. (2008) and with the crystallographic structure.
The CD spectrum of the medial tail (residues 906–991) also indicates high α-helix content (Supplementary Figure 3A), but its melting curve (Supplementary Figure 3B) is gradual and non-cooperative. This is consistent with a stable single α-helical domain, as proposed by Spink et al. (2008). The CD spectrum and melting curve for construct 834–991 is the sum of the individual spectra and melting curves of the proximal and medial tail, but with the cooperative transition shifted to approximately 67°. This may indicate that interactions between the 3-helix bundle and the extended α-helix of the medial tail further stabilize the bundle. We confirmed that, as expected, this construct is monomeric using analytical ultracentrifugation (data not shown).
To induce dimerization, we added GCN4 to the end (834-991-GCN4), mirroring our MVI-HMM construct, confirmed the construct was a dimer, and examined its CD spectra and melting. [GCN4 melts at >90°C, outside of the temperature range we are probing (Akey et al., 2001).] What we observe in the case of this dimer is that the melting is gradual and non-cooperative; the steep transition that we ascribe to melting of the 3-helix bundle is no longer present. Thus even in the absence of the myosin head, dimerization of the tail following residue 991 is propagated along the helical segment to the 3-helix bundle and induces unfolding of the bundle. However, the α-helical content is not decreased by formation of the dimer (Supplementary Figure 3A). The data also rule out the presence of a large segment of coiled coil. However, there appears to be a small degree of cooperative melting centered on ∼77°C. This may represent melting of a short segment of coiled coil structure that has formed in addition to the short segment of coiled coil in GCN4.
To provide definitive evidence for dimerization-induced unfolding of the three-helix bundle, we introduced two cysteine residues within the 3-helix bundle at positions T845 and A880 into “cys-lite” constructs (Shih et al., 2000), in which reactive cysteines were removed. This was done for both a monomeric construct (MVI-917) and our zippered dimer (MV1-991-GCN4), which were then labeled with TMR (tetramethylrhodamine) to measure the fluorescence. From the crystal structure of the 3-helix bundle, the distance between these two residues in an intact 3-helix bundle is about 18Å (Figure 4D). This distance should place the two rhodamines close enough to stack with each other and quench the fluorescence via exciton coupling (Okoh et al., 2006). However, if the bundle unfolds, the distance could increase up to 59Å (Figure 4C), which would allow fluorescence to occur unquenched from both fluorophores. The results are summarized in Table 4. Indeed, the MVI-917 (T845C, A880C) monomer with both cysteines labeled displays low fluorescence, consistent with the two rhodamines being in close proximity as expected in a folded bundle, as compared to a control MVI-917 (T845C) with one cysteine labeled (Table 4). On the contrary, in the zippered dimer with the same two cysteines labeled, a high fluorescence signal was observed, indicative of fluorescence from two rhodamines that are now too far apart to allow stacking. This fluorescence was the same in the presence or absence of actin, demonstrating that upon dimerization the 3-helix bundle unfolds.
Table 4.
Evidence for Unfolding of the Three-Helix Bundle. (Fluorescence observed by 5-TMR labeling of one or two cysteine residues inserted into the three-helix bundle of monomers and dimers as shown in Figure 4D).
| CONSTRUCT | Fluorescence Ratioa | Molar Ratio of labeling per myosin headb | ||
|---|---|---|---|---|
| without Actin or ATP | with Actin | with Actin +ATP | ||
| MVI-917 T845C | 266.2±28.1 | 213.3±15.8 | 232.8±11.4 | 1.02 |
| MVI-917 T845C, A880C | 24.5±4.6 | 18.3±7.8 | 23.1±6.2 | 2.06 |
| MVI-991-GCN4 A880C | 248.4±34.9 | 253.7±19.2 | 230.5±30.3 | 1.14 |
| MVI-991-GCN4 T845C, A880C | 196.7±20.7 | 215.6±.23.2 | 224.9±.14.6 | 2.30 |
Fluorescence was analyzed by a ratio of the emission values to that of the absorption values for each construct from at least four independent measurements. Mean values (±S.D.) are reported.
The molar ratio was calculated by comparing the myosin concentration to the concentration of the incorporated TMR.
A leucine zipper immediately following the 3-helix bundle allows large steps
In the publication of Spink et al. (2008) addition of GCN4 immediately following the 3-helix bundle resulted in non-processive molecules with measured stroke sizes of ∼23nm. They offered this as evidence that the region beyond the 3-helix bundle is responsible for extending the step of myosin VI. However, we previously noted (Rock et al., 2005) that GCN4 alone does not maintain dimers when diluted to the concentrations of single molecule assays. We cited this as evidence that there must be some additional dimerization domain prior to residue 991 that in conjunction with GCN4 allows our myosin VI HMM to remain dimeric at pM concentrations. Thus Spink et al. (2008) must have been examining the behavior of monomers, not dimers.
To address this point, we created a nearly identical construct (myosin VI truncated at Gln919 followed by GCN4 and GFP). In ATPase assays with this construct, with or without clustering on actin, there was no gating present. The Vmax was 7.2/sec/head, virtually identical to the monomeric form of MVI-940 (Table 2). This does not necessarily imply loss of dimerization, and indeed native gels (not shown) revealed that the construct was a dimer. Thus the construct had lost communication between the heads for unknown structural reasons. As we have previously demonstrated, a loss of gating not only results in an increase in ATPase activity, but also decreases actin filament sliding velocities in in vitro motility assays (Morris et al., 2003). This is consistent with the decreased velocity reported by Spink et al. (2008) for their nearly identical construct.
Loss of gating will not abolish processive movement if the motor possesses a high duty ratio unless both heads cannot simultaneously bind to actin due to structural constraints. Thus we next examined the construct in single molecule stepping assays (using FIONA). Without actin clustering, no molecules exhibited processive movement. However, this would be expected since GCN4 alone cannot maintain dimerization at sub-nanomolar concentrations. In order to increase the effective concentration, we bound the MVI-919-GCN4 molecules to actin filaments in the absence of ATP. Following this clustering on actin in rigor, addition of ATP revealed processive molecules. These molecules displayed a shorter average step size (decreased by ∼4nm) than either MVI-940 or the zippered HMM (Figure 3B&C). Interestingly the distribution of step sizes was just as broad as for the longer constructs, but the mean step size was reduced due to a large increase in the number of extremely small steps (∼12–15 nm), which might be expected if the 3-helix bundles remain folded. This could be an indication of dynamic folding and unfolding of the 3-helix bundles due to inappropriate structure and/or inappropriate steric hindrance of the flanking residues between GCN4 and the last helix of the bundle. It also could indicate that dynamic folding and unfolding of the bundle normally occurs and in part is responsible for the large distribution of myosin VI step sizes. The local structural perturbation due to insertion of GCN4 may have simply shifted the equilibrium more toward the folded conformation.
Deletion of the last two helices of the bundle greatly reduces the step size
To provide a final piece of evidence for unfolding of the bundle, we deleted the second and third helices and their connecting loops (deletion of residues 849–909 within the full-length molecule). The rationale was that if the 3-helix bundle does not unfold, then this deletion would have minimal impact on the step size, whereas if it does unfold, the step size would be greatly decreased. As shown in the model in Figure 4E, the step size would be predicted to decrease from 30–36nm to ∼15nm (three actin monomers). Stepping data in single molecule (FIONA) assays revealed the average step size to be 16.1±7.0 nm (n=61), in good agreement with the model (Figure 3B&C). The step size distribution is still broad, however. There are a large number of smaller steps, centered on a step size indicative of stepping two actin monomers, as well as occasional larger steps, which may be possible if the weak dimerization occasionally dissociates. Dissociation of the weak dimerization may also underlie the extremely large steps occasionally seen for the native molecule (Park et al. 2006).
DISCUSSION
Myosin VI contains a dimerization domain outside of the cargo-binding region
Spink et al. (2008) concluded that dimerization of the full-length molecule is exclusively due to cargo-binding domain interactions. However, this cannot explain our previously reported data that constructs truncated at Arg991 can be induced to dimerize, as evidenced by gating in ATPase assays, rotary shadowing, or single molecule processivity (Park et al., 2006). This dimerization is weak, and is only seen if the monomers are held in close proximity, either by binding to actin in rigor or by being bound to an antibody, creating extremely high effective concentrations. Furthermore, once removed from actin or from antibody, the dimers dissociate within minutes to hours, as previously noted (Rock et al., 2005) making true estimates of the percentage of dimers difficult, especially in EM. However, in ATPase assays where the molecules remain bound to actin, decreases in the bulk actin-activated ATPase to levels of the zippered HMM assays are indicative of a very high percentage of dimers that are gating.
Herein we have extended the earlier work and demonstrate that dimerization must occur between Leu913 (last residue of the three helix bundle) and Arg940. The construct truncated at Arg940 can be induced to dimerize and display gated ATPase activity, dimers in EM and processive stepping with steps identical to those of the full-length molecule. As previously noted (Knight et al., 2005; Spink et al., 2008), most of the myosin VI medial tail sequence (from Leu913 to Arg 980) is characterized by alternating clusters of positively and negatively charged amino acids that are predicted to form stable single α-helices. However, there is a short sequence between amino acids Leu913 and Lys936 that may contain a sufficient number of appropriately spaced hydrophobic and polar amino acids to form a short coiled coil. Furthermore, the experiment described above, involving deletion of the last two helices of the bundle (deletion of residues 849–909), provides strong evidence that the sequence immediately following the 3-helix bundle participates in dimerization. It is also possible that this putative coiled coil extends into the third helix of the unfolded bundle. Additional work is needed to define the exact nature of the dimerization in this region. Between Arg937 and Arg980, the sequence consists exclusively of alternating positive and negative clusters of four amino acids, which would be predicted to form a SAH.
Role of the cargo-binding domain in dimerization
The conclusion of Spink et al. (2008) that dimerization is solely via the cargo-binding domain was based on dimerization of a construct beginning at 835 and thus containing not only the cargo-binding domain (residues 1035–1285), but the 3-helix bundle (which they call the proximal tail), the region that they call the medial tail (residues 907–980), and residues 980–1035 (which they refer to as the distal tail). They also demonstrated that constructs missing the cargo-binding domain, but containing residues 907–980 did not dimerize at µM concentrations, which is consistent with our data. We only see dimerization of MVI-940 upon actin clustering, which greatly increases the effective concentration. Thus it is possible that two cargo-binding domains may interact and dimerize, which then promotes dimerization within the region from amino acids 907–940. We previously noted that inclusion of the sequence between 991 and 1050 greatly increased the percentage (from 10 to 90%) of processive dimers (Park et al., 2006). The region from 991 to 1050 also inhibits the ATPase activity of the monomer to the same degree as seen in the full-length construct (Park et al., 2006) suggesting that head-tail interactions inhibit ATPase activity. Head-tail interactions may also inhibit dimerization in the absence of cargo as suggested by Spink et al. (2008). In our model of cargo-mediated dimerization, it is the cargo itself that brings the two myosin VI monomers together (likely breaking interactions between the cargo-binding domain and the rest of the myosin VI molecule), promoting internal dimerization. It is likely that interactions within either the cargo-binding domains, or the sequence just proximal to it, initiate internal dimerization that propagates to the region immediately distal to the 3-helix bundle, triggering its unfolding.
Unfolding of a 3-helix bundle extends the reach of myosin VI
The structure of the myosin VI fragment containing amino acids 835–913 confirms the prediction of Spink et al. (2008) that the region that immediately follows the lever arm, formerly referred to as the proximal tail, is in fact a 3-helix bundle. However, in the functional dimer, this bundle must unfold to form the extension of the lever arm (Figure 4). From a detailed examination of the structure, we conclude that the bundle is lacking in stabilizing interactions, especially toward the distal end (Figure 2C).
Based on CD (Supplementary Figure 3) and fluorescence data (Table 4), we demonstrate that dimerization disrupts the bundle. Dimer formation could either recruit residues immediately following or perhaps even within the last helix, which would destabilize the 3-helix bundle. Alternatively, the bundle could be induced to unfold by steric hindrance when the two bundles are placed in close proximity. Either mechanism would provide a means for the bundle to be stable in a monomer, but unfold in a dimer. That this is the case is evident from the fluorescence data (Table 4) which shows ∼10-fold fluorescence quenching between the two rhodamines (indicative of a folded bundle structure) in monomeric MVI-917 compared to the unfolded bundle in the dimeric MVI-991-GCN4. As described above, the working model is that dimerization initiates distally upon cargo binding. Either due to steric constraints or by participation of residues of the last helix of the bundle in dimerization, the bundles are forced to unfold, remaining as extended α-helices connected by flexible hinges, as diagrammed in Figure 4. Maintenance of the α-helices would provide a much stiffer structure than a random coil. Indeed, Spink et al. (2008) demonstrated that perturbing the sequence of this region, likely generating a random coil, resulted in a greatly diminished myosin VI step size in a zippered dimer.
Both the construct truncated at Gln919 with a leucine zipper appended and the full-length construct in which the last two helices of the bundle (residues 849–909) are deleted provide further evidence for this. First, the construct truncated at Gln919 dimerizes very poorly, likely due to the fact that the leucine zipper is too close to the bundle to efficiently initiate dimerization. However, the few molecules that do dimerize can step processively with as broad a distribution of steps as the longer constructs (Figure 3C). An important difference is that MVI-919-GCN4 displays a large increase in the number of extremely small steps that could represent steps made with the bundles folded. There are 7 amino acids (913–919, inclusive) on each heavy chain to act as spacers between the bundles. This may be enough of a spacer for the bundles to dynamically fold and unfold during processive movement. This may imply that these amino acids normally contribute to a coiled coil but are not in proper phasing with GCN4. Deletion of the last two helices of the bundle greatly reduced the step size of the full-length molecule (Figure 3C) to that predicted in a model in which dimerization occurs immediately following the first helix of the bundle (Figure 4E), also indicating that dimerization occurs immediately following the 3-helix bundle in the native molecule.
Model for function of the myosin VI dimer
Our current model of the myosin VI dimer, shown in Figure 4C requires only that there be a short dimerization region at the end of the lever arm extension that forms upon distal coupling (cargo binding) of two myosin VI monomers, leading to unfolding of the bundle. The remainder of the medial tail may indeed be SAH, with additional interactions occurring between segments of the distal tail and cargo-binding domains. These distal interactions may be critical to initiate the weak dimerization adjacent to the 3-helix bundle, triggering its unfolding to form the lever arm extension. Note that in this model, the SAH domains are primarily spacers between the functional motors and cargo and have no impact on step size, consistent with our data. In this manner the SAH would play a role more like that suggested in caldesmon (Wang et al., 1991), rather than as a force-bearing extension of the lever arm.
Why such a design?
A relevant cell biological question is why design a myosin motor that forms a multi-folded, compact monomer that only unfolds and dimerizes upon cargo binding? In the case of myosin VI, the answer may lie in where it must function. For many of its functions, its cargo (or binding target) is located at the plasma membrane. Myosin VI must diffuse through a network of cortical actin in order to reach these targets, and thus a compact structure will diffuse more readily. This may be one reason that a “normal” lever arm consisting of multiple calmodulin/light chain binding sites is not used by myosin VI, even though we have demonstrated that single molecules of engineered myosin VI can function with myosin V lever arms in in vitro experiments (Park et al., 2007). Diffusion through the actin network would likely be extremely difficult if the myosin VI had to diffuse as an active dimer with the lever arm extensions unfolded. Not only would the size pose a problem, but as soon as the myosin VI encountered an actin filament, it would walk processively away from the plasma membrane. This could account for the inability of a forced dimer of myosin VI to stabilize actin during spermatid individualization in Drosophila melanogaster (Noguchi et al., 2009). The fact that the full-length monomer is not inactive, but maintains actin-activated ATPase activity may be advantageous, since reversible actin interactions of the monomer could create facilitated diffusion of myosin VI, concentrating it in regions where F-actin is most abundant.
In summary, the effective lever arm of myosin VI consists of its two CaM-containing lever arm and a lever arm extension that is formed by the unfolding of a 3-helix bundle. While most of the region following this bundle (medial tail) is likely an extended α-helix, there likely is a region near to, and perhaps even encompassing at least part of the last helix of the 3-helix bundle, that dimerizes. Dimerization, presumably initiated by cargo binding in a cell, destabilizes the bundles and extends the myosin VI lever arms. In the monomer, the 3-helix bundle remains folded, and may contribute interactions that participate in the compact folding of the full-length (monomeric) molecule. Whether myosin VI ever functions as a monomeric motor in the cell, or whether this form simply allows for optimal diffusion in the cell, remains to be elucidated. It would appear that if monomers bind to cargoes in close proximity to each other, then they will dimerize.
The idea of a motor being monomeric until it interacts with cargo also has been proposed for a kinesin superfamily member, Unc104 (Al-Bassam et al., 2003), and thus may be a recurring design within a subset of the myosin and kinesin superfamilies. A common feature of the myosins VI, VIIa and X that contain SAH regions may be their ability to form tightly folded monomers in the absence of being bound to cargo, and to unfold and dimerize upon cargo binding. Spink et al. (2008) present a model for the folding of the myosin VI monomer that is consistent with their small-angle X-ray scattering (SAXS) data and previous EM data (Lister et al., 2004) in which the SAH provides many of the interactions necessary for intramolecular folding. More recently myosin VIIa has been shown to form a compact folded monomer (Yang et al., 2009). Thus it is possible that the myosin SAH domains serve as scaffolds on which to fold the proteins. It is unclear if in some cases they can also serve as lever arm extensions. While this latter role has been proposed for the SAH of myosin X (Knight et al., 2005), and could be the case if myosin VI ever functions as a monomer, the myosin VI dimer uses the unprecedented mechanism of triggering a 3-helix bundle unfolding to extend its reach on actin.
Experimental Procedures
Protein Constructs, Expression and ATPase assays
A series of truncations of porcine myosin VI cDNA were generated. As depicted in Figure 1, C-terminal truncations were made corresponding to amino acids Gln-919 and Arg-940. Each of these had a Flag tag (encoding GDYKDDDDK) appended to the C-terminus to facilitate purification as described previously (Sweeney et al., 1998). The construct that was used to crystallize the FLA was the construct truncated at Lys-917 followed by myc and Flag tags. As previously described (De La Cruz et al., 2001), a “zippered” dimer myosin VI construct was created by truncation at Arg-991, followed by a leucine zipper (GCN4; Lumb et al., 1994) to ensure dimerization. These constructs were used to create a baculovirus for expression in SF9 cells (Sweeney et al., 1998). ATPase assays were performed as previously described (De La Cruz et al., 2001).
Crystallization, data collection and structure determination
Crystals of the full lever arm (FLA) of myosin VI (residues 770 to 917 with two bound calmodulins) grew spontaneously at 4 °C by the vapour diffusion method in hanging drops with a 1:1 mixture of 15 mg/ml protein and precipitant solution (16 % PEG 2K, 50 mM tris pH 8.5 and 100 mM MgCl2) equilibrated against 0.5 ml precipitant. Crystals were improved by addition of 2 % PEG 20K. Prior to freezing and data collection, the crystals were transferred stepwise into a final cryoprotectant solution containing 30% ethylene glycol. X-ray data sets were collected at 100 K on the ID23-1 beamline of the European Synchrotron Radiation Facility. Data sets were integrated with XDS package (Kabsch, 1988) and scaled with SCALA (CCP4, 1994 ; Evans, 2006). The structure was solved by molecular replacement with Phaser (McCoy, 2007) using the structure of the Ins2 (insert2) and the IQ motif of myosin VI bound to their respective calmodulins (CaM) (from the nucleotide free structure, 2BKH) as a model. Model building and refinement was performed with Coot (Emsley and Cowtan, 2004) and Phenix (Adams et al, 2002). Note that all figures were computed using Pymol (DeLano, 2002).
Dimerization Initiation Procedures
Dimerization was initiated in the truncated myosin constructs by either antibody binding to the C-terminal Flag tag or by saturating F-actin filaments as previously described (Park et al., 2006). By these methods, we create a high local concentration of myosin monomers bound to either actin or antibody, allowing weak dimerization to take place that would not occur at the bulk solution concentration.
Electron Microscopy (Rotary Shadowing)
Electron Microscopy was carried out as described previously in Park et al. (2006). Details can be found in the Supplementary Materials. Myosin dimers are removed from actin by centrifugation in the presence of ATP prior to rotary shadowing EM.
Single-molecule FIONA assays
To selectively label only one of the two myosin VI CaMs, we incubated labeled (Cy3) CaM with myosin VI, and then raised and lowered the free calcium concentration, as previously described (Yildiz et al., 2004). This procedure resulted in 10–20% labeling of the IQ motifs of myosin VI, so that when dimers were created no more than one head carried label. Single-molecule FIONA (Fluorescence Imaging with One Nanometer Accuracy) assays were performed as previously described (Park et al., 2006; Yildiz et al., 2003). To assess the ability of monomeric constructs to dimerize when held in close proximity (mimicking cargo binding), the actin clustering technique was applied. The microscope chamber was coated with 5mg/ml casein to block non-specific binding and PCA/PCD/trolox was used as an oxygen scavenging system (Aitken et al., 2008). Under these conditions, non-specific binding of myosin to the surface is minimal. The optics used were as previously described (Yildiz et al., 2004).
Absorbance and Fluorescence Measurements
Cysteine residues were introduced to replace T845 and A880 (Figure 4D) in MVI-917 and MVI-991-GCN4 constructs with no reactive cysteines, referred to as “cys-lite” constructs (Shih et al., 2000). Control constructs contained one reactive cysteine, T845C or A880C. The proteins (1mg each) were labeled with a 10-fold molar excess of tetramethylrhodamine 5-iodoacetamide (5-TMRIA; Anaspec, San Jose, CA) per cysteine (from a stock concentration of 20mM in dimethylformamide) at 4°C for 1–3 hours (Okoh et al., 2006). Unbound rhodamine was removed by gel filtration and overnight dialysis. Absorption spectra were measured in a HP Diode Array Spectrophotometer and fluorescence spectra were obtained in a PTI QM3 luminescence spectrofluorometer. The excitation and emission spectra were measured at 552nm and 575nm, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Anna Li and Xiaoyan Liu for protein purification. We also thank Jérome Cicolari for assistance with crystallization and X-ray data collection, and Dr. E. Michael Ostap for the use of his spectrofluorometer. This work was supported by grants from the NIH (HLS and PRS), the CNRS (AH), the ANR blanche BLAN07-3_193368 (AH) and the ACI BCMS (AH). MM is supported by a fellowship from the AHA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBER
The coordinates for the myosin VI lever arm have been deposited in the PDB with accession number 3GN4.
REFERENCES
- Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Cryst. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophysical J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey DL, Malashkevich VN, Kim PS. Buried polar residues in coiled-coil interfaces. Biochemistry. 2001;40:6352–6360. doi: 10.1021/bi002829w. [DOI] [PubMed] [Google Scholar]
- Al-Bassam J, Cui Y, Klopfenstein D, Carragher BO, Vale RD, Milligan RA. Distinct conformations of the kinesin Unc104 neck regulate a monomer to dimer motor transition. J Cell Biol. 2003;163:743–753. doi: 10.1083/jcb.200308020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman D, Sweeney HL, Spudich JA. The mechanism of myosin VI translocation and its load-induced anchoring. Cell. 2004;116:737–749. doi: 10.1016/s0092-8674(04)00211-9. [DOI] [PubMed] [Google Scholar]
- Bahloul A, Chevreux G, Wells AL, Martin D, Nolt J, Yang Z, Chen LQ, Potier N, Van Dorsselaer A, Rosenfeld S, Houdusse A, Sweeney HL. The unique insert in myosin VI is a structural calcium-calmodulin binding site. Proc Natl Acad Sci USA. 2004;101:4787–4792. doi: 10.1073/pnas.0306892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss F, Spudich G, Kendrick-Jones J. Myosin VI: Cellular Functions and Motor Properties. Annu Rev Cell Dev Biol. 2004;20:649–676. doi: 10.1146/annurev.cellbio.20.012103.094243. [DOI] [PubMed] [Google Scholar]
- Bryant Z, Altman D, Spudich JA. The power stroke of myosin VI and the basis of reverse directionality. Proc Natl Acad Sci USA. 2007;104:772–777. doi: 10.1073/pnas.0610144104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System on World Wide Web. 2002 http://www.pymol.org.
- De La Cruz EM, Ostap EM, Sweeney HL. Kinetic mechanism and regulation of myosin VI. J Biol Chem. 2001;276:32373–32381. doi: 10.1074/jbc.M104136200. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Frank DJ, Noguchi T, Miller KG. Myosin VI: a structural role in actin organization important for protein and organelle localization and trafficking. Curr Opin Cell Biol. 2004;16:189–194. doi: 10.1016/j.ceb.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Holmes KC, Geeves MA. The structural basis of muscle contraction. Philos Trans R Soc Lond B Biol Sci. 2000;355:419–431. doi: 10.1098/rstb.2000.0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KC, Schröder RR, Sweeney HL, Houdusse A. The structure of the rigor complex and its implications for the power stroke. Philos Trans R Soc Lond B Biol Sci. 2004;359:1819–1828. doi: 10.1098/rstb.2004.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houdusse A, Silver M, Cohen C. A model of Ca2+-free calmodulin binding to unconventional myosins reveals how calmodulin acts as a regulatory switch. Structure. 1996;4:1475–1490. doi: 10.1016/s0969-2126(96)00154-2. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Automatic indexing of rotation diffraction patterns. J. Appl. Cryst. 1988;21:67–72. [Google Scholar]
- Knight PJ, Thirumurugan K, Xu Y, Wang F, Kalverda AP, Stafford WF, 3rd, Sellers JR, Peckham M. The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J Biol Chem. 2005;280:34702–34708. doi: 10.1074/jbc.M504887200. [DOI] [PubMed] [Google Scholar]
- Lister I, Schmitz S, Walker M, Trinick J, Buss F, Veigel C, Kendrick-Jones J. A monomeric myosin VI with a large working stroke. EMBO J. 2004;23:1729–1738. doi: 10.1038/sj.emboj.7600180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumb KJ, Carr CM, Kim PS. Subdomain folding of the coiled coil leucine zipper from the bZIP transcriptional activator GCN4. Biochemistry. 1994;33:7361–7367. doi: 10.1021/bi00189a042. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta AD, Rock RS, Rief M, Spudich JA, Mooseker MS, Cheney RE. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- Ménétrey J, Bahloul A, Wells AL, Yengo CM, Morris CA, Sweeney HL, Houdusse A. The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature. 2005;435:779–785. doi: 10.1038/nature03592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménétrey J, Llinas P, Mukherjea M, Sweeney HL, Houdusse A. The structural basis for the large powerstroke of myosin VI. Cell. 2007;131:300–308. doi: 10.1016/j.cell.2007.08.027. [DOI] [PubMed] [Google Scholar]
- Morris CA, Wells AL, Yang Z, Chen LQ, Baldacchino CV, Sweeney HL. Calcium functionally uncouples the heads of myosin VI. J Biol Chem. 2003;278:23324–23330. doi: 10.1074/jbc.M208957200. [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Homma K, Komori Y, Iwaki M, Wazawa T, Hikikoshi Iwane A, Saito J, Ikebe R, Katayama E, Yanagida T, Ikebe M. Class VI myosin moves processively along actin filaments backward with large steps. Biochem Biophys Res Commun. 2002;290:311–317. doi: 10.1006/bbrc.2001.6142. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Frank DJ, Isaji M, Miller KG. Coiled-coil-mediated dimerization is not required for myosin VI to stabilize actin during spermatid individualization in Drosophila melanogaster. Mol Biol Cell. 2009;20:358–367. doi: 10.1091/mbc.E08-07-0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007;8:R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoh MP, Hunter JL, Corrie JET, Webb MR. A biosensor for inorganic phosphate using a rhodamine-labeled phosphate binding protein. Biochem. 2006;45:14764–14771. doi: 10.1021/bi060960j. [DOI] [PubMed] [Google Scholar]
- Park H, Ramamurthy B, Travaglia M, Safer D, Chen LQ, Franzini-Armstrong C, Selvin PR, Sweeney HL. Full-Length Myosin VI Dimerizes and Moves Processively along Actin Filaments upon Monomer Clustering. Mol Cell. 2006;21:331–336. doi: 10.1016/j.molcel.2005.12.015. [DOI] [PubMed] [Google Scholar]
- Park H, Li A, Chen LQ, Houdusse A, Selvin PR, Sweeney HL. The unique insert at the end of the myosin VI motor is the sole determinant of directionality. Proc Natl Acad Sci USA. 2007;104:778–783. doi: 10.1073/pnas.0610066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock RS, Rice SE, Wells AL, Purcell TJ, Spudich JA, Sweeney HL. Myosin VI is a processive motor with a large step size. Proc Natl Acad Sci USA. 2001;98:13655–13659. doi: 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock RS, Ramamurthy B, Dunn AR, Beccafico S, Rami BR, Morris C, Spink BJ, Franzini-Armstrong C, Spudich JA, Sweeney HL. A Flexible Domain Is Essential for the Large Step Size and Processivity of Myosin VI. Mol Cell. 2005;17:603–609. doi: 10.1016/j.molcel.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Shih WM, Gryczynski Z, Lakowicz JR, Spudich JA. A FRET-based sensor reveals large ATP hydrolysis-induced conformational changes and three distinct states of the molecular motor myosin. Cell. 2000;102:683–694. doi: 10.1016/s0092-8674(00)00090-8. [DOI] [PubMed] [Google Scholar]
- Sivaramakrishnan S, Spink BJ, Sim AY, Doniach S, Spudich JA. Dynamic charge interactions create surprising rigidity in the ER/K {alpha}-helical protein motif. Proc Natl Acad Sci USA. 2008;105:13356–13361. doi: 10.1073/pnas.0806256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spink BJ, Sivaramakrishnan S, Lipfert J, Doniach S, Spudich JA. Long single alpha-helical tail domains bridge the gap between structure and function of myosin VI. Nat Struct Mol Biol. 2008;15:591–597. doi: 10.1038/nsmb.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney HL, Rosenfeld SS, Brown F, Faust L, Smith J, Stein L, Sellers J. Kinetic tuning of myosin via a flexible loop adjacent to the nucleotide-binding pocket. J. Biol. Chem. 1998;273:6262–6270. doi: 10.1074/jbc.273.11.6262. [DOI] [PubMed] [Google Scholar]
- Sweeney HL, Houdusse A. What can myosin VI do in cells? Curr Opin Cell Biol. 2007;19:57–66. doi: 10.1016/j.ceb.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Terrak M, Rebowski G, Lu RC, Grabarek Z, Dominguez R. Structure of the light chain-binding domain of myosin V. Proc Natl Acad Sci USA. 2005;102:12718–12723. doi: 10.1073/pnas.0503899102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CL, Chalovich JM, Graceffa P, Lu RC, Mabuchi K, Stafford WF. A long helix from the central region of smooth muscle caldesmon. J Biol Chem. 1991;266:13958–13963. [PMC free article] [PubMed] [Google Scholar]
- Wells AL, Lin AW, Chen LQ, Safer D, Cain SM, Hasson T, Carragher BO, Milligan RA, Sweeney HL. Myosin VI is an actin-based motor that moves backwards. Nature. 1999;401:505–508. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- Yang Y, Baboolal TG, Siththanandan V, Chen M, Walker ML, Knight PJ, Peckham M, Sellers JR. A FERM domain autoregulates Drosophila myosin 7a activity. PNAS. 2009 doi: 10.1073/pnas.0808682106. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A, Park H, Safer D, Yang Z, Chen LQ, Selvin PR, Sweeney HL. Myosin VI steps via a hand-over-hand mechanism with its lever arm undergoing fluctuations when attached to actin. J Biol Chem. 2004;279:37223–37226. doi: 10.1074/jbc.C400252200. [DOI] [PubMed] [Google Scholar]
- Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.