Abstract
The human left and right cerebral hemispheres are anatomically and functionally asymmetric. To test whether human cortical asymmetry has a molecular basis, we studied gene expression levels between the left and right embryonic hemispheres using Serial Analysis of Gene Expression (SAGE), and identified and verified 27 differentially expressed genes, suggesting that human cortical asymmetry is accompanied by early, striking transcriptional asymmetries. LMO4 is consistently more highly expressed in the right perisylvian human cerebral cortex than in the left, and is essential for cortical development in mice, suggesting that human left-right specialization reflects asymmetric cortical development at early stages.
One of the most remarkable aspects of the human cerebral cortex is that the two hemispheres are specialized for distinct cognitive and behavioral functions. Whereas the right cerebral cortex regulates movement of the left side of the body and vice versa, approximately 90% of the human population is naturally more skilled with the right hand than with the left (1). This motor asymmetry is strongly correlated with language dominance: language function is predominantly localized to a distributed network in the left perisylvian cortex in 97% of right-handers and about 60% of left-handers (2, 3). Functional asymmetries exist in mathematical ability, and spatial and facial recognition as well. These functional asymmetries have been related to anatomical asymmetries of the cortex that are somewhat more subtle (2, 4). For example, the posterior end of the Sylvian fissure, is higher in the right hemisphere than in the left (5). The planum temporale, a region in the posterior portion of the superior temporal sulcus in which Wernike’s area resides, is larger in the left than in the right in more than 65% of examined adult and 56-79% of fetuses or infant brains, so that the anatomical asymmetries are less striking than the functional ones (6, 7). Although genetic factors connecting cerebral asymmetry and functional dominance have been supported (8), no molecular correlate of cerebral asymmetry has been identified.
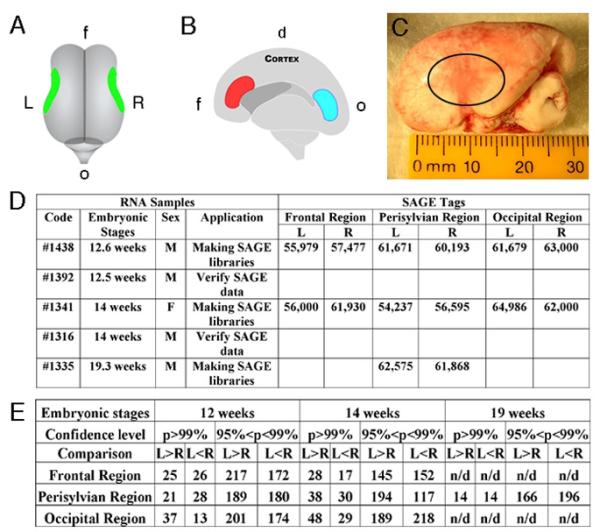
Here we directly tested the hypothesis that left-right cortical asymmetry in humans results from differential gene expression at early embryonic stages, long before the onset of organized cerebral cortical function. By applying Serial Analysis of Gene Expression (SAGE), we measured gene expression levels between the left and right hemispheres in early (12-14 weeks) embryonic human brains, during periods of neuronal proliferation and migration, and later (19 weeks), after these processes are largely completed (9). Brain tissues were first dissected from matching perisylvian regions in two hemispheres (Fig. 1, A to C). The cortex was then separated at the midline. On the medial side of the hemisphere, tissues were also dissected from the ventricular zone in the frontal and occipital regions (Fig. 1B). Total RNA was isolated and 14 SAGE libraries were generated (Fig. 1D). To detect genes with differential expression levels, we compared tag frequency for each gene between two SAGE libraries generated from the frontal, perisylvian and occipital regions in the left-right hemispheres. To verify the statistical significance of differences in each comparison, we performed a Monte Carlo test and verified this using the Chi-square test. Using the chi squared distribution with degree of freedom of 1 and confidence levels (p), we sorted genes within each comparison (e.g. left-right). A higher chi-value indicates a greater statistically significant difference.
Fig. 1.
Dissection of human embryonic brain tissues and generation of human SAGE libraries. (A) A top view of a 14-week human embryonic brain. Tissues were dissected from perisylvian regions in the left (L) and right (R) hemispheres. (B) A side view of a 14-week human embryonic right hemisphere. Tissues in the frontal (f) (red) and occipital (o) (blue) ventricular regions containing dividing cells were dissected. The dorsal cortex (d) is on the top. (C) The left side view of a real 14-week human embryonic brain. The perisylvian region is circled. (D) Summary of human brain SAGE libraries. The male (M) and female (F) brains are listed. At least 55,000 tags were sequenced in each library. (E) Summary of differentially expressed genes detected by SAGE analysis between the left-right hemispheres. Numbers of genes that are highly expressed in the left (L>R) or right (L<R) are listed with confidence levels p>99% and 95%<p<99%.
In all, 49 differentially expressed genes were identified by SAGE with p>99% (chi-value>6.63) between the left-right perisylvian regions of a 12-week embryonic human cortex. Among them 21 genes were highly expressed in the left, while 28 genes were highly expressed in the right (Fig. 1E). Moreover, 68 genes were identified with p>99% between the left-right perisylvian regions of a 14-week cortex (Fig. 1E). By combining analyses, we generated a list of statistically differentially expressed genes between the left-right hemispheres in the perisylvian regions (Fig. 1E, table S1 to S5), frontal and occipital regions (table S9 to S12) of human embryonic brains at 12, 14 and 19 weeks. Differential gene expression levels detected by SAGE suggested an early transcriptional asymmetry between the left-right hemispheres in human embryonic brains.
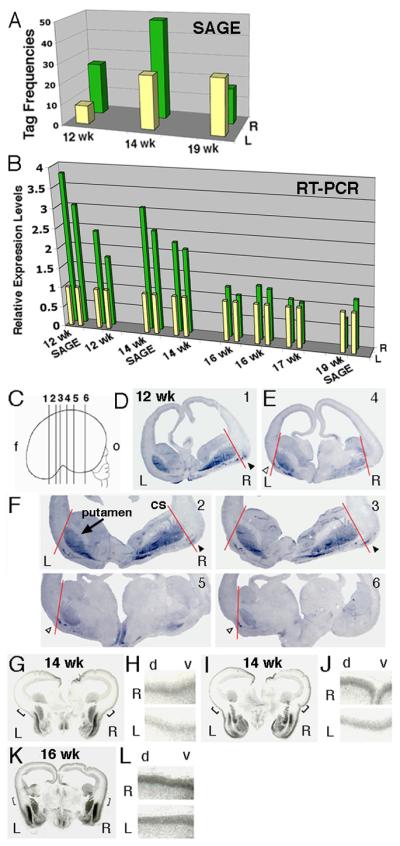
One of the genes reproducibly asymmetrically expressed was the transcription factor Lim Domain Only 4 (LMO4). Using SAGE analysis, we found that the human LMO4 is more highly expressed in the perisylvian regions of the right hemisphere than in the left at both 12 and 14 weeks (Fig. 2A). In contrast, LMO4 expression levels did not show significant differences between the left and right perisylvian regions at 19 weeks (Fig. 2A). We then quantified LMO4 expression levels using real-time SYBR-Green Reverse Transcription (RT)-PCR and confirmed higher LMO4 expression in the right perisylvian regions than the left of embryonic 12 and 14 weeks but not 19 week brains using the same RNA samples for SAGE analysis (Fig. 2B). Moreover, we confirmed higher levels of LMO4 expression in the right perisylvian region versus the left in a second 12-week brain, a second 14-week brain, and observed modest differences in two 16-week brains and one 17-week human embryonic brain (Fig. 2B).
Fig. 2.
Human LMO4 was highly expressed in the right hemisphere as detected by SAGE, real-time RT-PCR and in situ hybridization. (A) The human LMO4 expression levels in the perisylvian regions measured by SAGE (tag frequencies) in 12 (12 wk), 14 (14 wk) and 19 week (19 wk) brains. (B) The LMO4 expression levels between the left (L) and right (R) hemispheres were verified by real-time RT-PCR in eight human embryonic brains (12-19 weeks). Two data points from duplicated experiments for each sample are illustrated. (C-F) LMO4 expression in coronal sections cut from the frontal (f) to occipital (o) lobes of a human embryonic 12-week brain. The medial-lateral extent of LMO4 expression in the cortical plate between the left (white arrowheads) and right (black arrowheads) hemispheres was defined by a red line connecting the corticostriatal sulcus (cs) and the lateral border of the putamen (arrow). Numbers of sections illustrated in (C) are labeled in sections shown. Human LMO4 was more highly expressed in the cortical plates in the right hemispheres than the left in coronal sections of a 14-week brain (G-J) and 16-week brain (K-L). (H, J and L) are high power views of selected areas in panel G, I and K, respectively. The dorsal (d) and ventral (v) cortex is labeled.
We next performed nonradioactive in situ hybridization on human embryonic brains, and noted right-left differences in the extent of LMO4 expression at 12 weeks. We serially sectioned the cortex in the frontal plane and performed in situ hybridization on at least 54 sections from this series, covering most of the frontal to occipital extent of the cerebral cortex (Fig. 2C). Consistent with the early expression of Lmo4 in mice (see below), LMO4 in this 12 week human brain was expressed in the ventral lateral cortical plate in a patchy fashion (Fig. 2, D to F). LMO4 was also expressed highly and symmetrically in non-cortical telencephalic structures, notably the putamen (Fig. 2F). We analyzed the medial-lateral extent of LMO4 expression in the cortical plate in relation to the lateral border of the basal ganglia, defined by connecting the corticostriatal sulcus and the lateral border of the putamen (Fig. 2F). In this brain, LMO4 expression was observed further dorso-lateral in the cortical plate in the right hemisphere than the left, particularly in sections near the future perisylvian region (Fig. 2F).
We then confirmed asymmetric LMO4 expression in several additional human fetal brains, focusing in the perisylvian region, using [35S]-labeled radioactive in situ hybridization. At 14 weeks, LMO4 was highly expressed in the cortical plate around the entire perimeter of the cortex. Although levels of LMO4 expression in the right hemisphere were comparable to the left in broad areas of dorso-medial neocortex, paralleling the fact that anatomical asymmetries of these medial areas are not known, it was consistently more highly expressed over broad areas of the right perisylvian cortex than the left (Fig. 2, G to J). At 16 weeks, asymmetric cortical expression was still observed, although it was diminished relative to earlier timepoints, consistent with the real-time RT-PCR analysis (Fig. 2, B, K and L). Striking asymmetries in LMO4 expression were seen in the perisylvian region of a human 17 week cortex studied by nonradioactive in situ hybridization (fig. S1), even more apparent then the RT-PCR results (Fig. 2B). In a 19 week brain, consistent with RT-PCR results, left-right differences in LMO4 expression were not obvious (fig. S1). Overall, the in situ hybridization analysis mirrored the SAGE and RT-PCR analysis, with clearer but variable asymmetries in expression among individuals at earlier stages and no clear asymmetry at the latest stage examined (19 weeks).
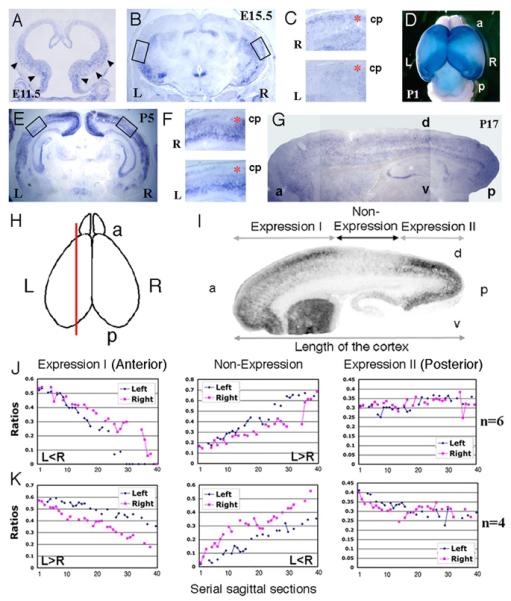
To better understand the dynamic change in Lmo4 expression during cortical development, we analyzed Lmo4 expression in mouse brains. Similar to its expression in the 12-week human embryonic brain, Lmo4 was weakly expressed in the ventral cortex in the embryonic day 11.5 (E11.5) mouse brain and its expression increased during development (Fig. 3A). Lmo4 expression boundaries were fairly sharp at postnatal day 1 (P1) with expression being high in the anterior and posterior portions of the cortex, but with a large zone of non-expression that overlaps the presumptive parietal cortex in between (Fig. 3D). However, this non-expression zone disappeared at P17 (Fig. 3G). In coronal sections of E15.5 and P5 mouse cortices, Lmo4 was expressed in the medial and lateral cortical areas as well (Fig. 3, B and E). Interestingly, Lmo4 in the mouse showed apparent asymmetries in the cortical area in which it was highly expressed and the expression pattern was quite dynamic (Fig. 3, A to G).
Fig. 3.
The dynamic and asymmetric expression of Lmo4 in mouse brains. The patchy and asymmetric expression patterns of Lmo4 in the coronal sections in the left (L) and right (R) hemispheres of E11.5 (A), E15.5 (B) and P5 (E) mouse brains. Its expression appears uniform in the cortical plate in a sagittal section of a P17 brain (G). (C) and (F) High power views of Lmo4 expression in the cortical plates (cp) (red stars) in selected areas in panel (B) and (E), respectively. The dorsal cortex is on the left side. (D) The dorsal view of whole mount in situ hybridization of a P1 mouse brain. (I) Lmo4 expression was divided into the anterior (a) (Expression I), the posterior (p) (Expression II) and the Non-expression region in a sagittal section (H) of a P1 mouse cortex. The dorsal (d) and ventral (v) cortex is labeled. (J) and (K) Asymmetric expression of Lmo4 in the left-right hemispheres in representative P1 mouse cortices (totally 6 brains were similar to that in J and 4 brains to that in K).
Since the levels of coronal sections may affect in situ hybridization signal, we mapped the Lmo4 expression on serial sagittal sections to provide an accurate gene expression pattern (Fig. 3H). We divided the cortical expression of Lmo4 in P1 brain into three regions: anterior (expression I), intermediate (non-expression) and posterior (expression II) (Fig. 3I). The ratios of the length of each Lmo4 expression region versus the full length of the cortex were calculated from the medial to lateral cortical regions and analyzed using histograms (Fig. 3, J and K). The Lmo4 expression areas in the anterior were smaller in the left hemisphere than in the right in 6 tested brains (demonstrated by one representative brain in Fig. 3J) but larger in the left than the right in 4 tested brains (demonstrated by one representative brain in Fig. 3K). Correspondingly, the Lmo4 non-expression areas (intermediate) were larger in the left hemisphere than those in the right in the same 6 tested brains (Fig. 3J) but smaller in the same 4 tested brains (Fig. 3K). However, we did not detect asymmetric Lmo4 expression in the posterior cortex between the left-right hemispheres in any of the tested brains (Fig. 3, J and K). Thus, although Lmo4 expression in mouse cortex was moderately asymmetrical in every individual brain tested so far, it was not consistently lateralized to the right or left side. This may relate to behavioral and anatomical studies in mice, in which sensory-motor asymmetries, like paw preference, are observed in individual mice, but are not biased on a population level to either the right or left hemisphere as is hand preference in humans (10-14). The differences in mice and humans suggest the intriguing possibility that paw preference in rodents might reflect an early, perhaps stochastic, developmental asymmetry that is established perinatally, prior to paw usage, implying a transcriptional asymmetry that is not consistently lateralized to the left and right. Evolution of mechanisms that bias or entrain a modest and random asymmetry in lower organisms may have allowed the development of more consistent functional asymmetries in the human cortex (15).
To identify other differentially expressed genes between the left-right hemispheres, we focused on genes showing different expression levels measured by SAGE in the perisylvian regions of human embryonic 12 weeks brains. Using RNA samples for generating SAGE libraries, we verified 76 genes using real-time RT-PCR (table S6 and S7) and found 39 genes (51%) showing consistent differential expression as measured by SAGE (table S6). To further test the reproducibility of verifying SAGE data, we verified expression levels of these 76 genes using a second 12 week old brain. We found that 27 genes (36%) consistently showed differential expression (either left-high or left-low) in both brain samples (table S6). We also tested 17 genes with lower chi values (<1.9) but which have been implicated in cortical development and found 7 genes showing relatively significant differences of gene expression levels, though they were not detected by SAGE (table S8).
The left-right differences in LMO4 expression in humans could potentially reflect either a differing topographic mapping in the two hemispheres, or a difference in the tempo of cortical development with the right hemisphere’s development leading over the left. In mice, Lmo4 expression marks anterior and posterior regions in the mouse cortex, and is shifted in cortices of Pax6 and Fgf8 mutants, consistent with the suggestion that Lmo4 expression at P0-P5 reflects overall cortical topographic mapping (fig. S2). On the other hand, there is some evidence for the appearance of the several cortical sulci and gyri at earlier ages in human right hemisphere than in the left, for instance the rolandic sulcus (17-20 weeks) and the superior temporal fissure (23 weeks) (2, 7, 16). Thus, higher LMO4 expression in the right than the left hemisphere could reflect the arrival of corresponding developmental stages sooner in the right than the left hemisphere. Either model however, implies molecular events that greatly precede morphological asymmetries, and provide potential insight into mechanism of generation of asymmetry. Furthermore, the molecular events that regulate LMO4 expression in human may be secreted molecules and/or transcription factor gradients in the ventricular zone at earlier stages, likely including PAX6 and FGF8, as in mice . Indeed, some factors with potential roles in cortical development, such as FGFR3, ID2, NUMB and NEUROD6, were asymmetric in SAGE and/or RT-PCR analyses in human embryonic 12-week brains (table S6 and S8). Understanding the factors that regulate LMO4 expression may ultimately identify earlier events in left-right brain asymmetry. On the other hand, our results generally confirm earlier suggestion that genes previously implicated in visceral asymmetries are not detectably implicated in cerebral asymmetries(3, 17). For instance, mutations that result in “situs inversus” in humans do not appear to disrupt the left hemisphere localization of language, mathematical and hearing abilities and handedness (18). Except possibly for the FGF signaling pathway, we did not detect significant differences of expression levels of genes regulating body asymmetry in the human embryonic SAGE libraries, though asymmetries at earlier developmental stages cannot be ruled out (3, 19).
Defects of cerebral cortical asymmetry have been reported in a wide array of neuropsychiatric disorders, such as autism, schizophrenia and dyslexia (20-23). The presence of early asymmetries in gene transcription in the two cerebral hemispheres, thus provides potential pathways through which a number of developmental disorders may ultimately converge on the abnormal development of human cerebral asymmetry.
Supplementary Material
Acknowledgments
We thank Y.F. Chen, J. Meschter, V. Petkova and P. Grosu for technical support, S. Blackshaw, K. Polyak and E. Fox for advice on SAGE. We are grateful to R. Vigorito, R. Johnson and B. Poulos at the NIH funded Brain and Tissue Banks. We also thank S. Dymecki for supplying Pax6 mutant brains and E. Meyers and G. Martin for Fgf8 mutant brains. This work was supported by the McKnight Endowment Foundation, NINDS RO1 R37 NS35129 (C.A.W), MH60233 (D.H.G) and the James S. McDonnell Foundation (D.H.G). T.S. is supported by the Advanced Postdoctoral Fellowship from the National Multiple Sclerosis Society and a NIH training grant from the Children’s Hospital, Boston. C.A.W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
One-sentence summaries: The left-right asymmetry of the human brain is accompanied by early transcriptional asymmetry of gene expression.
References
- 1.Corballis MC. Behav Brain Sci. 2003;26:199–208. doi: 10.1017/s0140525x03000062. discussion 208-60. [DOI] [PubMed] [Google Scholar]
- 2.Galaburda AM, LeMay M, Kemper TL, Geschwind N. Science. 1978;199:852–6. doi: 10.1126/science.341314. [DOI] [PubMed] [Google Scholar]
- 3.Geschwind DH, Miller BL. Am J Med Genet. 2001;101:370–81. [PubMed] [Google Scholar]
- 4.Toga AW, Thompson PM. Nat Rev Neurosci. 2003;4:37–48. doi: 10.1038/nrn1009. [DOI] [PubMed] [Google Scholar]
- 5.LeMay M, Culebras A. N Engl J Med. 1972;287:168–70. doi: 10.1056/NEJM197207272870404. [DOI] [PubMed] [Google Scholar]
- 6.Geschwind N, Levitsky W. Science. 1968;161:186–7. doi: 10.1126/science.161.3837.186. [DOI] [PubMed] [Google Scholar]
- 7.Chi JG, Dooling EC, Gilles FH. Arch Neurol. 1977;34:346–8. doi: 10.1001/archneur.1977.00500180040008. [DOI] [PubMed] [Google Scholar]
- 8.Geschwind DH, Miller BL, DeCarli C, Carmelli D. Proc Natl Acad Sci U S A. 2002;99:3176–81. doi: 10.1073/pnas.052494999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sidman RL, Rakic P. In: Histology and Histopathology of the Nervous System. Haymaker W, Adams RD, editors. C. C. Thomas Pub.; Springfield Illinois: 1982. [Google Scholar]
- 10.Biddle FG, Coffaro CM, Ziehr JE, Eales BA. Genome. 1993;36:935–43. doi: 10.1139/g93-123. [DOI] [PubMed] [Google Scholar]
- 11.Barneoud P, Van der Loos H. Proc Natl Acad Sci U S A. 1993;90:3246–50. doi: 10.1073/pnas.90.8.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riddle DR, Purves D. J Neurosci. 1995;15:4184–95. doi: 10.1523/JNEUROSCI.15-06-04184.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Signore P, et al. Physiol Behav. 1991;49:701–4. doi: 10.1016/0031-9384(91)90305-8. [DOI] [PubMed] [Google Scholar]
- 14.Collins RL. Brain Res. 1991;564:194–202. doi: 10.1016/0006-8993(91)91455-a. [DOI] [PubMed] [Google Scholar]
- 15.Palmer AR. Science. 2004;306:828–33. doi: 10.1126/science.1103707. [DOI] [PubMed] [Google Scholar]
- 16.Chi JG, Dooling EC, Gilles FH. Ann Neurol. 1977;1:86–93. doi: 10.1002/ana.410010109. [DOI] [PubMed] [Google Scholar]
- 17.Capdevila J, Vogan KJ, Tabin CJ, Belmonte J. C. Izpisua. Cell. 2000;101:9–21. doi: 10.1016/S0092-8674(00)80619-4. [DOI] [PubMed] [Google Scholar]
- 18.Kennedy DN, et al. Neurology. 1999;53:1260–5. doi: 10.1212/wnl.53.6.1260. [DOI] [PubMed] [Google Scholar]
- 19.Abu-Khalil A, Fu L, Grove EA, Zecevic N, Geschwind DH. J Comp Neurol. 2004;474:276–88. doi: 10.1002/cne.20112. [DOI] [PubMed] [Google Scholar]
- 20.Herbert MR, et al. Brain. 2005;128:213–26. doi: 10.1093/brain/awh330. [DOI] [PubMed] [Google Scholar]
- 21.Falkai P, et al. Schizophr Res. 1992;7:23–32. doi: 10.1016/0920-9964(92)90070-l. [DOI] [PubMed] [Google Scholar]
- 22.Hugdahl K, et al. Scand Audiol Suppl. 1998;49:26–34. doi: 10.1080/010503998420621. [DOI] [PubMed] [Google Scholar]
- 23.Galaburda AM, Menard MT, Rosen GD. Proc Natl Acad Sci U S A. 1994;91:8010–3. doi: 10.1073/pnas.91.17.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.