Abstract
Semiconductor QDs are being developed as fluorescent tags for biomedical applications such as imaging, targeting, therapeutic carriers, drug delivery, nanomedicine, and in vitro and in vivo biological labeling. However, potential toxicity and clearance of semiconductor QDs in biological systems are of concerned. We have tested toxicity and clearance in vitro and in vivo (subcutaneous)of CdTe and CdHgTe semiconductor QDs in human breast cancer MCF7 and MBA-MD-231 cells and prostate cancer PC3 cells over a period of 40 days. Our results show that both CdTe and CdHgTe QDs are cytotoxic to human breast and prostate cancer cells. CdHgTe ODs were cleared rapidly from the site of injection, while CdTe were still detectable 18 days after injection, but were cleared by 30 days.
Keywords: Semiconductor QDs, CdTe, CdHgTe, Toxicity, Clearance
1. INTRODUCTION
Semiconductor QDs have been developed as fluorescence tags in biological and medical applications such as in vitro and in vivo biological labeling, imaging,1–6 targeting,7,8 therapeutic carriers and drug delivery.9–13 To avoid QD aggregation, improve water-solubility and biocompatibility, and exert specific surface chemistry for targeting and delivery, various conformations of semiconductor QDs, such as core-shell structure, organic molecule-modified geometry, and functional polymer capped structure have been developed14–16. Semiconductor QDs are shown to have advantages of efficient luminescence, high photobleaching threshold, flexible surface chemistry, and good water-solubility for biological and medical applications. However, the issues of toxicity and clearance of QDs in bio-systems are of concern,17–24 as these two issues are not fully understood and must be carefully assessed if QDs are to move from scientific curiosity to biomedical application.
Several reports have examined the toxicity and clearance of QDs.8,18–32 One common understanding for toxicity of cadmium-containing semiconductor QDs is that their toxicity is closely related to the concentration of free Cd2+.17,21,22,28,29 Derfus et al observed that the cytotoxicity of CdTe QDs was correlated to the liberation of free Cd2+.29 and Zhang et al recently presented similar evidence showing that the cytotoxicity of QDs behaves in a concentration- and size-dependent manner17. Another common assumption is that surface capping layers of QDs synthesized under different methods may contribute to the level of toxicity following different mechanisms such as oxidation and breakdown of nanostructures.21,22,28 The distribution and damage of QDs to organs are highly variable, and the clearance of QDs in bio-system also varies according to the concentration and structure of QDs.28, 33, 34.
In this paper, we assess the toxicity and clearance of semiconductor CdTe and CdHgTe QDs in vitro in human breast and prostate cancer cells and in vivo following subcutaneous injection in mice.
2. MATERIALS AND METHODS
2.1 Synthesis of Semiconductor CdTe and CdHgTe QDs
The preparation of CdTe QDs has been reported elsewhere.35–38 The QDs were stabilized by thioglycolic acid (TGA) on their surfaces, enhancing water solubility, and facilitating conjugation with ligands. The average sizes of QDs used was 6–8 nm. The procedures for making CdTe QDs are briefly described as follows: CdTe QDs were created by the reaction of precursors containing cadmium perchlorate hydrate [Cd(ClO4)2*H2O] and hydrogen telluride (H2Te) through vigorous stirring. The Cd2+ solution was prepared by dissolving 731 mg of Cd(ClO4)2*H2O in 125 mL of water. TGA (0.396 mL) was then added to the solution. 0.1M NaOH solution was also added to adjust the pH to approximately 11. The solution was then purged with nitrogen for at least 30 minutes. H2Te gas was produced by the chemical reaction of excess aluminum telluride with 0.5 M sulfuric acid in an inert atmosphere (nitrogen) and was combined with the above Cd2+ solution using a set-up described previously36. After completion of the reaction a yellow solution of CdTe QD nuclei was obtained. This solution was then refluxed at 100 °C to promote crystal growth. The QDs were extracted and stored at 4 °C in the dark. The near infrared emitting CdHgTe QDs were obtained by adding 2.5 ml of 0.1 M mercury perchlorate solution to 50 ml of CdTe QD solution to gradually form CdHgTe QDs.
2.2 Fluorescent Imaging
Red CdTe and Infrared CdHgTe QDs with fluorescence emission peaks in the 650 nm and 900 nm ranges, respectively were used. Imaging was performed using a multispectral Maestro Fluorescent Imaging System (Cri, Waltham, MA) with a green filter. For in vivo fluorescent imaging experiments CdTe and CdHgTe QDs (50 μl) were injected subcutaneously into mice at two locations. A mouse was anesthetized with 80 μl of a mixture of ketamine, xylazine, and acepromazine. Fluorescent images were acquired immediately after injection of CdTe and CdHgTe QDs with 100 ms exposure. CdTe and CdHgTe clearance and toxicity were monitored over a period of 40 days.
2.3 Toxicity
Human breast cancer MCF7 and MDA-MD-231 cells and prostate cancer PC3 cells were seeded in 96-well plates (Costar, Corning, NY) at a concentration of 1 × 104 cells in 100 μl of medium per well. Each treatment condition was assessed in groups of 8. After 24 hours, the medium was aspirated and new medium containing the QDs were added. At the indicated time, total cell number was determined using a crystal violet assay. Briefly, the medium was aspirated and 1% glutaraldehyde (100 μl; Sigma, St. Louis, MO) in PBS was added for incubation for 15 minutes. After removing glutaraldehyde, 0.5% crystal violet (Sigma) was incubated for 15 minutes, and the plates were washed with water (twice) and soaked in water for 10 min before drying at room temperature. Once dry, 100 μl of Sorenson’s solution (a solution of 9 g tri-sodium citrate in 305 ml of distilled water with 195 ml 0.1 N HCl and 500 ml 90% ethanol) was added to elute the crystal violet. After 30 minutes, it was read at 540 nm using an ELX800 microplate reader (Bio-Tek Instruments, Winooski, VT).
3. RESULTS AND DISCUSSION
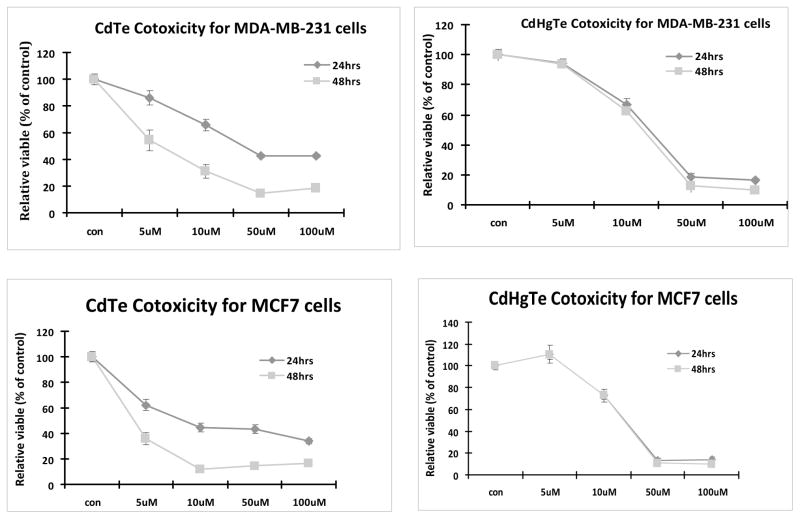
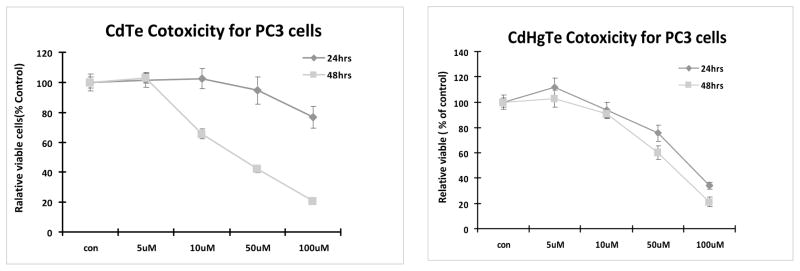
We tested the concentration and time-dependent cytotoxicity of CdTe and CdHgTe QDs in human breast cancer MCF7 and MBA-MD-231 cells and prostate cancer PC3 cells. Both CdTe and CdHgTe QDs showed concentration-dependent cytotoxicity (Figs. 1 and 2) for each cell type. Intriguingly, cytotoxicity of CdTe QDs increased with exposure time, but this was not seen for CdHgTe for any of the tumor cell lines. For 24 hrs exposure, there was minimal cytotoxicity up to 5 μM, but it increased significantly with higher concentrations.
Figure 1.
Cytotoxicity of CdTe and CdHgTe QDs in breast cancer MDA-MB-231 and MCF7 Cells
Figure 2.
Cytotoxicity of CdTe and CdHgTe QDs in prostate cancer PC3 cells
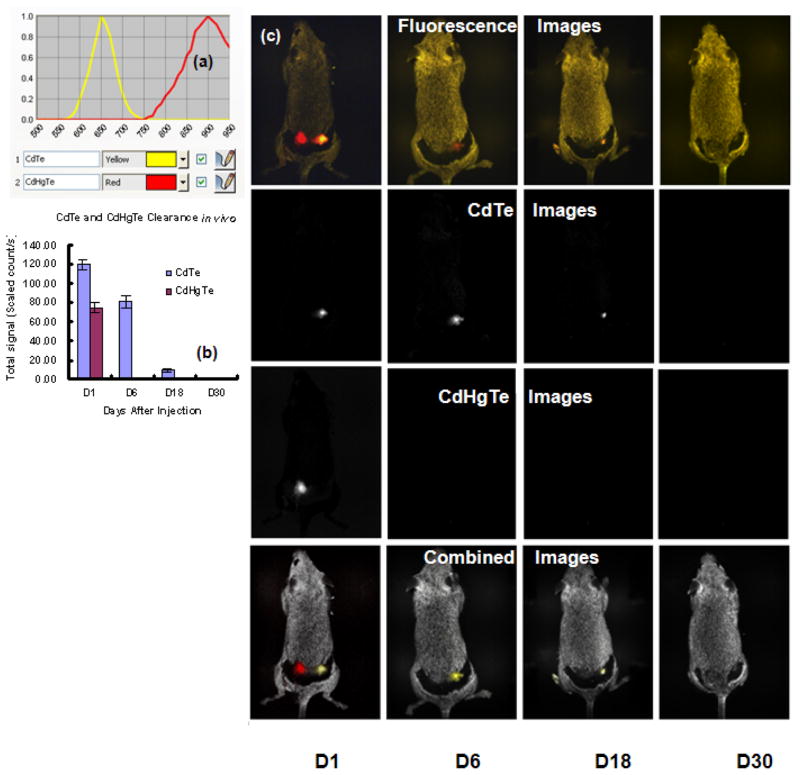
When CdTe and CdHgTe QDs (50 μL) were injected subcutaneously into a group of 6 mice at two locations all survived and fluorescent images were acquired immediately and up to 40 days. Figure 3(a) shows the in vivo fluorescence spectra of CdTe and CdHgTe QDs at 650 nm and 900 nm. Figure 3(b) indicates the change of fluorescence signals for CdTe and CdHgTe QDs over 30 days. The fluorescent signals from CdTe QDs decayed more slowly than those of CdHgTe QDs. CdTe QDs still showed 15% fluorescent intensity after 18 days, but signal disappeared by 30 days. The fluorescent signals from CdHgTe QDs decayed rapidly and no fluorescent signal was detectable by the 6th day. The first row in Figure 3(c) shows the fluorescence images of a mouse 1, 6, 18 and 30 days following subcutaneous injection of 50 μL CdHgTe (left fluorescence spot) and CdTe (right fluorescence spot) QDs. Fluorescence from CdTe QDs could be observed clearly at 18 days, while the fluorescence from CdHgTe QDs persisted less than 6 days. The discrepancy in fluorescence decay may be due to the lower chemical-stability of CdHgTe in comparison with CdTe QDs. Spectral unmixing showed the fluorescent images individually for CdTe and CdHgTe QDs, respectively (Figure 3c, second and third rows), Combined images of QDs overlaid on auto fluorescence (Figure 4d).
Figure 3.
(a) Fluorescence spectra of CdTe and CdHgTe QDs; (b) CdTe and CdHgTe clearance in vivo; (c) First row: Fluorescence images of grey mouse following subcutaneous injection of 50 μL CdHgTe (left fluorescent spot) and CdTe (right fluorescent spot) QDs observed at 1, 6, 18 and 30 days; Second row: Spectrally unmixed images to show CdTe; Third row: Spectrally unmixed images showing CdHgTe QDs. Fourth row: Combined images of CdTe and CdHgTe overlaid on autofluorescence.
CONCLUSION
Both CdTe and CdHgTe QDs could be detected by fluorescent imaging in vitro and in vivo. Both CdTe and CdHgTe QDs showed some cytotoxicity in human breast MCF7 and MBA-MD-231 and prostate cancer PC3 cells at 5 μM. CdTe QD toxicity increased with exposure time, but this was not seen in CdHgTe QDs. CdTe QDs could be observed in vivo following SC administration in mice for up to 18 days, while the fluorescent signal in CdHgTe disappeared within 6 days, possibly due to instability.
Acknowledgments
Supported by the Department of Energy (DE-FG02-05CH11280) and Southwestern Small Animal Imaging Research Program (SW-SAIRP), funded in part by NCI U24 CA126608. J. Zhang would like to thank financial aid from the NSFC (Grants No. 20601012), Inner Mongolia University “513 program” (No. 206043) and “Nanolab program”, and the Educational Department of Inner Mongolia (NJZY07011).
References and Notes
- 1.Chan WCW, Nie S. Science. 1998;281:2016. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 2.Jamieson T, Bakhshi R, Petrova D, Pocock R, Imani M, Seifalian AM. Biomaterials. 2008;28:4717. doi: 10.1016/j.biomaterials.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 3.Hoshino A, Manabe N, Fujioka K, Suzuki K, Yasuhara M, Yamamoto K. Journal of Artificial Organs. 2007;10:149. doi: 10.1007/s10047-007-0379-y. [DOI] [PubMed] [Google Scholar]
- 4.Gao X, Dave SR. Bio-applications of Nanoparticles. 2007;620:57. [Google Scholar]
- 5.Zhang J, Sun J, Liu L, Huang Y, Mason RP. Journal of Nanoscience and Nanotechnology. 2008;8:1155. [PubMed] [Google Scholar]
- 6.Su J, Zhang J, Liu L, Huang Y, Mason RP. Journal of Nanoscience and Nanotechnology. 2008;8:1174. [PubMed] [Google Scholar]
- 7.Gao X, Cui Y, Levenson RM, Chung LWK, Nie S. Nature Biotechnology. 2004;22:969. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 8.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Nature Materials. 2005;4:435. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 9.Chen W. Journal of Nanoscience and Nanotechnology. 2008;8:1019. doi: 10.1166/jnn.2008.301. [DOI] [PubMed] [Google Scholar]
- 10.Chen W, Zhang J. Journal of Nanoscience and Nanotechnology. 2006;6:1159. doi: 10.1166/jnn.2006.327. [DOI] [PubMed] [Google Scholar]
- 11.Takeda M, Tada H, Higuchi H, Kobayashi Y, Kobayashi M, Sakurai Y, Ishida T, Ohuchi N. Breast Cancer. 2008;15:145. doi: 10.1007/s12282-008-0037-0. [DOI] [PubMed] [Google Scholar]
- 12.Yezhelyev MV, Qi L, O’Regan RM, Nie S, Gao X. Journal of The American Chemical Society. 2008;130:9006. doi: 10.1021/ja800086u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu W, Choi HS, Zimmer JP, Tanaka E, Frangioni JV, Bawendi MG. Journal of The American Chemical Society. 2007;129:14530. doi: 10.1021/ja073790m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo W, Li J, Wang Y, Peng X. Journal of The American Chemical Society. 2003;125:3901. doi: 10.1021/ja028469c. [DOI] [PubMed] [Google Scholar]
- 15.Kim S, Bawendi MG. Journal of The American Chemical Society. 2003;125:14652. doi: 10.1021/ja0368094. [DOI] [PubMed] [Google Scholar]
- 16.Dubertret B, Skourides P, Norris DJ, Noireaux V, Brivanlou AH, Libchaber A. Science. 2002;298:1759. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Chen W, Zhang J, Liu J, Chen G, Pope C. Journal of Nanoscience and Nanotechnology. 2007;7:497. doi: 10.1166/jnn.2007.125. [DOI] [PubMed] [Google Scholar]
- 18.Male KB, Lachance B, Hrapovic S, Sunahara G, Luong JHT. Analytical Chemistry. 2008;80:5480. doi: 10.1021/ac8004555. [DOI] [PubMed] [Google Scholar]
- 19.Byrne SJ, Williams Y, Davies A, Corr SA, Rakovich A, Gun’ko YK, Rakovich YR, Donegan JF, Volkov Y. Small. 2007;3:1152. doi: 10.1002/smll.200700090. [DOI] [PubMed] [Google Scholar]
- 20.Chang E, Thekkek N, Yu W, Colvin VL, Drezek R. Small. 2006;2:1412. doi: 10.1002/smll.200600218. [DOI] [PubMed] [Google Scholar]
- 21.Fischer HC, Liu L, Pang K, Chan WCW. Advanced Functional Materials. 2006;16:1299. [Google Scholar]
- 22.Fisher HC, Chan WCW. Current Opinion in Biotechnology. 2007;18:565. doi: 10.1016/j.copbio.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 23.Choi HS, Liu W, Misra P, Tanaka E, Zimmer JP, Ipe BI, Bawendi MG, Frangioni JV. Nature Biotechnology. 2007;25:1165. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai W, Hsu AR, Li Z, Chen X. Nanoscale Research Letters. 2007;2:265. doi: 10.1007/s11671-007-9061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo G, Liu W, Liang J, He Z, Xu H, Yang X. Materials Letters. 2007;61:1641. [Google Scholar]
- 26.Han M, Gao X, Su JZ, Nie S. Nature Biotechnology. 2001;19:631. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- 27.Ballou B, Ernst LA, Waggoner Current Medicinal Chemistry. 2005;12:795. doi: 10.2174/0929867053507324. [DOI] [PubMed] [Google Scholar]
- 28.Ballou B, Lagerholm BC, Ernst LA, Bruchez MP, Waggoner AS. Bioconjugation Chemistry. 2004;15:79. doi: 10.1021/bc034153y. [DOI] [PubMed] [Google Scholar]
- 29.Derfus AM, Chan WCW, Bhatia SN. Nano Letters. 2004;4:11. doi: 10.1021/nl0347334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fountaine TJ, Wincovitch SM, Geho DH, Garfield SH, Pittaluga S. Modern Pathology. 2006;19:1181. doi: 10.1038/modpathol.3800628. [DOI] [PubMed] [Google Scholar]
- 31.Smith AM, Ruan G, Rhyner MN, Nie S. Annals of Biomedical Engineering. 2006;34:3. doi: 10.1007/s10439-005-9000-9. [DOI] [PubMed] [Google Scholar]
- 32.Fu A, Gu W, Larabell C, Alivisatos AP. Current Opinion in Neurobiology. 2005;15:568. doi: 10.1016/j.conb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 33.Voura EB, Jaiswal JK, Mattoussi H, Simon SM. Nature Medicine. 2004;10:993. doi: 10.1038/nm1096. [DOI] [PubMed] [Google Scholar]
- 34.Lovric J, Bazzi HS, Cuie Y, Fortin GR, Winnik FM, Maysinger D. Journal of Molecular Medicine. 2005;83:377. doi: 10.1007/s00109-004-0629-x. [DOI] [PubMed] [Google Scholar]
- 35.Morgan NY, English S, Chen W, Chernomordik V, Russo A, Smith PD, Gandjbakhche A. Academic Radiology. 2005;12:313. doi: 10.1016/j.acra.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 36.Gaponik N, Talapin DV, Rogach AL, Hoppe K, Shevchenko EV, Kornowski A, Eychmuller A, Weller H. J Phys Chem B. 2002;106:7177. [Google Scholar]
- 37.Joly AG, Chen W, McCready DE, Malm J, Bovin J. Phys Rev B. 2005;71:165304. [Google Scholar]
- 38.Chen W, Zhang J, Westcott SL, Joly AG, Malm J, Bovin J. J Appl Phys. 2006;99:034302. [Google Scholar]