Abstract
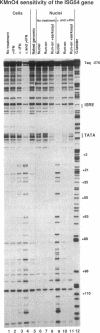
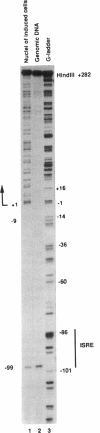
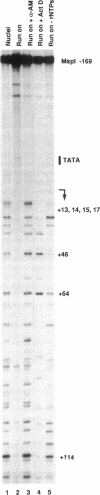
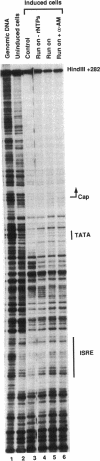
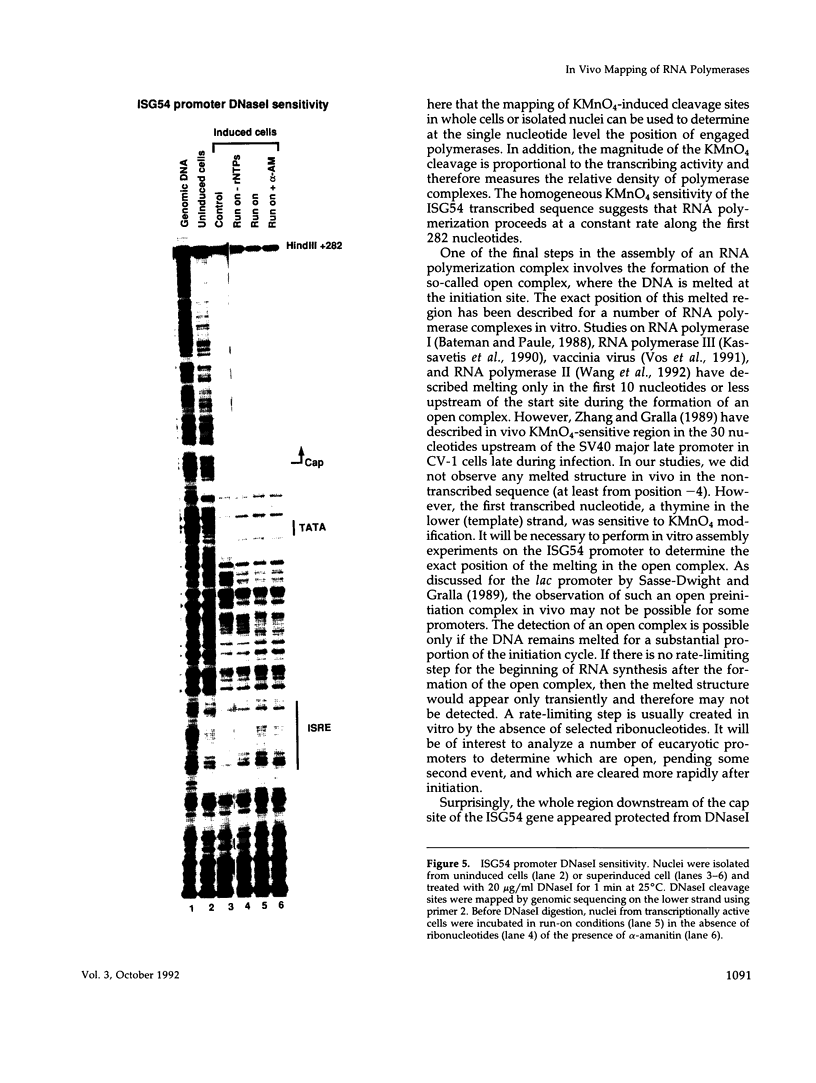
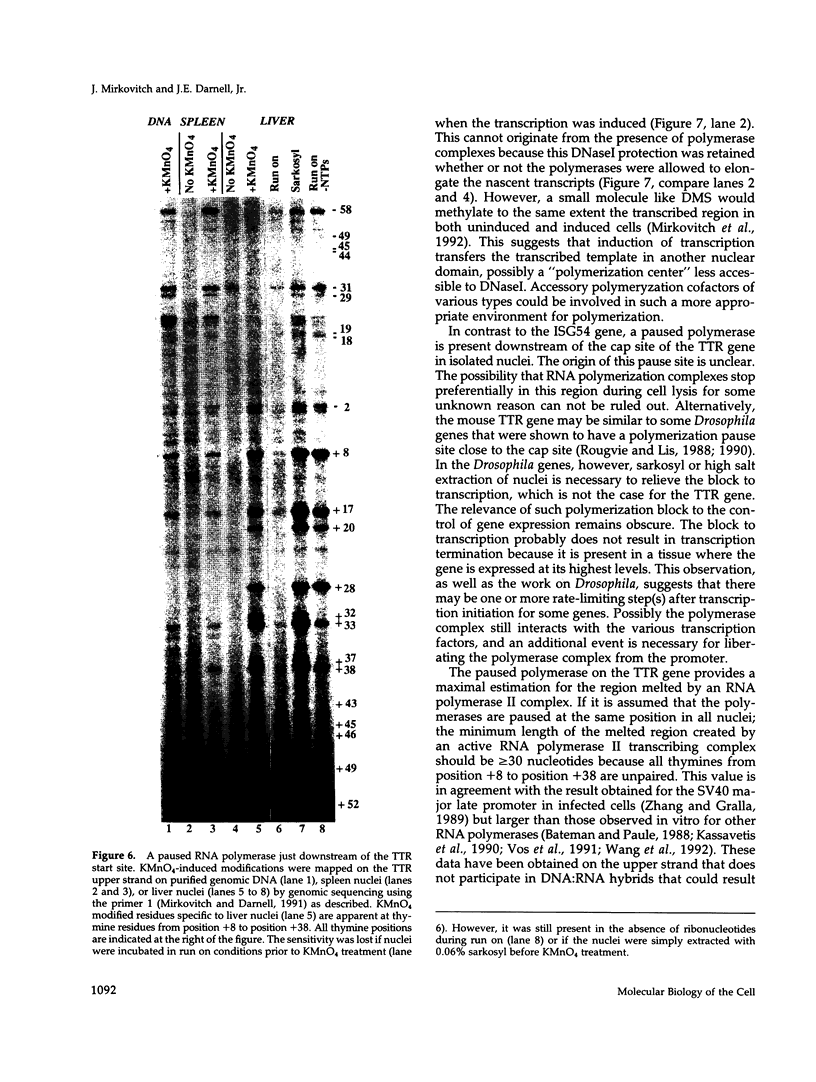
The assembly of an RNA polymerase II initiation complex at a promoter is associated with the melting of the DNA template to allow the polymerase to read the DNA sequence and synthesize the corresponding RNA. Using the specific single-stranded modifying reagent KMnO4 and a new genomic sequencing technique, we have explored the melted regions of specific genes in genomic DNA of whole cells or of isolated nuclei. We have demonstrated for the first time in vivo the melting in the promoter proximal transcribed region that is associated with the presence of RNA polymerase II complexes. An interferon-inducible gene, ISG-54, exhibited KMnO4 sensitivity over approximately 300 nucleotides downstream of the RNA initiation site in interferon-treated cells when the gene was actively transcribed but not in untreated cells where the gene was not transcribed. The extent of KMnO4 modification was proportional to transcription levels. The KMnO4 sensitivity was retained when nuclei were isolated from induced cells but was lost if the engaged polymerases were further allowed to elongate the nascent RNA chains ("run-on"). The sensitivity to KMnO4 in isolated nuclei was retained if the run-on incubation was performed in the presence of alpha-amanitin, which blocks progress of engaged polymerases. A similar analysis identified an open sequence of only approximately 30 bases just downstream of the start site of the transthyretin (TTR) gene in nuclei isolated from mouse liver, a tissue where TTR is actively transcribed. This abrupt boundary of KMnO4 sensitivity, which was removed completely by allowing engaged polymerases to elongate RNA chains, suggests that most polymerases transcribing this gene paused at about position +20. The possibility of mapping at the nucleotide level the position of actively transcribing RNA polymerases in whole cells or isolated nuclei opens new prospects in the study of transcription initiation and elongation.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bateman E., Paule M. R. Events during eucaryotic rRNA transcription initiation and elongation: conversion from the closed to the open promoter complex requires nucleotide substrates. Mol Cell Biol. 1988 May;8(5):1940–1946. doi: 10.1128/mcb.8.5.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengal E., Flores O., Krauskopf A., Reinberg D., Aloni Y. Role of the mammalian transcription factors IIF, IIS, and IIX during elongation by RNA polymerase II. Mol Cell Biol. 1991 Mar;11(3):1195–1206. doi: 10.1128/mcb.11.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec J. A., Hurwitz J. Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J. 1988 Oct;7(10):3149–3158. doi: 10.1002/j.1460-2075.1988.tb03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec J. A., Zhang L., Sasse-Dwight S., Gralla J. D. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J Mol Biol. 1987 Jul 5;196(1):101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- Buratowski S., Sopta M., Greenblatt J., Sharp P. A. RNA polymerase II-associated proteins are required for a DNA conformation change in the transcription initiation complex. Proc Natl Acad Sci U S A. 1991 Sep 1;88(17):7509–7513. doi: 10.1073/pnas.88.17.7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darst S. A., Edwards A. M., Kubalek E. W., Kornberg R. D. Three-dimensional structure of yeast RNA polymerase II at 16 A resolution. Cell. 1991 Jul 12;66(1):121–128. doi: 10.1016/0092-8674(91)90144-n. [DOI] [PubMed] [Google Scholar]
- Falvey E., Schibler U. How are the regulators regulated? FASEB J. 1991 Mar 1;5(3):309–314. doi: 10.1096/fasebj.5.3.2001790. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., Roeder R. G. Functional steps in transcription initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J Biol Chem. 1987 Mar 15;262(8):3452–3461. [PubMed] [Google Scholar]
- Hawley D. K., Roeder R. G. Separation and partial characterization of three functional steps in transcription initiation by human RNA polymerase II. J Biol Chem. 1985 Jul 5;260(13):8163–8172. [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Braun B. R., Nguyen L. H., Geiduschek E. P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990 Jan 26;60(2):235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. RNA polymerase: regulation of transcript elongation and termination. FASEB J. 1991 Oct;5(13):2833–2842. doi: 10.1096/fasebj.5.13.1916107. [DOI] [PubMed] [Google Scholar]
- Kessler D. S., Levy D. E., Darnell J. E., Jr Two interferon-induced nuclear factors bind a single promoter element in interferon-stimulated genes. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8521–8525. doi: 10.1073/pnas.85.22.8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larner A. C., Chaudhuri A., Darnell J. E., Jr Transcriptional induction by interferon. New protein(s) determine the extent and length of the induction. J Biol Chem. 1986 Jan 5;261(1):453–459. [PubMed] [Google Scholar]
- Levy D. E., Kessler D. S., Pine R., Darnell J. E., Jr Cytoplasmic activation of ISGF3, the positive regulator of interferon-alpha-stimulated transcription, reconstituted in vitro. Genes Dev. 1989 Sep;3(9):1362–1371. doi: 10.1101/gad.3.9.1362. [DOI] [PubMed] [Google Scholar]
- Levy D. E., Kessler D. S., Pine R., Reich N., Darnell J. E., Jr Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev. 1988 Apr;2(4):383–393. doi: 10.1101/gad.2.4.383. [DOI] [PubMed] [Google Scholar]
- Levy D. E., Lew D. J., Decker T., Kessler D. S., Darnell J. E., Jr Synergistic interaction between interferon-alpha and interferon-gamma through induced synthesis of one subunit of the transcription factor ISGF3. EMBO J. 1990 Apr;9(4):1105–1111. doi: 10.1002/j.1460-2075.1990.tb08216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D., Larner A., Chaudhuri A., Babiss L. E., Darnell J. E., Jr Interferon-stimulated transcription: isolation of an inducible gene and identification of its regulatory region. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8929–8933. doi: 10.1073/pnas.83.23.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn S. C., Luse D. S. RNA polymerase II elongation complexes paused after the synthesis of 15- or 35-base transcripts have different structures. Mol Cell Biol. 1991 Mar;11(3):1508–1522. doi: 10.1128/mcb.11.3.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K. J. The interactions of transcription factors and their adaptors, coactivators and accessory proteins. Bioessays. 1991 Oct;13(10):499–503. doi: 10.1002/bies.950131003. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
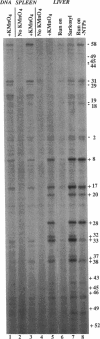
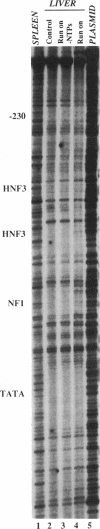
- Mirkovitch J., Darnell J. E., Jr Rapid in vivo footprinting technique identifies proteins bound to the TTR gene in the mouse liver. Genes Dev. 1991 Jan;5(1):83–93. doi: 10.1101/gad.5.1.83. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Decker T., Darnell J. E., Jr Interferon induction of gene transcription analyzed by in vivo footprinting. Mol Cell Biol. 1992 Jan;12(1):1–9. doi: 10.1128/mcb.12.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Steigerwald S. D., Mueller P. R., Wold B., Riggs A. D. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989 Nov 10;246(4931):810–813. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- Powell D. J., Friedman J. M., Oulette A. J., Krauter K. S., Darnell J. E., Jr Transcriptional and post-transcriptional control of specific messenger RNAs in adult and embryonic liver. J Mol Biol. 1984 Oct 15;179(1):21–35. doi: 10.1016/0022-2836(84)90304-8. [DOI] [PubMed] [Google Scholar]
- Reich N., Evans B., Levy D., Fahey D., Knight E., Jr, Darnell J. E., Jr Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6394–6398. doi: 10.1073/pnas.84.18.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder R. G. The complexities of eukaryotic transcription initiation: regulation of preinitiation complex assembly. Trends Biochem Sci. 1991 Nov;16(11):402–408. doi: 10.1016/0968-0004(91)90164-q. [DOI] [PubMed] [Google Scholar]
- Rougvie A. E., Lis J. T. Postinitiation transcriptional control in Drosophila melanogaster. Mol Cell Biol. 1990 Nov;10(11):6041–6045. doi: 10.1128/mcb.10.11.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie A. E., Lis J. T. The RNA polymerase II molecule at the 5' end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988 Sep 9;54(6):795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Rubin C. M., Schmid C. W. Pyrimidine-specific chemical reactions useful for DNA sequencing. Nucleic Acids Res. 1980 Oct 24;8(20):4613–4619. doi: 10.1093/nar/8.20.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman A. G., Weinmann R. Promoter specificity and modulation of RNA polymerase II transcription. FASEB J. 1989 Apr;3(6):1723–1733. doi: 10.1096/fasebj.3.6.2649403. [DOI] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J Biol Chem. 1989 May 15;264(14):8074–8081. [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Role of eukaryotic-type functional domains found in the prokaryotic enhancer receptor factor sigma 54. Cell. 1990 Sep 7;62(5):945–954. doi: 10.1016/0092-8674(90)90269-k. [DOI] [PubMed] [Google Scholar]
- Sawadogo M., Sentenac A. RNA polymerase B (II) and general transcription factors. Annu Rev Biochem. 1990;59:711–754. doi: 10.1146/annurev.bi.59.070190.003431. [DOI] [PubMed] [Google Scholar]
- Spencer C. A., Groudine M. Transcription elongation and eukaryotic gene regulation. Oncogene. 1990 Jun;5(6):777–785. [PubMed] [Google Scholar]
- Szentirmay M. N., Sawadogo M. Transcription factor requirement for multiple rounds of initiation by human RNA polymerase II. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10691–10695. doi: 10.1073/pnas.88.23.10691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke M. W., Roeder R. G., Sawadogo M. Physical analysis of transcription preinitiation complex assembly on a class II gene promoter. Science. 1988 Sep 9;241(4871):1335–1338. doi: 10.1126/science.3413495. [DOI] [PubMed] [Google Scholar]
- Vos J. C., Sasker M., Stunnenberg H. G. Promoter melting by a stage-specific vaccinia virus transcription factor is independent of the presence of RNA polymerase. Cell. 1991 Apr 5;65(1):105–113. doi: 10.1016/0092-8674(91)90412-r. [DOI] [PubMed] [Google Scholar]
- Wang W., Carey M., Gralla J. D. Polymerase II promoter activation: closed complex formation and ATP-driven start site opening. Science. 1992 Jan 24;255(5043):450–453. doi: 10.1126/science.1310361. [DOI] [PubMed] [Google Scholar]
- Weber J., Jelinek W., Darnell J. E., Jr The definition of a large viral transcription unit late in Ad2 infection of HeLa cells: mapping of nascent RNA molecules labeled in isolated nuclei. Cell. 1977 Apr;10(4):611–616. doi: 10.1016/0092-8674(77)90093-9. [DOI] [PubMed] [Google Scholar]
- Zhang L., Gralla J. D. In situ nucleoprotein structure at the SV40 major late promoter: melted and wrapped DNA flank the start site. Genes Dev. 1989 Nov;3(11):1814–1822. doi: 10.1101/gad.3.11.1814. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]