Abstract
Proteins play essential roles in all aspects of cellular processes, such as biosynthesis, division, growth, motility, metabolism, signaling, and transmission of genetic information. Proteins, however, could deform under mechanical forces, thus altering their biological functions. Here we present protein deformation as a possible molecular basis for mechanosensing and mechanotransduction, elucidate the important features of protein mechanics including protein deformation mode and dynamics, illustrate how protein deformation could alter biological function, and describe the important roles of protein deformation in force-sensing, force transducing and mechanochemical coupling in cells. The experimental and modeling challenges in protein mechanics are discussed.
Keywords: Protein, Deformation, Biomechanics, Conformational change, Mechanotransduction
Introduction
The biological cell constitutes the basic unit of life and performs a large variety of functions through synthesis, sorting, storage and transport of biomolecules; expression of genetic information; recognition, transmission and transduction of signals; and converting different forms of energy [1]. Many of the cellular processes involve mechanical force, or deformation, at the cellular, subcellular and molecular levels [2, 3]. For example, biomolecular motors and machines convert chemical energy into mechanical motion in performing their diverse range of functions [4, 5]. During cell migration, contractile forces must be generated within the cell in order for the cell body to move forward [6]. Adhesion of cells to extracellular matrix (ECM) through focal adhesion complexes is sensitive to the stiffness of the substrate [7, 8]. All living cells on Earth are constantly under physical force (gravitational and other forms of force), and many normal and diseased conditions of cells are dependent upon or regulated by their mechanical environment. Some cells, such as bone and endothelial cell, are subjected to specific forces as part of their ‘native’ physiological environment. Some other cells, such as muscle and cochlear outer hair cells [9], perform a mechanical function by converting an electrical or chemical stimulus into a mechanical motion.
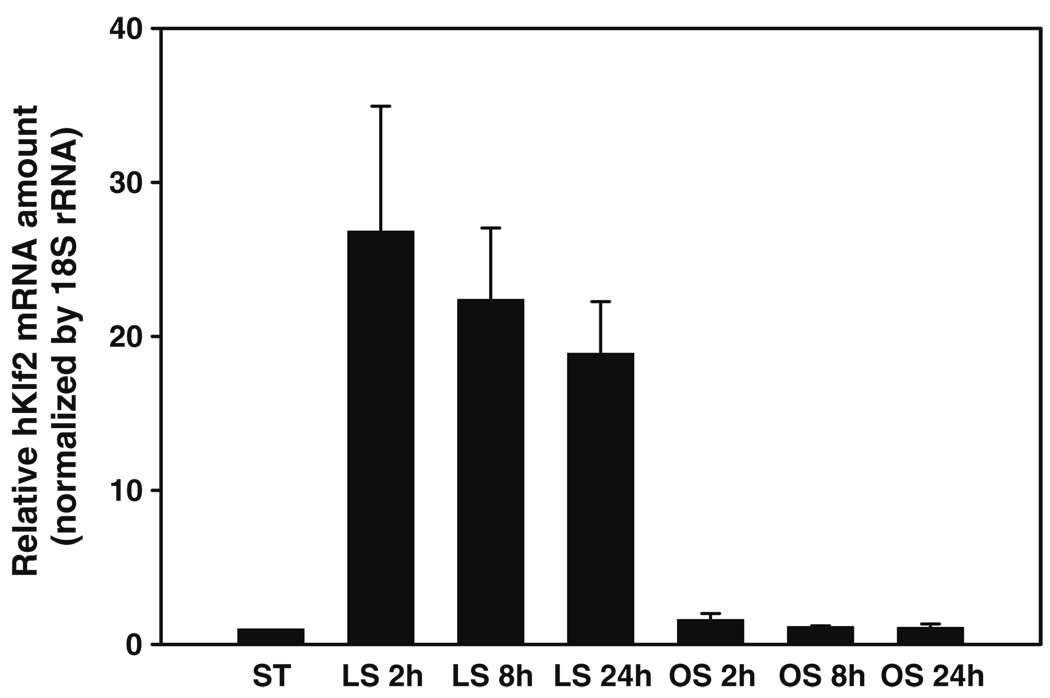
Of particular importance is the ability of cells to sense mechanical force or deformation, and transduce this mechanical signal into a biological response [10–14]. It is well established that vascular smooth muscle cells in the arterial wall remodels when subject to hemodynamic forces [15]. Bone keeps adjusting its structure to adapt to its mechanical environment [16]. Stem cells, on the other hand, can sense the elasticity of the substrate and differentiate into different phenotypes accordingly [17–19]. Endothelial cells can recognize the magnitude, mode (steady or pulsatile), type (laminar or turbulent) and duration of applied shear flow, and respond accordingly, maintaining healthy endothelium or leading to vascular diseases including thrombosis and atherosclerosis [20]. As an example, Fig. 1 shows shear stress-induced changes in the expression level of the Krüppel-like factor 2 (KLF2) mRNA in human umbilical vein endothelial cells (HUVEC) under static, laminar shear and oscillatory shear conditions, with applied shear stress of 15 dyn/cm2. These and other examples clearly demonstrate that cells can sense and respond to its local mechanical environment, including applied mechanical force and deformation, the stiffness of the substrate and surrounding media, vibration, and even sound waves (hair cells in the mammalian cochlea are believed to sense and amplify incoming sound waves).
Fig. 1.
Shear stress-induced changes in the expression level of Krüppel-like factor 2 (KLF2) mRNA in HUVEC under static, laminar flow and oscillatory flow conditions for different shear durations, with applied shear stress of 15 dyn/cm2. It reveals a significant increase of KLF2 mRNA level under laminar shear (LS); however the KLF2 expression remains at the control (static) level under oscillatory shear (OS)
But how do cells sense mechanical force (or deformation)? How do cells transduce mechanical signal into biochemical or biological responses? To date, the molecular mechanisms of force sensing and mechanotransduction remain elusive. It has been speculated that it is the specialized ‘force sensors’ or stretch-activated ion channels on the cell membrane that sense and transduce the mechanical signal [21], or it is the interaction between the cell and the extracellular matrix (ECM) through integrin molecules that serves as a force sensor and transducer [22]. However the underlying molecular mechanism of mechanotransduction is not well understood, and there is a definite need to identify such a mechanism that is applicable to most, if not all, cell types [18].
In the following sections, we present protein conformational change under force as a possible molecular basis for mechanotransduction, elucidate the important features of protein mechanics, and illustrate how protein deformation could alter biological function, and its essential roles in force-sensing, force transducing and mechanochemical coupling functions in cells. The experimental and modeling challenges in protein mechanics are discussed.
Protein Deformation and its Biological Consequences
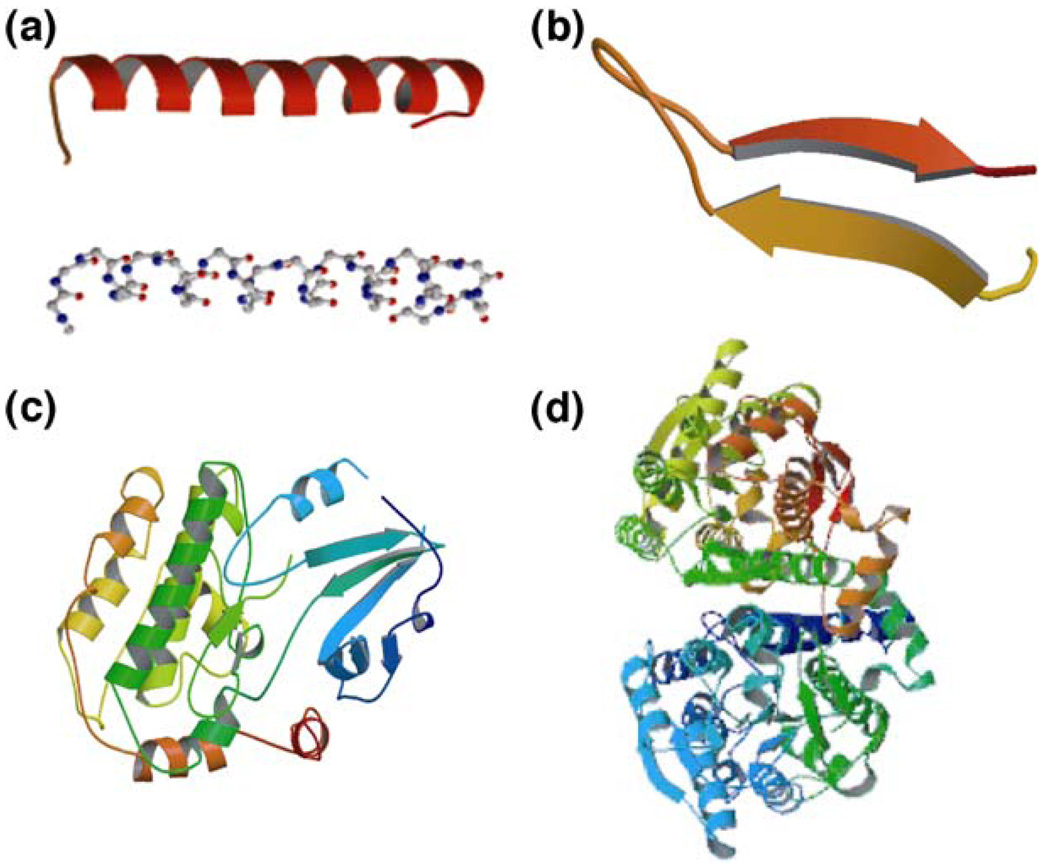
Proteins perform a broad range of tasks in living cells, including metabolic and catalytic functions (enzymes), signal transduction (signaling proteins), transport of biomolecules (transporters), transmission of genetic information (nucleoprotein machines), and structural support (filaments and their networks) [1]. With a specific polypeptide sequence of up to 20 different amino acids, most proteins (globular proteins) form by folding the polypeptide into a tertiary structure [Fig. 2(d)] of single or multiple globular domains [Fig. 2(c)] consisting of α-helices [Fig. 2(a)], β-sheets [Fig. 2(b)] and polypeptide loops. Proteins may also have ‘rod’ or ‘wire’-like tertiary structures (fibrous proteins). Polypeptide chains of more than ∼200 amino acids usually fold into two or more globular domains. These structurally independent domains have an average size of ∼2.5 nm and possess unique three-dimensional geometries. Most protein domains have specific functions such as the binding of small molecules (ligands) and other protein domains. The ligand-binding sites in multi-domain proteins are often located in the clefts between domains to facilitate the interactions with different ligands [1, 23].
Fig. 2.
Basic structural features of proteins. Proteins are polypeptides constructed from 20 different amino acids. The polypeptide folds into secondary structures such as α-helices (a) and β-sheets (b) which, together with polypeptide loops, form globular domains (c). Multidomain globular proteins typically have a tertiary structure (d)
It has been well established that the three-dimensional conformation of a protein largely determines its function [1]. However, the conformation of a protein can be altered by applied mechanical force, resulting in changes of the functional states of the protein and inducing down-stream biochemical and biological effects. Therefore, protein deformation, or protein conformational change under mechanical force, is an excellent candidate for the molecular mechanism of mechanotransduction [10, 11, 24].
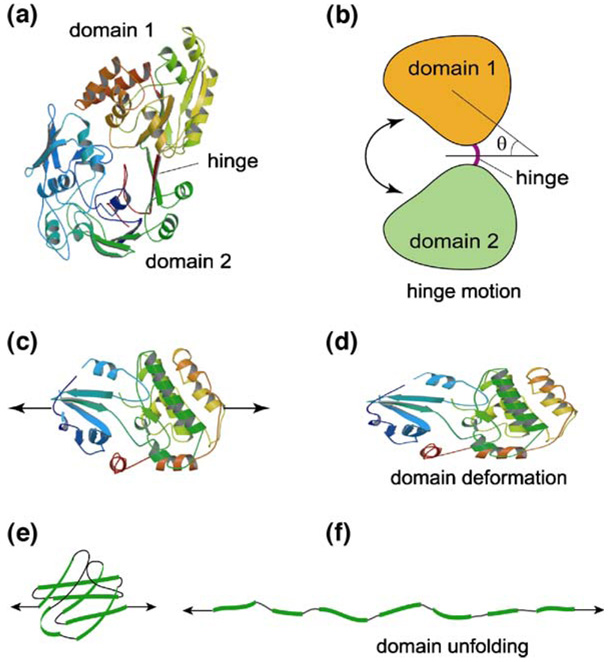
Similar to well-known physical (e.g., temperature increase) or chemical (e.g., changes in pH) events that alter protein conformation [23, 25], which may transform protein from a native or biologically active state to a denatured or inactive state, the application of mechanical forces to a protein can lead to protein conformational changes including domain motion [Fig. 3(a), (b)], domain deformation [Fig. 3(c), (d)] and domain unfolding [Fig. 3(e), (f)], all of which can have significant biological implications. In general, protein domain motion includes domain hinge motion and domain sliding motion. In domain hinge motion [Fig. 3(b)], the individual globular domains have very limited deformation; the motion largely consists of rotations of domains around a flexible hinge (e.g., loops, α-helices or β-sheets that join the domains together). In domain sliding motion, two protein globular domains have relative (in-plane or out-of-plane) sliding (‘shear’) motion. It is also possible to have mixed mode domain motions, such as a combination of rotation about a hinge and twist about an axis of the domain. Protein domain motions can be driven by applied torque as well as Brownian forces or moments (∼1 kBT) [26], which are equivalent to forces on the order of 0.5–10 pN (depending on the degree of motion). With large tensile forces in the range of 50–200 pN, a protein globular domain may unfold [27–30], as illustrated in Fig. 3(e) and (f).
Fig. 3.
Protein deformation modes. Protein conformational changes under force including domain motion (a, b), domain deformation (c, d) and domain unfolding (e, f). These deformation modes occur according to different levels of the applied force
Under applied tensile (compressive) forces, a protein globular domain may elongate (contract), as illustrated in Fig. 3(c) and (d). This type of protein conformational change is referred to as domain deformation, which is distinct from pure domain motion (little or none conformational change in the domain itself) and domain unfolding (opening of a globular domain). Domain deformation typically requires forces in the range of 1–100 pN. Although the three protein deformation modes shown in Fig. 3 have different characteristics and force ranges, it is possible that they are coupled. For example, before domain unfolding occurs, there could be a significant domain deformation. Under mechanical forces, some protein domains may unfold gradually, others may unfold abruptly. It is also true that during domain motion, individual protein domains may have small conformational changes. The amount of coupling between different modes of protein deformation depends not only on the applied forces but also on protein structure, as is expected.
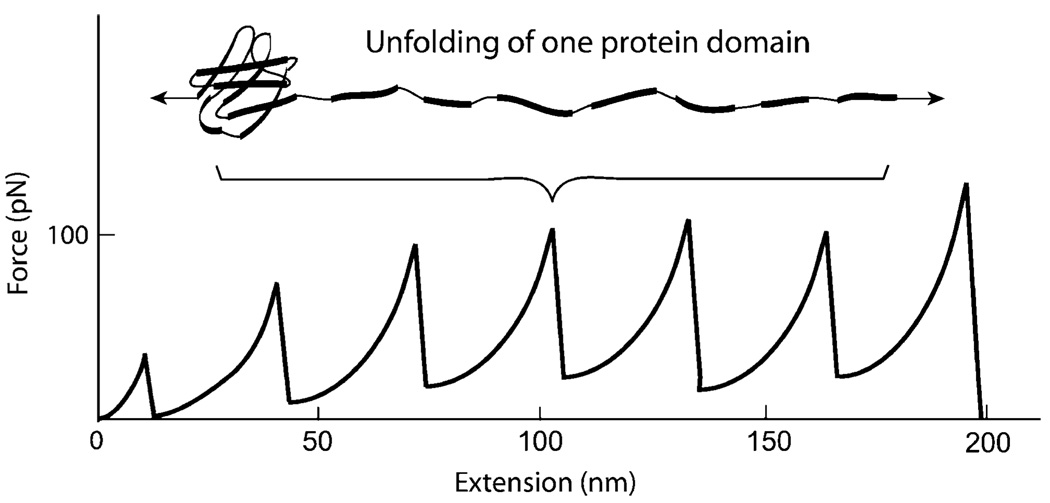
There is ample experimental evidence to suggest that protein domains can sustain deformation or even unfolding under normal physiological forces. It has been revealed that, under stretching with AFM or optical tweezers, the immunoglobulin domains of muscle molecule titin may deform and unfold, with a characteristic sawtooth pattern [27, 28], as shown schematically in Fig. 4. Clearly, a force of ∼100 pN is required for domain unfolding of titin. The FN-III domains of the ECM protein tenascin unfold upon stretch in a similar fashion [30]. Single-molecule biomechanics studies, both experimental and theoretical, have been performed for the forced unfolding of ECM molecule fibronectin, which regulates many cellular functions through its binding to integrin [31–33]. Experimental evidence suggests that, during cell adhesion and locomotion, the FN-III domains of fibronectin may unfold and refold due to the tensile and contractile forces the cells apply to the ECM [32]. This unfolding and refolding may have significant implications to cell signaling, since the resulting structural changes of the RGD loop seems to regulate the binding between fibronectin and integrin, as revealed by molecular dynamics simulations [33–35].
Fig. 4.
An example of protein domain deformation and unfolding. Under applied tensile force, multiple immunoglobulin domains of muscle molecule titin deform and unfold, showing a characteristic sawtooth pattern
Protein unfolding could occur at multiple scales when subjected to a pulling force. For example, a coiled multimeric protein (such as vWF) could be uncoiled under mechanical forces before domain unfolding occurs. A multi-domain protein with repeated globular domains could unfold sequentially, starting from the ‘weakest link’, i.e., a domain that requires the smallest force to unfold. As the pulling force increases, more domains unfold, resulting in a see-saw like unfolding response. The sequence of unfolding of repeated multi-domains could also be stochastic, since thermal forces may play some role in forced unfolding. Proteins with multiple globular domains may unfold completely, i.e., all the domains are unfolded. They may have partial unfolding, i.e., one or a few domains may unfold but other remain intact. This is most likely due to the differences in domain structure, and the tertiary structure of the multi-domain protein. It is also possible that a domain partially unfolds, i.e., some secondary structures (α-helixes and β-sheets) are being stretched out, but others remain folded together.
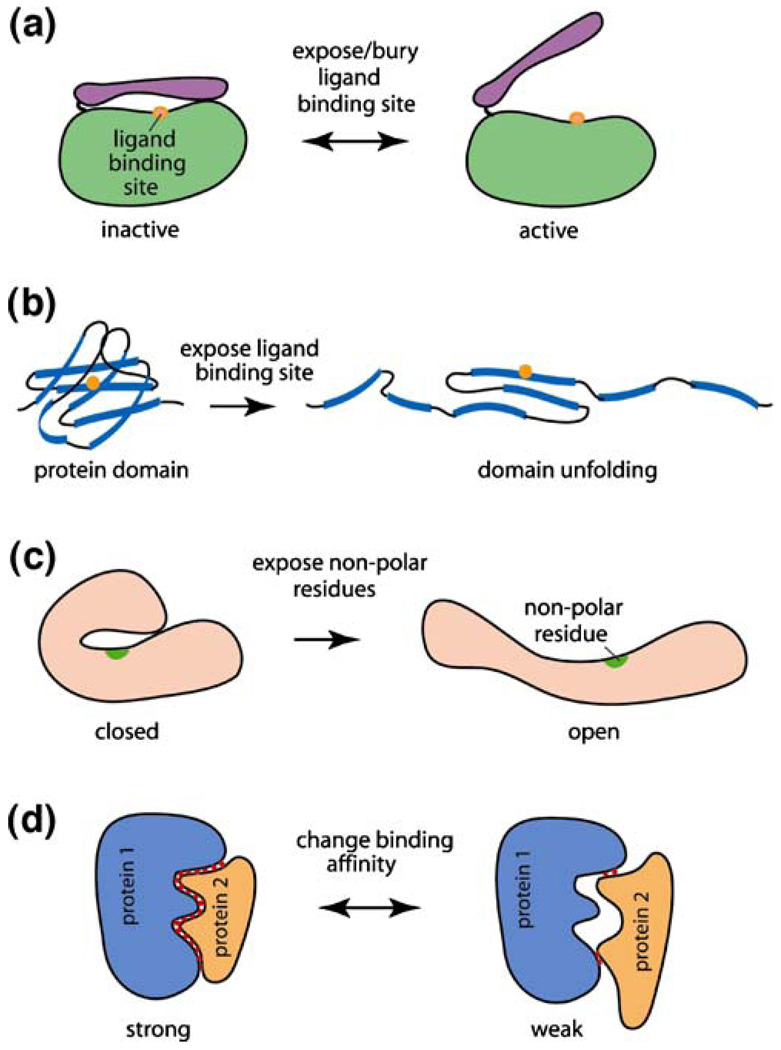
There are two important issues concerning protein deformation, or protein conformational change under mechanical force. The first is the biochemical and biological consequences of such deformation; the second is how protein deformation is related to protein structures (discussed in the next section). As with protein conformational change due to other chemical or physical events, protein conformational change under mechanical force could result in a wide range of biological consequences. Shown in Fig. 5 are some examples of the effect of protein deformation in a living cell. Many proteins have specific ligand binding sites buried initially by a protein domain or a peptide (a ‘lid’). As illustrated in Fig. 5(a), upon applying mechanical forces to such a protein, the ‘lid’ opens, exposing the ligand binding site. The reverse is also true: protein deformation can close the ‘lid’ that is initially open, thereby bury the ligand binding site. Alternatively, a protein globular domain could unfold under mechanical force, exposing the ligand binding site that is buried inside the globular domain [Fig. 5(b)]. A specific example is the uncoiling of the glycoprotein von Willebrand Factor (vWF) under blood-flow induced shear stress. It has been suggested that the unfolding of the A1 domain of vWF is responsible for the increased interaction between circulating vWF and the glycoprotein GP1bα on platelets, causing platelets to adhere to the vessel wall [36]. This has significant implications to platelet adhesion, thrombus formation and coagulation.
Fig. 5.
Examples of biological consequences of protein deformation. Mechanical forces can (a) switch a ‘lid’ in a protein from ‘close’ to ‘open’ position, or (b) unfold a protein domain, thus exposing the ligand binding site. Protein deformation could also (c) expose the nonpolar residues, causing non-specific interaction between the protein domain and other biomolecules; or (d) induce a change in binding affinity, altering protein-protein interactions
Of the 20 amino acids that form a protein, ten (including cysteine, glycine and alanine) are non-polar, hydrophobic residues (side chains); these residues are usually buried inside a globular or filamentous domain of a protein to avoid direct contact with water [1, 23]. Mechanical forces, however, could unfold the domain and thus expose the nonpolar residues [Fig. 5(c)] which may cause non-specific interaction between the protein domain and other biomolecules, and thus alter protein function. It has been demonstrated recently that, when red blood cells were subjected to shear stress, the sterically shielded cysteine residues of the cytoskeletal protein spectrin could be labeled with fluorophores, suggesting that specific domains of spectrin have undergone forced unfolding, exposing the otherwise buried cysteine residues [37]. Other non-polar residues could be exposed in a similar fashion. It is also possible that domain motion or unfolding of protein domains could expose phosphorylation sites that are initially buried.
It is well known that proteins interact with each other based on conformational matches: good conformational match leads to high binding specificity and affinity between two proteins, while poor conformational match does the reverse [1]. As shown schematically in Fig. 5(d), when proteins 1 and 2 have good conformational match, they have strong interactions to realize their functions, for example, to activate a signaling cascade, or facilitate an enzymatic activity. However, when one of the proteins, say, protein 2, sustains a force-induced conformational change, the interaction between proteins 1 and 2 becomes weak due to the poor conformational match, thus altering the function of protein 2. The reverse is also true: deformation of a protein could increase its affinity to another protein that other wise would not interact due to the poor conformational match in its native state. This concept is not limited to protein–protein interactions; protein-DNA, protein–RNA and protein–small molecule interactions can be altered by force-induced protein conformational change as well.
Force-induced protein conformation, or protein deformation, could have many effects on the biology of a cell. In certain cases, the biological consequence of protein deformation is similar to that of protein conformational change due to biochemical events such as allosteric regulation. In some other cases, the biological consequence of protein deformation may be similar to that of protein misfolding. In fact, protein deformation and protein misfolding share many common features including: (a) no change in amino acid sequence; (b) resulting in two or more stable conformations; (c) altering protein binding/aggregation; and (d) possibly inducing toxic activity or loss of normal biological function [38]. More molecular-level experimental evidence is needed to confirm that protein deformation in a living cell indeed causes gain or loss of protein function.
A distinctive feature of protein deformation in living cells is the spatial and temporal regulation of forces that alter protein conformation. When a cell is subjected to mechanical forces (or displacements), the intracellular distribution of the forces varies with location and time, depending on not only the applied forces but also the intracellular structure (such as cell cytoskeleton) and its dynamics, as well as cell–ECM and cell–cell interactions. As such, proteins that are well connected to the force-bearing structures of a cell may be easier to deform, whereas proteins that are not ‘wired’ to the force-bearing structures may rely on viscous forces in order to deform. Such viscous forces may be due to the deformation of cells that induces shear flow of the highly viscous cytosol and nucleoplasm, transmitting shear stress to proteins. Of course the deformability of a protein is critically dependent on its structure, as will be discussed later. It is possible that cells utilize force-induced domain deformation and unfolding in proteins at specific locations and in a time-dependent manner to transduce a mechanical signal into a biochemical reaction. This adds to the richness (and complexity) of the spatial and temporal regulation of mechanotransduction in a cell.
Protein Mechanics
It is clear that protein deformation may play important roles in biology and medicine in general, and mechanotransduction in particular. However, the understanding of the mechanics of proteins is still in its infancy, and many important questions remain open. For example, how a protein deforms under different applied forces or displacements? How the deformation of a protein is dependent on its 3D structure? How protein deformation affects protein function? What is the dynamic behavior of proteins? Even at the very fundamental level, protein mechanics is different from the classical mechanics of deformable bodies in terms of the following aspects: (a) thermal energy (∼1 kBT) and entropic elasticity may become dominant; (b) local quantities such as stress and strain (or stretch ratio) may not be the best parameters to characterize the mechanical behavior of a protein; (c) mechanics and chemistry of a protein are intrinsically coupled. Unlike a typical engineering material of which the mechanical behavior does not depend on the chemistry of its surrounding media; for a protein, it is critical to include the aqueous solution (and its ionic strength) in the modeling and experimental studies of conformational change under force. Similar to all polymeric materials, protein molecules are viscous in nature, and their deformations are rate and temperature dependent.
Protein Constitutive Behavior
To describe how a material deforms under mechanical forces, one typically develops a constitutive model that links deformation to the applied forces. For example, for a linear elastic material that is homogeneous and isotropic, the one-dimensional relationship between stress σ and strain ɛ is given by σ=Eɛ where E is elastic modulus. For three-dimensional deformations, more complicated constitutive models are required.
To describe the mechanical behavior of a protein, that is, how a protein deforms under mechanical force, a constitutive model is also required. Perhaps the simplest model so far is the worm-like chain (WLC) model [39] for a protein under uniaxial stretching [30]:
| (1) |
where f is the applied force, ξ the flexural persistence length, kB is Boltzmann’s constant, T is temperature, x the extension, L the ‘contour’ length of the protein (length of polypeptide under stretching). The WLC model seems to work well for domain deformation of protein molecules containing multiple, individually folded domains with β-sandwich structures [40]. Although new experimental data are needed in order to develop constitutive models for a wide class of proteins, it is possible that, for a class of globular proteins with single or multiple domains, their domain deformation and unfolding under uniaxial stretching can be described by:
| (2) |
where ϕ is a non-dimensional function, α1, α2, …αn are parameters that may reflect the specific structure of the protein, rate effect, and the chemical environment such as ionic strength. Both the form of the function ϕ and its dependence on the parameters αi can be determined by single-molecule experiments of stretching globular proteins.
As a specific example of the parameters αi in equation (2), consider the loading rate ḟ, or the rate by which a force f is applied to a protein. It has been revealed that the strengths of receptor–ligand bonds (such as biotin–streptavidin bond) are critically dependent on loading rate [41]. Therefore, it is expected that the deformation of a protein under uniaxial stretching depends on loading rate. To determine the specific : dependence of ϕ on ḟ,it will be necessary to perform single-molecule stretching experiments of a protein with : different loading rate ḟ, and quantify the corresponding protein deformation and unfolding. Alternatively, in order to determine the functional form between the applied force and deformation of the protein, protein stretching experiments could be formed by applying different stretching rate λ̇ and measuring the corresponding force f.
There is a lack of understanding of how proteins deform in living cells under physiological conditions. Given that most cells generate contractile forces during locomotion, division, and adhesion, it is reasonable to believe that certain proteins deform under stretching or compression in living cells, assume the native conformation upon unloading (relaxation), and such stretching and relaxation could be one- or multi-dimensional. However, protein deformation under compression is most likely very different from that under stretching, since the globular domains are much more resistant to compression than to tension. It is also possible that certain proteins in a living cell sustain shear or torsional deform due to, for example, forces generated by motors and other molecular machines; however currently there is a lack of experimental data at single-molecule level to provide a basis for constitutive modeling for proteins undergoing shear or torsional deformation.
Protein Domain Hinge Motion
Many protein molecules undergo hinge motion in cells, which has many biological implications [42] (see also http://bioinfo.mbb.yale.edu/MolMovDB/). In these proteins, one or more domains rotate about a hinge due to chemical reaction (such as ATP hydrolysis), Brownian force, applied force (torque), or molecular interactions (such as ligand binding). While some motor molecules rely on hinge motion to convert chemical energy into mechanical force, other proteins may utilize hinge motion to transduce signals, regulate interactions, and facilitate enzymatic activities. As an example, the structure of E. coli periplasmic dipeptide binding protein is displayed in Fig. 2(a). It has two domains linked by a hinge consisting of two β-sheets; its hinge motion has been well documented (http://bioinfo.mbb.yale.edu/MolMovDB/).
Some insight can be gained by performing a Langevin dynamics analysis of protein hinge motion in which proteins having two domains linked by a hinge is considered [Fig. 3(b)]. The angle of rotation θ of a domain about the center of the hinge is assumed to obey the stochastic differential equation [26]:
| (3a) |
| (3b) |
where I0 is the moment of inertial of the domain, ηr is the rotational viscous drag coefficient, k the elastic constant of the hinge, F(t) the applied torque, and the kernel in the integral in equation (3b) represents the time rate of torque due to rotational viscous drag, with α(t) being a dissipation function. The Brownian torque R(t) in equations (3a and 3b) is a random function of time and represents the collective effect of the random collisions of the surrounding molecules in the aqueous solution with the protein. The ensemble average of R(t) is zero.
An underlying assumption in equation (3a), as in most Langevin dynamics analyses, is that the values of the Brownian torque R(t) at different times are completely uncorrelated, i.e., R(t) is a ‘white noise’ random process with the autocorrelation function:
| (4a) |
where the noise strength 2kBTηr comes from the equipartition theory of classical thermodynamics [43, 44]. As an alternative, colored noise may be considered in conjunction with equation (3b), and the autocorrelation function of R(t) may obey an exponential-decay function with time:
| (4b) |
where 1/β is the correlation time of the random process, which is an intrinsic property of the aqueous solution. Although equations (3a and 4a) are simpler, when the aqueous solution surrounding the protein is very viscous, such as inside a living cell where viscosity of cytoplasm can be three orders of magnitude higher compared with water, equations (3b and 4b) should be used according to the fluctuation–dissipation theory [45].
Domain hinge motion has a direct relevance to mechanochemical coupling and mechanotransduction. For example, in pumping protons across an insulating membrane or manufacturing ATP from ADP and phosphate, ATP synthase replies on hinge motion of the β domains of the F1-ATPase to generate mechanical torque from the free energy of ATP binding, or produce ATP using torques generated by the protonmotive force [46]. Protein domain hinge motion can also affect receptor–ligand binding. As mentioned before, many proteins have the ligand binding site located near the hinge between two globular domains. Therefore, depending on the magnitude and rate of domain hinge motion, receptor–ligand binding may be altered or even blocked. Specifically, if the rate of hinge motion is much faster than the time required for certain ligands to diffuse into the binding pocket, it is likely that binding of these ligands is prohibited. This may serve as a ‘selection’ mechanism for specific receptor–ligand binding.
Receptor–Ligand Binding
Perhaps one of the most important aspects of protein deformation under force is its effect on receptor–ligand binding, an essential process in cells [47], because specific molecular recognition and interactions rely on conformational matches and charge complementarity between the receptor and the ligand. In general, protein–protein and protein–DNA interactions are determined by many factors, including electrostatic double-layer force, van der Waals force, and ‘steric’ repulsion forces, hydrogen-bonding, and hydrophobic contacts. Most of these factors operate within short ranges [48]. Therefore, the three-dimensional geometry local to the binding pocket of the receptor and ligand contributes significantly to the characteristics of their binding.
When mechanical forces are applied to a receptor (and ligand), the receptor may deform, thereby altering the conformational match between the receptor and ligand. The specific effect of mechanical forces on receptor–ligand binding may depend on the mode and the magnitude of the deformation of a receptor or a ligand (or both), which are in turn determined by the structure of the molecules involved, the solvent surrounding, and loading history (including rate). It is possible that deformation only alters the kinetic rates [41, 49]. However, in certain cases, only protein deformation can expose the binding site, thus switching between the ‘on’ and ‘off’ states of the receptor. It is also possible that protein deformation can change the specificity of a receptor/ligand pair, i.e. binding to ligand B instead of ligand A upon deformation. The latter two effects are both well known for molecules such as integrins that can be activated biochemically [50]. It is likely that force-induced protein deformation may yield similar results.
The Mechanics of Molecular Motors
Motor molecules, a special class of proteins, play an essential role in many cellular processes, including muscle contraction, cell movement, cell division, vesicle transport, signal transduction, and DNA replication, condensation and transcription [1]. Specific motor molecules include the kinesin and dynein superfamily [51], the myosin superfamily [52], ATP synthase, DNA polymerase and RNA polymerase. These motor molecules directly convert chemical energy into mechanical work via conformational changes induced by ATP hydrolysis. As more detailed 3D structures of motor molecules (e.g. [53]) and images of their movements (e.g. [54]) become available, the structure-function relationship of molecular motors has begun to emerge. It is highly likely that molecular motors utilize protein conformational changes to store energy, generate motion, and control/regulate motor function [55]. However, for most of the motor proteins, their mechanics, i.e. how they move, how their conformational changes are related to ATP hydrolysis, how force-generation is related to their structural rigidity, is still largely unknown.
Kinesin is a linear motor with two heads which transports vesicles along microtubules over a distance of several micrometers. A force on the order of 5–7 pN is needed to block the motion of kinesin [56]. Myosin is also a double-headed linear motor which produces sliding between actin and myosin thick filament in muscle. ATP synthase is a special enzyme that can either pump protons across a membrane using ATP hydrolysis, or manufacture ATP from ADP and phosphate using the energy derived from a transmembrane protonmotive gradient. These biomotors directly convert chemical energy derived from ATP hydrolysis into mechanical force or motion, with extremely high efficiency.
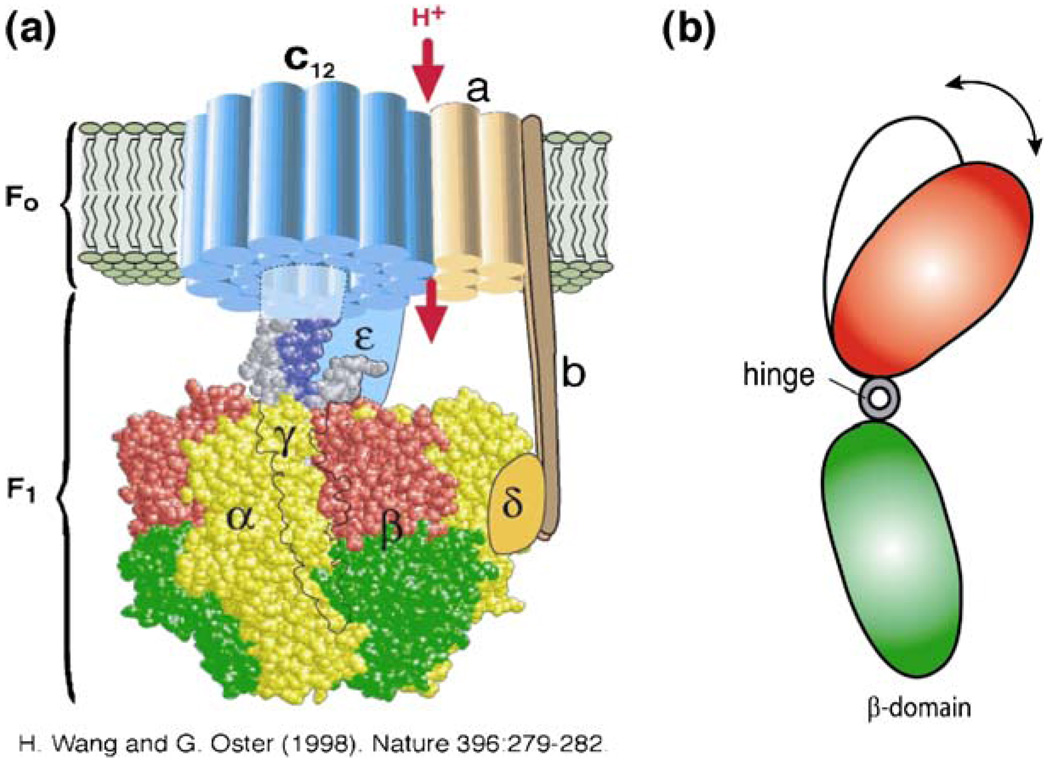
Figure 6 shows an ATP synthase consisting of a transmembrane component F0, a proton channel and a soluble component, F1-ATPase. F1-ATPase has catalytic sites for ATP hydrolysis and synthesis, which is coupled with the proton flow in the F0 domain through a shaft consisting of γ-, δ- and ɛ-subunits [46]. The γ-subunit is surrounded by three α- and three β-subunits, arranged alternately [Fig. 6(a)]. Sequential ATP hydrolysis on the three β-subunits induces rotation of the γ-subunit through hinge motion. When ATP is supplied, the F1-ATPase rotates 120° step-wise, with one ATP molecule hydrolyzed per step [57]; it can rotate at ∼130 rps with saturating ATP concentration without load. The torque generated by the F1-ATPase motor is ∼20–40 pN nm [58, 59].
Fig. 6.
Molecular motor ATP synthase. (a) The structure of ATP synthase (from ref. [59]). It has a transmembrane portion, F0, and a soluble component, F1, which contains catalytic sites located at the αβ interfaces. (b) Hydrolysis of ATP induces hinge motion of the β domain, which in turn drives the rotation of F1-ATPase
As perhaps the world’s smallest rotary engine, ATP synthase carries out both its synthetic and hydrolytic cycles through conformational changes of its β-domains in the F1 component. For example, it has been suggested that in pumping protons, the rotation of the γ-subunit in the F1 component is due to the hinge motion of the β-domain driven by ATP hydrolysis [Fig. 6(b)]. In analyzing energy transduction in ATP synthase, it is critical to understand how the protein enzyme converts the free energy of nucleotide binding into elastic energy. Here, protein deformation serves as the key mechanism for mechanochemical coupling, generating mechanical torque from the free energy of ATP binding, or producing ATP using torques generated by the protonmotive force ([60, 61]).
The Structural Basis for Protein Deformation
From mechanics point of view, deformation of a material is critically dependent on its structure, be it atomic or macroscopic. For protein deformation, this dependence could be even more profound: through evolution, the structure of a specific protein that has mechanical functions (load sensing, bearing and generating) may have been optimized, and the resulting structural features may provide insight for better design of engineering materials. In fact, most of the fibrous proteins that have ‘rod’ or ‘wire’-like tertiary structures, including keratins, collagens and elastins for constructing connective tissues, tendons, bone matrix and muscle fiber, are optimized for load bearing functions. However, it remains an important task to establish the structural features of globular proteins that have force-sensing and mechanotransduction functions.
Proteins can be classified into different classes according to their structural features. For example, in the Structural Classification of Proteins (SCOP) database developed by Alexey G. Murzin and his associates, proteins are divided into seven major classes based on general “structural architecture” of the protein domains (see: http://scop.mrclmb.cam.ac.uk/scop/): (a) All alpha proteins; (b) All beta proteins; (c) Alpha and beta proteins (a/b); (d) Alpha and beta proteins (a+b); (e) Multi-domain proteins (alpha and beta); (f) Membrane and cell surface proteins and peptides; (g) Small proteins. Each class of proteins is further grouped into family (with some sequence similarity), superfamily (with sufficient structural and functional similarity) and fold (with similar arrangement of regular secondary structures). This database, SCOP, provides a broad survey of all known protein folds, detailed information about the close relatives of any particular protein, and a framework for future research and classification.
Table 1 shows the classes and structural features of some key proteins that may have force sensing, force bearing and mechanotransduction functions. Interestingly, most of the proteins listed in Table 1 are concentrated in two classes: von Willebrand factor A1 and A3 domains and many integrins and integrin domains (Integrin molecules α1β1, α2β1, αxβ2, CD11a/CD18; Integrin αM and βA domains) belong to the class of ‘alpha and beta proteins (a/b)’, with vWA-like domain(s) consisting of mainly parallel beta sheets (beta-alpha-beta units). On the other hand, adhesion molecules VCAM-1, ICAM-1, ICAM-2, ECM molecules Fibronectin type III domain and Tenascin, muscle molecule Type I titin and Integrin β2 subunit belong to the class of ‘all beta proteins’, with immunoglobulin-like beta-sandwich domain consisting of a sandwich of seven β-strands in two sheets. The most striking feature revealed by Table 1 is that many of the known mechanically-stressful globular proteins such as integrins, adhesion molecules, ECM molecules, vWF and titin have domains that are rich in β-sheets. This might be due to the fact that these domains are quite compact thus resisting compression and shear; at the same time, it could be deformed and open under tension, thus exposing ligand binding sites. Although still very preliminary, the structural features shown in Table 1 provide some important insight into the mechanics of proteins.
Table 1.
Structural features of key force sensing or bearing proteins
| Protein | Class | Superfamily | Fold |
|---|---|---|---|
| Fibronectin type III domain, tenascin, type I titin, integrin beta-4 subunit |
All beta proteins | Fibronectin type III | Immunoglobulin-like beta-sandwich: sandwich; seven strands in two sheets |
| VCAM-1, ICAM-1, ICAM-2 | All beta proteins | Immunoglobulin | Immunoglobulin-like beta-sandwich: sandwich; seven strands in two sheets |
| von Willebrand factor A1 domain, von Willebrand factor A3 domain |
Alpha and beta proteins (a/b) | vWA-like | vWA-like: mainly parallel beta sheets (beta-alpha-beta units) |
| Integrin alpha1-beta1, integrin alpha2- beta1, integrin alpha-x beta2, integrin beta A domain, integrin alpha M, integrin CD11a/CD18 |
|||
| Integrin beta-3 | Small proteins | Plexin repeat | Trefoil/plexin domain-like: disulfide- rich fold; common core is alpha+beta with two conserved disulfides |
To establish a deeper understanding of the relationship between protein structure and protein deformation, a more systematic structural analysis of proteins that have mechanical load sensing, bearing and generation functions is needed. This includes the analysis of protein structural domains based on similarities of their amino acid sequences and three-dimensional structures using, for example, SCOP. It is also necessary to perform single-molecule experiments [40] and detailed structural analysis using computational tools such as steered molecular dynamics in order to understand why load sensing and bearing proteins have some common structural features, as revealed by Table 1. It is anticipated that, by systematically simulating protein conformational dynamics under force with sufficient details, the structural basis for protein deformation will emerge.
Modeling and Experimental Challenges in Protein Mechanics
The development of protein mechanics is the key to definitively establishing a molecular mechanism for mechanotransduction in living cells. This requires a quantitative description of the constitutive behavior of proteins, including how proteins deform under different mechanical loads; what are the modes and time scales of protein motions and deformations, and how such deformations are related to protein structural features. It also requires a better understanding of how protein deformation affects molecular interactions in cells, including protein–protein, protein–RNA and protein–DNA interactions.
Despite the significant progress over the last decade or so in developing tools for single-molecule biomechanics studies, including AFM, optical tweezers, magnetic tweezers, micropipette force spectroscopy, and surface force apparatus, experimental probing the deformation of single proteins is severely limited by at least three factors. First, it is difficult to image or measure the deformation of protein molecules by force due to the small (0.1–1 nm) geometric changes. Therefore, advances in protein mechanics require further progress in bioimaging technology. Second, it is difficult to hold, position and manipulate single protein molecules by conventional means. As a result, attaching a protein to devices that impose force or deformation in a controlled manner without affecting its native conformation is a major experimental challenge. Finally, the characteristic time for motion and relaxation of proteins and nucleic acids, which spans the wide range of nanoseconds to seconds, may render experimental studies of the underlying deformation mechanisms a rather difficult task. Since fluorescence resonance energy transfer (FRET) between donor and acceptor fluorophores is extremely sensitive to their relative distance (<10 nm), it is possible that the conformational dynamics of proteins under force can be studied using FRET. In fact, FRET has been used for imaging fibronectin extension and unfolding in cell culture [62, 63].
Theoretical modeling and numerical simulation of the deformation and dynamics of proteins is a very important area, and the steered molecular dynamics simulations of fibronectin and integrin have provided much insight into how globular proteins unfold under tensile forces [34, 35]. However, currently MD simulations can be performed only for a short time period, up to one hundred nanoseconds, while most biological processes in cells occur on a time scale of one millisecond. Consequently, the results of MD simulations are often qualitative rather than quantitative. Further, the force fields used in MD simulations (i.e., the potential functions that characterize the atomic and molecular interactions) are usually obtained based on experimental measurements of the interaction of limited pairs of atoms; they may not be applicable to many biomolecular interactions. Therefore, it is necessary to seek a new numerical approach to simplify the simulations while still preserve the basic features of the molecular interactions, to increase our computing power, and to develop more accurate force fields.
Conclusion
There are two major issues in cell and molecular biomechanics. The first is how mechanical forces are being ‘sensed’ by cells, and the second is how such a mechanical signal is being transduced into biochemical and biological responses. As a candidate for the molecular mechanism of mechanotransduction, protein deformation, or protein conformational change under mechanical force, may provide an answer for both. Cells may ‘sense’ mechanical forces by deforming specific proteins at specific locations; such deformation alters (induces or prohibits) molecular interactions such as receptor–ligand binding, causing changes in the signaling pathways and other cellular processes. For example, the receptor-mediated signaling processes in cells can be regulated by deformation of ECM molecules such as fibronectin, resulting in the exposure of an RGD (arginine–glycine–aspartate) sequence for binding to integrin, whose functions include both adhesion and signal transduction. In addition to the ability of switching between resting and active states of proteins upon ligand occupancy, it is conceivable that cells utilize more continuous conformational changes in response to various forces to produce more gradual exposure of the functional domain or affect the binding rate for the downstream signaling molecules, thereby transducing mechanical forces and deformations into biochemical responses. Therefore, it is essential to study protein mechanics in order to understand mechanotransduction, mechanochemical coupling and the mechanical functions of proteins in living cells. The ultimate goal is to understand the roles of protein mechanics in disease processes, and utilize this understanding in disease detection, treatment and prevention.
Acknowledgements
This work was supported by the National Heart Lung and Blood Institute of the NIH as a Program of Excellence in Nanotechnology (HL80711) and the NIH Roadmap Initiative in Nanomedicine through a Nanomedicine Development Center award (EY018244). The author would like to thank Dr. Wonjong Rhee for generating the results shown in Fig. 1.
References
- 1.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Molecular biology of the cell. 4th edn. New York, NY: Garland; 2002. [Google Scholar]
- 2.Wang N, Butler JP, Ingber DE. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 1993;260:1124–1127. doi: 10.1126/science.7684161. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- 3.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 4.Howard J. Mechanics of motor proteins and the cytoskeleton. Sunderland, MA: Sinauer; 2001. [Google Scholar]
- 5.Block SM, Goldstein LS, et al. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 1990;348:348–352. doi: 10.1038/348348a0. doi: 10.1038/348348a0. [DOI] [PubMed] [Google Scholar]
- 6.Stossel TP. On the crawling of animal cells. Science. 1993;260:1086–1094. doi: 10.1126/science.8493552. doi: 10.1126/science.8493552. [DOI] [PubMed] [Google Scholar]
- 7.Pelham RJ, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Discher DE, Janmey P, Wang Y. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 9.Brownell WE, Spector AA, Raphael RM, Popel AS. Micro-and nanomechanics of the cochlear outer hair cell. Annu Rev Biomed Eng. 2001;3:169–194. doi: 10.1146/annurev.bioeng.3.1.169. doi: 10.1146/annurev.bioeng.3.1.169. [DOI] [PubMed] [Google Scholar]
- 10.Bao G. Mechanics of biomolecules. J Mech Phys Solids. 2002;50:2237–2274. doi: 10.1016/S0022-5096(02)00035-2. [Google Scholar]
- 11.Zhu C, Bao G, Wang N. Cell mechanics: mechanical response, cell adhesion, and molecular deformation. Annu Rev Biomed Eng. 2000;2:189–226. doi: 10.1146/annurev.bioeng.2.1.189. doi: 10.1146/annurev.bioeng.2.1.189. [DOI] [PubMed] [Google Scholar]
- 12.Bershadsky AD, Balaban NQ, Geiger B. Adhesion-dependent cell mechanosensitivity. Annu Rev Cell Dev Biol. 2003;19:677–695. doi: 10.1146/annurev.cellbio.19.111301.153011. doi: 10.1146/annurev.cellbio.19.111301.153011. [DOI] [PubMed] [Google Scholar]
- 13.Bershadsky A, Kozlov M, Geiger B. Adhesion-mediated mechanosensitivity: a time to experiment, and a time to theorize. Curr Opin Cell Biol. 2006;18:472–481. doi: 10.1016/j.ceb.2006.08.012. doi: 10.1016/j.ceb.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Jalali S, del Pozo MA, Chen K, Miao H, Li Y, Schwartz MA, Shyy JY, Chien S. Integrin-mediated mechanotransduction requires its dynamic interaction with specific extracellular matrix (ECM) ligands. Proc Natl Acad Sci USA. 2001;98:1042–1046. doi: 10.1073/pnas.031562998. doi: 10.1073/pnas.031562998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fung YC. Biomechanics: motion, flow, stress, and growth. New York: Springer; 1990. [Google Scholar]
- 16.Fung YC. Biomechanics: mechanical properties of living tissues. 2nd edn. New York: Springer; 1993. [Google Scholar]
- 17.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 18.Ali MH, Schumacker PT. Endothelial responses to mechanical stress: where is the mechanosensor? Crit Care Med. 2002;30:S198–S206. doi: 10.1097/00003246-200205001-00005. doi: 10.1097/00003246-200205001-00005. [DOI] [PubMed] [Google Scholar]
- 19.Wootton DM, Ku DN. Fluid mechanics of vascular systems, diseases, and thrombosis. Annu Rev Biomed Eng. 1999;1:299–329. doi: 10.1146/annurev.bioeng.1.1.299. doi: 10.1146/annurev.bioeng.1.1.299. [DOI] [PubMed] [Google Scholar]
- 20.Fisher AB, Chien S, Barakat AI, Nerem RM. Endothelial cellular response to altered shear stress. Am J Physiol Lung Cell Mol Physiol. 2001;281:L529–L533. doi: 10.1152/ajplung.2001.281.3.L529. [DOI] [PubMed] [Google Scholar]
- 21.Hu S, Sachs F. Stretch-activated ion channels in the heart. J Mol Cell Cardiol. 1997;29:1511–1523. doi: 10.1006/jmcc.1997.0392. doi: 10.1006/jmcc.1997.0392. [DOI] [PubMed] [Google Scholar]
- 22.Ingber DE. Mechanical signaling and the cellular response to extracellular matrix in angiogenesis and cardiovascular physiology. Circ Res. 2002;91:877–887. doi: 10.1161/01.res.0000039537.73816.e5. doi: 10.1161/01.RES.0000039537.73816.E5. [DOI] [PubMed] [Google Scholar]
- 23.Creighton TE. Proteins. New York, NY: Freeman; 1993. [Google Scholar]
- 24.Bao G, Suresh S. Cell and molecular mechanics of biological materials. Nat Matters. 2003;2:715–726. doi: 10.1038/nmat1001. doi: 10.1038/nmat1001. [DOI] [PubMed] [Google Scholar]
- 25.Voet D, Voet JG. Biochemistry. 2nd edn. New York, NY: Wiley; 1995. [Google Scholar]
- 26.McCammon JA, Gelin BR, Karplus M, Wolynes PG. The hinge-bending mode in lysozyme. Nature. 1976;262:325–326. doi: 10.1038/262325a0. doi: 10.1038/262325a0. [DOI] [PubMed] [Google Scholar]
- 27.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub H. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 28.Kellermayer MSZ, Smith SB, Granzier HL, Bustamante C. Folding–unfolding transitions in single titin molecules characterized with laser tweezers. Science. 1997;276:1112–1116. doi: 10.1126/science.276.5315.1112. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 29.Tskhovrebova L, Trinnick J, Sleep JA, Simmons RM. Elasticity and unfolding of single molecules of the giant muscle protein titin. Nature. 1997;387:308–312. doi: 10.1038/387308a0. doi: 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]
- 30.Oberhauser AF, Marszalek PE, Erickson HP, Fernandez JM. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 1998;393:181–185. doi: 10.1038/30270. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 31.Krammer A, Lu H, Isralewitz B, Schulten K, Vogel V. Forced unfolding of the fibronectin type III module reveals a tensile molecular recognition switch. Proc Natl Acad Sci USA. 1999;96:1351–1356. doi: 10.1073/pnas.96.4.1351. doi: 10.1073/pnas.96.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohashi T, Kiehart DP, Ericson H. Dynamics and elasticity of the fibronectin matrix in living cell culture visualized by fibronectin-green fluorescent protein. Proc Natl Acad Sci USA. 1999;96:2153–2158. doi: 10.1073/pnas.96.5.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Craig D, Krammer A, Schulten K, Vogel V. Comparison of the early stages of forced unfolding for fibronectin type III modules. Proc Natl Acad Sci USA. 2001;98:5590–5595. doi: 10.1073/pnas.101582198. doi: 10.1073/pnas.101582198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogel V, Thomas WE, Craig DW, Krammer A, Baneyx G. Structural insights into the mechanical regulation of molecular recognition sites. Trends Biotechnol. 2001;19:416–423. doi: 10.1016/S0167-7799(01)01737-1. doi: 10.1016/S0167-7799(01)01737-1. [DOI] [PubMed] [Google Scholar]
- 35.Puklin-Faucher E, Gao M, Schulten K, Vogel V. How the headpiece hinge angle is opened: new insights into the dynamics of integrin activation. J Cell Biol. 2006;175:349–360. doi: 10.1083/jcb.200602071. doi: 10.1083/jcb.200602071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruggeri ZM. von Willebrand factor. Curr Opin Hematol. 2003;10:142–149. doi: 10.1097/00062752-200303000-00008. doi: 10.1097/00062752-200303000-00008. [DOI] [PubMed] [Google Scholar]
- 37.Johnson CP, Tang HY, Carag C, Speicher DW, Discher DE. Forced unfolding of proteins within cells. Science. 2007;317:663–666. doi: 10.1126/science.1139857. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soto C. Protein misfolding and disease; protein refolding and therapy. FEBS Lett. 2001;498:204–207. doi: 10.1016/s0014-5793(01)02486-3. doi: 10.1016/S0014-5793(01)02486-3. [DOI] [PubMed] [Google Scholar]
- 39.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759–8770. doi: 10.1021/ma00130a008. [Google Scholar]
- 40.Schwaiger I, Schleicher M, Noegel AA, Rief M. The folding pathway of a fast-folding immunoglobulin domain revealed by single-molecule mechanical experiments. EMBO Rep. 2005;6:46–51. doi: 10.1038/sj.embor.7400317. doi: 10.1038/sj.embor.7400317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Energy landscapes of receptor–ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397:50–53. doi: 10.1038/16219. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- 42.Subbiah S. Protein motions. Austin, TX: Chapman & Hall; 1996. [Google Scholar]
- 43.van Kampen NG. Stochastic processes in physics and chemistry. New York, NY: North-Holland; 1981. [Google Scholar]
- 44.Risken H. The fokker-planck equation: methods of solution and applications. 2nd edn. New York, NY: Springer; 1984. [Google Scholar]
- 45.Uhlenbeck GE, Ornstein LS. On the theory of the Brownian motion. Phys Rev. 1930;36:823–841. doi: 10.1103/PhysRev.36.823. [Google Scholar]
- 46.Boyer PD. The binding change mechanism for ATP synthase—some probabilities and possibilities. Biochem Biophys Acta. 1993;1140:215–250. doi: 10.1016/0005-2728(93)90063-l. doi: 10.1016/0005-2728(93)90063-L. [DOI] [PubMed] [Google Scholar]
- 47.Lauffenburger DA, Linderman JJ. Receptors. New York: Oxford University Press; 1993. [Google Scholar]
- 48.Israelachvili J. Intermolecular and surface forces. San Diego, CA: Academic; 1992. [Google Scholar]
- 49.Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanchez-Mateos P, Cabanas C, Sanchez-Madrid F. Regulation of integrin function. Semin Cancer Biol. 1996;7:99–109. doi: 10.1006/scbi.1996.0015. doi: 10.1006/scbi.1996.0015. [DOI] [PubMed] [Google Scholar]
- 51.Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 52.Mermall V, Post PL, Mooseker MS. Unconventional myosins in cell movement, membrane traffic, and signal transduction. Science. 1998;279:527–533. doi: 10.1126/science.279.5350.527. doi: 10.1126/science.279.5350.527. [DOI] [PubMed] [Google Scholar]
- 53.Cramer P, Bushnell DA, Fu J, Gnatt AL, Maier-Davis B, Thompson NE, Burgess RR, Edwards AM, David PR, Kornberg RD. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- 54.Walker ML, Burgess SA, Sellers JR, Wang F, Hammer JA, 3rd, Trinick J, Knight PJ. Two-headed binding of a processive myosin to F-actin. Nature. 2000;405:804–807. doi: 10.1038/35015592. doi: 10.1038/35015592. [DOI] [PubMed] [Google Scholar]
- 55.Vale RD, Milligan RA. The way things move: looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 56.Meyhofer E, Howard J. The force generated by a single kinesin molecule against an elastic load. Proc Natl Acad Sci USA. 1995;92:574–578. doi: 10.1073/pnas.92.2.574. doi: 10.1073/pnas.92.2.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yasuda R, Noji H, Yoshida M, Kinosita K, Jr, Itoh H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature. 2001;410:898–904. doi: 10.1038/35073513. doi: 10.1038/35073513. [DOI] [PubMed] [Google Scholar]
- 58.Kinosita K, Jr, Yasuda R, Noji H. F1-ATPase: a highly efficient rotary ATP machine. Essays Biochem. 2000;35:3–18. doi: 10.1042/bse0350003. [DOI] [PubMed] [Google Scholar]
- 59.Soong RK, Bachand GD, Neves HP, Olkhovets AG, Craighead HG, Montemegno CD. Powering an inorganic nanodevice with a biomolecular motor. Science. 2000;290:1555–1558. doi: 10.1126/science.290.5496.1555. doi: 10.1126/science.290.5496.1555. [DOI] [PubMed] [Google Scholar]
- 60.Elston T, Wang H, Oster G. Energy transduction in ATP synthase. Nature. 1998;391:510–513. doi: 10.1038/35185. doi: 10.1038/35185. [DOI] [PubMed] [Google Scholar]
- 61.Wang H, Oster G. Energy transduction in the F1 motor of ATP synthase. Nature. 1998;396:279–282. doi: 10.1038/24409. doi: 10.1038/24409. [DOI] [PubMed] [Google Scholar]
- 62.Baneyx G, Baugh L, Vogel V. Coexisting conformations of fibronectin in cell culture imaged using fluorescence resonance energy transfer. Proc Natl Acad Sci USA. 2001;98:14464–14468. doi: 10.1073/pnas.251422998. doi: 10.1073/pnas.251422998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baneyx G, Baugh L, Vogel V. Fibronectin extension and unfolding within cell matrix fibrils controlled by cytoskeletal tension. Proc Natl Acad Sci USA. 2002;99:5139–5143. doi: 10.1073/pnas.072650799. doi: 10.1073/pnas.072650799. [DOI] [PMC free article] [PubMed] [Google Scholar]