Abstract
In eukaryotes, mitochondrial activity controls ATP production, calcium dynamics, and redox state, thereby establishing physiological parameters governing the transduction of biochemical signals that regulate nuclear gene expression. However, these activities are commonly assumed to fulfill a ‘housekeeping’ function: necessary for life, but an epiphenomenon devoid of causal agency in the developmental flow of genetic information. Moreover, it is difficult to perturb mitochondrial function without generally affecting cell viability. For these reasons little is known about the extent of mitochondrial influence on gene activity in early development. Recent discoveries pertaining to the redox regulation of key developmental signaling systems together with the fact that mitochondria are often asymmetrically distributed in animal embryos suggests that they may contribute spatial information underlying differential specification of cell fate. In many cases such asymmetries correlate with localization of genetic determinants (i.e., mRNAs or proteins), particularly in embryos that rely heavily on cell-autonomous means of cell fate specification. In such embryos the localized genetic determinants play a dominant role, and any developmental information contributed by the mitochondria themselves is likely to be less obvious and more difficult to isolate experimentally. Hence, ‘regulative’ embryos that make more extensive use of conditional cell fate specification are better suited to experimental investigation of mitochondrial impacts on developmental gene regulation. Recent studies of the sea urchin embryo, which is a paradigmatic example of such a system, suggest that anisotropic distribution of mitochondria provides a source gradient of spatial information that directs epigenetic specification of the secondary axis via Nodal-Lefty signaling.
1. Introduction
Mitochondria provide much of the energy that fuels eukaryotic life. In the process, they control many aspects of cell physiology, including redox state and calcium dynamics. Mitochondria also play leading roles in orchestrating the death of unhealthy cells. New fluorescent probes and technologies for live cell imaging have stimulated a resurgence of interest among cell biologists in the biochemical and molecular linkages between mitochondria, cell signaling, and gene expression. However, as of yet there has not been a similar revival in the field of developmental biology, particularly among researchers focused on the problem of cell fate specification. This stems partly from the common assumption that mitochondria fulfill ‘housekeeping’ functions that are causally irrelevant to development, but perhaps more importantly from the experimental impracticality of perturbing mitochondrial function without generally affecting cell viability. Since (in contrast) the targeted inactivation of regulatory genes typically produces specific developmental defects without affecting cell viability, it is understandable that most developmental biologists choose to ignore developmental physiology and focus their attention instead on the molecular genetics of development.
There are nonetheless two compelling reasons for suspecting that mitochondria may play causal roles in directing the developmental flow of genetic information. First, by controlling the rate of ATP production, by sequestering calcium, and by modulating intracellular redox state, mitochondria establish physiological parameters governing the transduction of biochemical signals to the cell nucleus. Second, the spatial distribution of maternal mitochondria in animal zygotes is typically anisotropic, leading to quantitative differences in mitochondrial density among cleaving blastomeres [1]. These differences often correlate with redox gradients that were described by C.M. Child and colleagues nearly a century ago, which prefigure regional differentiation in developing embryos from across the phylogenetic spectrum [1, 2]. Although Child’s gradients have long been dismissed as epiphenomena, the time appears to be ripe for reevaluating them in light of our modern understanding of the physiology of developmental gene regulation, using more specific experimental perturbations than were previously available.
In the following I review studies linking mitochondria to the cell physiology of signal transduction and the regulation of gene activity in early development. Particular emphasis is given to work in my laboratory concerning the influence of mitochondria on axis specification in sea urchin embryos. The evidence at hand indicates that the signaling systems involved in specifying axial polarities and cell fate in animal embryos are all linked to mitochondrial redox physiology, suggesting that mitochondria may have played causal roles in the evolutionary development of animal epigenesis.
2. Cell physiology and gene regulatory networks
2.1 Mitochondria and cell physiology
Life is contingent on a continuous flow of energy, much of which occurs via the currency of ATP. Eukaryotic cells obtain most of their ATP from mitochondria, wherein a proton gradient across the inner mitochondrial membrane provides the potential energy for ATP synthesis. The proton gradient is in turn established via energy obtained from aerobic respiration, a process whereby ‘high energy’ electrons extracted from food molecules flow down a redox energy gradient via the citric acid cycle (within the mitochondrial matix) and a series of electron transfer complexes (within the plane of the inner mitochondrial membrane) to reduce molecular oxygen and thus produce water. As with any form of work, this process is governed by the second law of thermodynamics and is hence less than 100% efficient; both heat and reactive oxygen species (ROS) are produced, the latter when oxygen is partially reduced by electron ‘leakage’ from intermediate steps in the electron transport chain.
ROS are highly reactive and therefore damaging to cells, and their production by mitochondria is countered by a complex array of enzymatic and non-enzymatic antioxidants [3]. For example, the mitochondrial enzyme superoxide dismutase 2 (SOD2) converts superoxide anion (the primary ROS produced by the electron transport chain) to H2O2, a less toxic ROS, while catalase reduces H2O2 to water. Reduced glutathione is a redox buffer that protects cellular components from oxidation by ROS. The ratio of reduced to oxidized glutathione (GSH/GSSG) is a major determinant of intracellular redox state, which in physiological conditions is highly reducing. Oxidative stress in response to ROS can become pathological when the antioxidant systems are compromised or overwhelmed; however, in physiological conditions ROS production is not only tolerated, but sometimes required as a mechanism of signal transduction [3]. This often involves enzymes such as the NADPH oxidases [4], which is outside the scope of this review; however, accumulating evidence indicates that mitochondrial H2O2 is also required as a signaling intermediate in a variety of contexts (e.g., [5, 6]).
As the major locus of aerobic respiration in eukaryotes, mitochondria are the key hub of intermediary metabolism in animals. The physiological balance between anabolism and catabolism—and ultimately of growth and differentiation of a cell—requires that the activity of mitochondria be tightly regulated. One control is the availability of the substrate for ATP synthesis, ADP, which increases as the use of ATP increases. Increasing availability of ADP stimulates an increase in the rate of mitochondrial respiration, and vice versa, a phenomenon referred to as ‘respiratory control’. When inhibited by a high ATP/ADP ratio, mitochondrial respiration is in a submaximal ‘resting state’ or State 4. Increase in ADP availability stimulates an increase in respiration, up to its maximum or State 3. Transitions from State 4 to State 3 are typically driven by energy demands associated with work. Oxygen can also limit the rate of mitochondrial respiration, as occurs in hypoxia. When substrate is plentiful (as is generally the case in early embryos, which are highly reducing) and respiration rate is submaximal due to either low ADP or hypoxia, the electron transport system tends to become more highly reduced, which increases ROS production.
Mitochondrial activity is also controlled by calcium. In living cells mitochondria are closely associated with the major intracellular source of calcium, the endoplasmic reticulum (ER). Because of this association mitochondria sequester calcium released from the ER in response to various signals, which stimulates respiration [7, 8]. In mouse zygotes calcium signaling stimulates oxidative phosphorylation, leading to an increase in ATP levels [9]. At the same time, the sequestration of calcium by actively respiring mitochondria modulates the strength of the calcium signal, decreasing frequency while increasing amplitude [10], probably by increasing the threshold of local Ca2+ feedback activation of IP3 receptors [11]. In vascular smooth muscle calcium uptake by mitochondria is inhibited by TGF-β via the down-regulation of IP3 receptors [12]. Thus, calcium signaling and respiratory control are coupled, and this linkage is regulated by intercellular signaling.
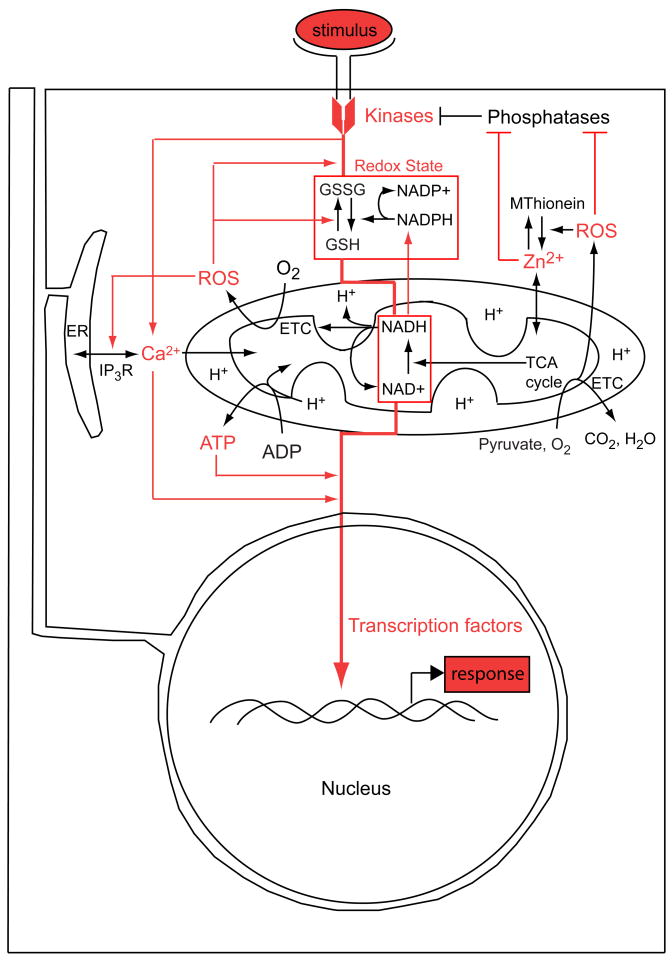
The transduction of extracellular and intracellular signals into specific genomic responses typically involves a complex interplay of intracellular calcium, redox state, and phosphorylation, leading to the (often transient) activation of a defined set of transcriptional regulatory proteins. Thus, by controlling calcium dynamics, redox state, and ATP production, mitochondria are a major source of the physiological parameters that govern signal transduction (Fig. 1). The question for developmental biology is whether these parameters also represent spatiotemporal variables that regulate developmental processes in healthy embryos, or are merely (as is commonly assumed) physiological boundary conditions that are not rate limiting, and hence causally irrelevant to the information flow of normal development.
Figure 1.
Mitochondrial influence on signal transduction in a metazoan cell. An idealized representation of redox-regulated information flow (red arrows) mediating the production of a transcriptional response to a signal (in this case, binding of an extracellular ligand to its transmembrane receptor). Note that signals impinging on gene expression might also originate intracellularly along these pathways.
2.2 Gene regulatory networks and development
Heritable aspects of animal development are determined by gene regulatory networks (GRNs), which encode the spatial and temporal contingency logic underlying the stereotypical progression of animal ontogeny [13]. GRN architecture is a function of the nucleotide sequence-specific cis- and trans- regulatory interactions within and between genes, which is mediated in large part by transcriptional regulatory proteins (‘transcription factors’). Many, perhaps most of the causally determinative aspects of animal ontogeny can be attributed to the regulatory logic encoded in the GRN of each species [14]. However, the GRN is necessary but not sufficient for the information flow of normal development; its operation depends on (and canalizes) epigenetic and often stochastic symmetry breaking processes (‘epigenesis’) [15] that are often susceptible to environmental perturbation.
Development is a unidirectional process whereby the specification of fate (i.e., actualization of one outcome among many alternatives) is accompanied by a loss of developmental potency (diminished degrees of freedom). Although this occurs very early in the ontogeny of some animals (e.g., in cases of autonomous specification following oogenetic segregation of molecular determinants), in others it is more gradual, a process of conditional specification contingent upon the extracellular environment of a cell. One way that developmental potency is maintained in such ‘regulative’ development is through the broad deployment of inactive transcription factors (or their precursors), which become locally activated by posttranslational modification in response to context-specific activation of a system of intercellular signal transduction. In metazoans this is governed by a few well-defined pathways [16]. The posttranslational modifications of transcription factors used in developmental signaling include phosphorylation, acetylation and/or proteolytic cleavage, or conformational changes in response to binding of a membrane permeable ligand (e.g., steroid hormone). Typically such modifications result in the nuclear accumulation of the active transcription factor, which then binds to specific sequences contained within the cis-regulatory domains of a battery of target genes. Genes that encode and/or respond to the components of intercellular signaling systems are thus key architectural elements of developmental GRNs, representing the major conduit into the GRN for the epigenetic information that drives conditional specification of cell fate.
3. Redox state and developmental signaling
3.1 Redox regulation of transcription factor activity
The redox state of a cell is a complex metabolic phenomenon dependent on the mutual coupling of numerous redox couples, the most prominent of which involve glutathione (GSH/GSSG) and nicotinamide adenine dinucleotide (NAD(P)+/NAD(P)H) (Fig. 1). The glutathione system functions as a redox ‘buffer’ that defends against oxidative damage (reviewed in [3]). It is normally maintained in a relatively reducing state (high GSH/GSSG), and perturbations that produce a more oxidizing state are generally associated with oxidative stress. Although oxidative stress is usually viewed as a pathological condition, oxidative metabolism normally induces a physiological (i.e. non-pathological) level of stress. The effect of such stress has recently been measured in mouse zygotes, wherein GSH levels were found to be depleted by about a third in two cell stage embryos compared to mature stage MII oocytes, which was accompanied by a concomitant depletion of NADPH [17].
The conformation and hence activity of proteins is often affected by the redox state of their cysteine residues, which depends (in complex ways involving numerous enzymes) on the redox state of the cell. Many (perhaps most) families of transcription factors are characterized by conserved cysteine residues that control their activity [1, 18]. Although the physiological relevance of these is often unclear due to the extreme conditions used in many of the in vitro DNA binding studies, in some cases these conditions have been calibrated to the measured glutathione (GSH/GSSG) redox state of cells; such studies indicate that the DNA binding activity of developmentally relevant transcription factors (AP-1 and NFκB) is sensitive to shifts in redox states that occur in physiological conditions [19]. In the case of NF-kB (p50), the DNA binding domain contains a critical cysteine residue that exists in an oxidized state in the cytoplasm, which is reduced by Ref-1 in the nucleus, thereby activating DNA binding [20].
The GSH/GSSG redox state is coupled to that of NAD(P)+/NAD(P)H, and in the early mouse embryo NADPH is required to maintain reduced glutathione [17]. NADH on the other hand is the major electron carrier in oxidative metabolism, and hence more directly linked to mitochondrial activity. Two compelling examples of transcription factor activities that are directly regulated by NAD+/NADH redox state are the NPAS proteins that regulate circadian rhythms [21], and the cell cycle phase dependent recruitment via Oct-1 of the H2B co-activator complex OCA-S [22]. An essential component of OCA-S is GAPDH, whose recruitment to the complex is regulated by the NAD+/NADH redox state [23, 24]. These examples show that some metazoan gene regulatory programs are linked to cyclic physiological variations in oxidative metabolism, a linkage that has been shown to govern the life cycles of lower eukaryotes [25, 26]. Redox state changes also accompany development, and it has been shown that NAD+/NADH redox state regulates differentiation of muscle cells via the activity of the Sir2 histone deacetylase, which inhibits differentiation and is activated by the high (more oxidized) ratio characteristic of undifferentiated cells [27]. Finally, recruitment of the CtBP co-repressor by transcriptional repressors has been shown to be stimulated by NADH [28].
Most of what is known about redox-regulated transcription factor activity comes from in vitro biochemistry and cell culture studies, and the relevance of these to the normal operation of GRNs that control early metazoan development is still largely unexplored. The best evidence that redox regulation of transcription factor activity is causally important for early ontogeny remains circumstantial, coming from in vivo DNA binding studies of the P3A2 transcription factor (a homologue of Nuclear Respiratory Factor-1) in sea urchin embryos [29], and from the fact that abnormal development is sometimes induced by environmental agents that cause oxidative stress, as in the case of thalidomide-mediated teratogenesis via redox effects on NF-κB [30].
3.2 Redox regulation of developmental signaling in animals
Metazoan development is governed by a relatively small number of evolutionarily conserved pathways of intercellular and intracellular signal transduction [16]. Several of these, including those involving Wnt/beta-catenin, Integrin, Receptor Tyrosine Kinase (RTK), JAK/STAT, and Notch signaling have been shown in at least some contexts to be responsive or otherwise linked to redox state (Table 1). Moreover, many of the biochemical networks engaged by these signals converge on or utilize Ca2+ signaling, which as mentioned above is coupled to redox signaling [31–35].
Table 1.
Examples of redox-regulated signal transduction pathways
| Pathway | Endogenous source of ROS | Redox sensors/linkages | References |
|---|---|---|---|
| Wnt/beta-catenin | N.D. | NRX/dsh | [37] |
| Integrin/Rac | Mitochondria | N.D. | [108] |
| RTK/Ras/MAPK | NADPH oxidases, Mitochondria | Tyrosine kinases*, MAPKs (p38, JNK, ERK)* | [38, 39, 109] |
| RTK/PI3K/AKT | Mitochondria | PTEN | [5] |
| JAK/STAT | N.D. | CaMKII, JAK2 | [32, 110, 111] |
| Notch | Mitochondria | PHD/HIF-1α, FIH-1/NICD | [68, 69] |
probably via redox-sensitive phosphatases
N.D., not determined
The beta-catenin pathway is fundamentally involved in the developmental control of axis specification, patterning, and cell proliferation in metazoans. The pathway is typically activated by binding of extracellular Wnt ligands to their transmembrane Frizzled receptors, which leads to the inactivation of GSK-3β via the scaffolding protein disheveled (Dsh), and consequently to stabilization and nuclear accumulation of beta-catenin [36]. In a cell culture model it was recently shown that this pathway is inhibited by the thioredoxin family member nucleoredoxin (NRX) via the latter’s binding to Dsh, an interaction that is disrupted by H2O2, thus activating beta-catenin signaling independently Wnt binding [37]. The redox regulation of beta-catenin signaling has interesting ramifications that merit further exploration in the context of primary axis specification in animal embryos, as discussed below (section 5.2).
Some of the best examples of redox signaling in early animal development come from the nematode Caenorhabditis elegans. In clk-1 mutants of this species ROS levels are quenched by elevated levels of demethoxyquinone, and these animals display delayed and otherwise defective development of the germline and vulva due to compromised Ras and IP3 signaling [38]. In vitro cell culture models have shown that Ras signaling promotes ROS production, which in some cases involves mitochondria [39], whereas IP3 signaling evokes Ca2+ release from the ER, which is redox regulated via both the IP3 receptor and sarco/endoplasmic reticulum Ca-ATPase (SERCA), as well as at the level of IP3 production [34, 40, 41]. Moreover, calcium oscillations in pancreatic cells have recently been shown to require mitochondrial ROS [35].
Another C. elegans developmental regulatory gene that is linked to redox signaling is skn-1, which encodes a transcription factor whose activity is localized to the EMS lineage of the embryo and required for initiating endomesoderm specification in that lineage [42–44]. SKN-1 is a homologue of mammalian Nrf2, a cap-n-collar (CNC) b-Zip transcription factor that responds to oxidative stress [45, 46]. Like its mammalian homologue, SKN-1 is required to mount defensive responses to oxidative stress, and skn-1 mutants have shortened life spans [47]. SKN-1 nuclear localization is antagonized by GSK-3β [48], and mammalian Nrf2 appears to be similarly regulated [49]. In early C. elegans embryos, GSK-3β antagonizes SKN-1 in the C-blastomere (an epidermal and body wall muscle precursor), thereby restricting the activity of the latter activity to the EMS lineage [43].
Nuclear localization of SKN-1 is activated by p38 MAPK [50], which was recently shown to antagonize GSK-3β in mouse thymocytes [51], and like other MAPKs is known to be activated by ROS [32, 52, 53]. For example, in HeLa cells, mitochondrial H2O2 activates c-Jun N-terminal kinase (JNK), which leads to the inactivation of GSK-3β via RSK3 [54]. Mitochondrial H2O2 also inactivates PTEN and thereby activates PI3K and hence Akt [5], another antagonist of GSK-3β. Thus it appears that GSK-3β inactivation, a key metabolic switch and initial event in metazoan endomesoderm specification, is a convergent effect of several pathways that are known to mediate redox signaling.
One of the best known paradigms of redox signaling in animals involves the hypoxia-inducible factor HIF-1α. Under normoxic conditions this transcription factor is targeted for destruction by VHL-mediated ubiquitination in response to its modification by a prolyl hydroxylase (PHD) [55, 56]. Hypoxia stabilizes HIF-1α by inhibiting the PHD via a poorly-understood process that requires mitochondrial ROS production [57–60]. Upon stabilization, HIF-1a heterodimerizes with a constitutively active HIF-1β subunit and translocates to the nucleus, where it activates hypoxia-response genes such as VEGF [61]. HIF-1α is required for various developmental processes in vertebrates, including limb development, vasculogenesis, and cardiogenesis [62–65]. HIF-1α activates expression of Twist [66, 67], a transcription factor with fundamental role in regulating mesoderm development and epithelial to mesenchymal transitions. Finally and perhaps most interestingly, HIF-1 appears to be mutually linked to the Notch-signaling pathway, synergizing with the latter in maintaining cells in an undifferentiated state in response to hypoxia [68, 69].
There is accumulating evidence that Zn2+ may be used as an intracellular signal [70] (Fig. 1). Cells contain very little free Zn2+; this ion is largely bound up in proteins, with the cysteine-rich protein metallothionein being the biggest reservoir [71]. The sulfhydryl side chains of the cysteines involved in coordinating zinc within metallothionein are highly sensitive to oxidation however, causing the protein to release free zinc in response to mild oxidative stress [72]. Zn2+ is a potent inhibitor of protein tyrosine phosphatases [73, 74], and is also known to modulate mitochondrial metabolism [75]. It thus seems likely that Zn2+ functions as an intracellular redox signaling intermediate. Very little is known about its role in development, although it is both an essential micronutrient required for normal embryogenesis [76, 77] as well as a potent environmental teratogen [78, 79].
4. Mitochondria and oral-aboral axis specification in sea urchin embryos
4.1 Axis specification via Nodal and Lefty
A perennial dialectic for developmental biology concerns the question of whether ontogeny occurs by way of ‘preformation’ or ‘epigenesis’. Although early preformationist conceptions of homunculi were long ago discarded, the determinism implicit in the term ‘preformation’ is in essence manifested by the spatial localization of maternal and/or zygotic of molecular genetic determinants (regulatory mRNAs and proteins) in the embryos of many species. Our understanding of one of the earliest steps in ontogeny—axis specification—is in fact heavily biased by studies of such species (i.e., fruit flies, nematodes, ascidians, and frogs), as well as by the fact that ‘determinative’ developmental processes are more mechanical and hence easier to study experimentally. Although sea urchin embryos have a maternally pre-formed primary (animal-vegetal) axis, the seminal experiments of Hans Driesch demonstrating the pluripotency of 2- and 4-cell stage sea urchin blastomeres [80] (reflecting the fact that the first and second cleavage planes in the sea urchin zygote are parallel with the primary axis) firmly established the sea urchin embryo as a paradigm of epigenesis. The secondary (oral-aboral) axis, which defines the plane of bilateral symmetry, is not predetermined; rather, it is progressively specified during cleavage stage via a conditional process that has only recently been elucidated.
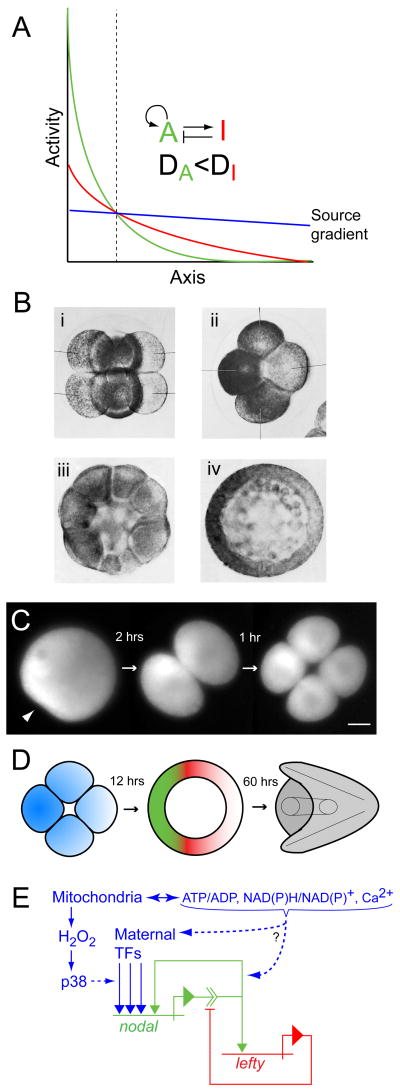
The genetic keystone of oral-aboral (OA) axis specification is nodal, which encodes a TGFβ family signaling ligand thus far found only in deuterostomes [81]. When it is first activated in the 32–60-cell embryo, nodal is expressed in a broad gradient, but by blastula stage it becomes confined to the prospective oral ectoderm [81, 82]. This localization, which is essential for OA axis specification, depends on a positive feedback signaling loop wherein Nodal activates its own expression [83, 84], as well as that of lefty, which encodes an extracellular Nodal antagonist. Although they are co-expressed in the same cells [81, 85], Lefty diffuses more rapidly than Nodal [86] and thus acts as a long-range nodal inhibitor [85]. Nodal and Lefty thus constitute an epigenetic system of short-range (auto)-activation and long-range inhibition [87], embodying a theory of pattern formation originally proposed by Alan Turing in 1952 [88] and further developed by Gierer and Meinhardt in 1972 [89] (Fig. 2A).
Figure 2.
Secondary axis specification in the sea urchin embryo. (A) Model of axis specification via a Gierer-Meinhardt type ‘reaction-diffusion’ system involving a short-range activator “A” that activates itself and its longer-range inhibitor “I” (redrawn from Gierer and Meinhardt, 1972: Figure 1c; [89]). “DA” and “DI” denote the respective rates of spatial redistribution (by diffusion or other means) of the activator and inhibitor. The final pattern of activator (green) and inhibitor (red) develops from an initially shallow source gradient of activator (blue). The vertical dashed line demarcates the boundary between final territories wherein the activator is ‘on’ and ‘off’. (B) Early sea urchin embryos labeled with indophenol blue, a specific indicator of cytochrome oxidase, showing asymmetry in mitochondrial distribution: (i) 8 cell embryo, lateral view; (ii) 8 cell embryo, polar view; (iii) 32 cell embryo, lateral view; (iv) mesenchyme blastula, polar view (from Czihak, 1963 [91]; reproduced with permission from Springer). (C) Time-lapse series of wide-field fluorescence micrographs showing the first two cleavages of a sea urchin zygote stained with MitoTracker Green FM. The arrowhead in the first panel points to the sperm entry point. An asymmetric distribution of mitochondria present in the egg produces a gradient of mitochondrial densities among the blastomeres, the high end of which specifies the oral pole of the embryo (from Coffman et al., 2004; [95]). (D) Schematic of oral-aboral axis specification in the sea urchin embryo showing relationships between mitochondrial gradient in the early embryo (blue), domains of dominance of Nodal (green) and Lefty (red) in blastula stage embryos, and the morphology of the pluteus larva (grey). The color scheme corresponds to that in (A). (E) Regulatory relationships of nodal and lefty that fulfill the activation and inhibition criteria shown in (A) (using the same color scheme), as discussed in the text. Mitochondrial inputs that contribute (or have the potential to contribute) to establishing the initial source gradient of nodal expression are shown in blue (modified from Coffman et al. [98]).
The fact that axis specification via nodal and lefty is a self-organizing process accounts for the regulative (i.e., conditional) aspects of OA axis specification in sea urchin embryos. However, it doesn’t account for the initial source of spatial information that specifies OA polarity. The fact that nodal is initially expressed in a broad gradient indicates that there must be an underlying source of graded spatial information in the early embryo. As shown by Gierer and Meinhardt, this ‘source gradient’ need not be especially pronounced; a subtle gradient is all that is needed for robust epigenesis via the self-organizing dynamics of the activator-inhibitor couple [89] (Fig. 2A).
4.2 A source gradient of mitochondria
The earliest known manifestation of OA polarity in the sea urchin embryo is a redox gradient, first observed by Child at blastula stage [2, 90] and later shown by Gerhard Czihak to be associated with cytochrome oxidase and present in the early cleavage stage embryo (Fig. 2B) [91]. That this gradient might be causally related to OA axis specification was first suggested by the work of Daniel Pease, who found that the polarity of the OA axis could be entrained in sand dollar embryos by exposing them to steep concentration gradients of respiratory inhibitors [92–94]. More recently it was shown that OA polarity is similarly entrained in immobilized clusters of embryos, wherein a mitochondrial respiration-dependent redox gradient is established along the inside-outside axis of the cluster [29].
The endogenous redox gradient is created by an asymmetric distribution of mitochondria in the early embryo, an anisotropy that is established maternally in S. purpuratus [95] (Fig. 2C). The mitochondrial asymmetry is causally implicated in specifying OA polarity, as evinced by the fact that OA polarity can be entrained by either centrifugation of eggs (which displaces mitochondria toward the centrifugal pole) or by injection of boluses of purified mitochondria [95]. These findings suggest that the mitochondrial gradient present in the early embryo might underlie the initial source gradient of nodal expression (Fig. 2D). Nodal is responsive to respiratory activity, as shown by the fact that its expression (and hence OA axis specification) is suppressed in hypoxically cultured embryos [95].
The expression of nodal is dependent on the activity of p38 MAPK, which is activated in the cleavage stage embryo [96]. As noted above, p38 is known to respond to oxidative stress, suggesting that it might provide a link between mitochondrial redox signaling and nodal expression. Recently we addressed this possibility by targeting catalase to the outer mitochondrial membrane using the C-terminal targeting sequence from the mammalian protein OMP-25 [97]. Expression of this mitochondrially-targeted catalase in sea urchin embryos was found to quench mitochondrial H2O2 emissions and down-regulate p38, inhibiting the initial activation of nodal at late cleavage stage [98]. Moreover, this perturbation entrained OA polarity toward aboral when confined to half an embryo by 2-cell stage blastomere injections. On the other hand, over-expression of the mitochondrial enzyme SOD2, which augments mitochondrial H2O2 production and up-regulates p38, failed to stimulate early nodal expression, and did not entrain OA polarity when localized by blastomere injections [98]. Thus, mitochondrial redox signaling via H2O2 and p38 appears to be only one of multiple rate limiting factors that contribute to establishing the source gradient of nodal activity.
As discussed above, the influence of mitochondria on cell signaling and gene expression is complex and physiologically integrated (Fig. 1), so it is not surprising that simply boosting mitochondrial H2O2 emission (and hence p38 activity) does not suffice to stimulate nodal activity or entrain OA polarity. The nodal cis-regulatory system has target sites for transcription factors from several different families [83, 84], and therefore mitochondria probably influence the activity of this system in a number of different ways; in addition to direct redox signals such as H2O2 this might involve indirect mitochondrial density-dependent effects on ATP production, redox state and Ca2+ signaling (Fig. 2E). Multiple parallel signals from mitochondria could impinge on the nodal cis-regulatory system, affecting not only its initial activation but potentially its positive and negative feedback regulation as well. Characterization of this system will continue to provide a fascinating subject for research.
5. Mitochondria and the evolutionary development of animals
5.1 Energy metabolism, growth, and differentiation
The gene expression programs that regulate the growth and development of lower (non-metazoan) eukaryotes are entrained by energy metabolism, which in budding yeast orchestrates cyclic programs of catabolism and anabolism involving much of the genome [25, 26]; and by the environment, which is a source of metabolic energy as well as stress. Growth rate is limited not only by the availability of energy, but also by environmental stress, which promotes gene expression programs that antagonize those associated with growth (as does cell differentiation in metazoans) [99]. Oxygen is a source of both energy and stress. Hence mitochondria, the principal source of aerobic metabolism, are a central nexus in the metabolic network that controls the balance between growth and stress response, and perhaps by extension, the balance between growth and differentiation.
Phenomenologically, development constitutes a trajectory leading from an initial stage of growth, characterized by an increasing rate of intrinsic flux, to a terminal state of differentiation (or senescence), characterized by a decreasing rate of intrinsic flux and loss of developmental potential [100]. The development rate can thus be defined as the rate at which this trajectory is traversed. Development rates are likely to vary stochastically among cells (and clones of cells) according to genetic, epigenetic, and environmental factors, with the quantity and quality of mitochondria in a given cell being a major determinant. In fact, there is abundant evidence that mitochondria play a major role in determining lifespan in multicellular organisms [101–103].
As noted above, development entails a progressive loss of developmental potency. Thus, differential rates of development among clonal colonies of cells will by itself produce heterogeneous populations manifesting different capabilities and developmental potencies. To the extent that these populations form cooperative (i.e., ecological) networks, they provide an evolutionary foundation for the higher level development of an organism.
5.2 Primordial embryogenesis: co-option of metabolic pathways for cell fate determination
Evolution is a conservative process, so it is reasonable to assume that the gene regulatory networks that control metazoan ontogeny are derived from regulatory systems that control the growth and development of lower eukaryotes. Moreover, development in the earliest metazoans was probably less genetically constrained and hence more reactive to environmental and epigenetic factors than it is in their modern descendents [104, 105]. Therefore, it seems likely that mitochondria played important causal roles in the epigenesis of early metazoans. Two such roles are suggested by the foregoing review.
The first involves transduction of the information established by oxygen gradients. The precursors of metazoans were undoubtedly clonal colonies of cells. A primary axis manifested in such colonies is the inside-outside axis, which produces an oxygen gradient, with the inside of the colony being more hypoxic than the outside. Given the relationship between metabolic rate and development rate, cells on the inside might be expected to develop more slowly than those on the outside (i.e., remain “pluripotent” longer), much as occurs with the inner cell mass in mammalian embryos today (and indeed, the pluripotency of embryonic stem cells is sensitive to oxygen; [106]). As reviewed in section 3.2, the gene regulatory systems that control metazoan endomesoderm specification are linked via multiple pathways to those that control the responses to hypoxia and oxidative stress (an initial response to hypoxia), suggesting that the former systems may be derived from the latter, having been initially co-opted for development via the epigenesis of an inside-outside axis in primitive metazoans [1].
The second role for mitochondria in metazoan epigenesis would be to provide spatial information through intracellular and intercellular variations in mitochondrial density. As reviewed in section 2.1, mitochondrial density is likely to modulate various cell physiological parameters, particularly redox and calcium signaling, which in turn modulate the expression of intercellular signaling systems. It seems probable that intercellular variations in mitochondrial density existed in early metazoans, much as it does in many modern embryos [1], and the studies of sea urchin embryos reviewed in section 4 show at such spatial information may indeed play a role in directing development.
In conclusion, the fundamental involvement of mitochondria in the cell physiology of eukaryotes does appear to be causally relevant to the developmental biology of metazoans, providing some vindication of the long discredited conceptualizations of C. M. Child [1, 2, 107]. Mitochondria are a source of both energy and information, and the information that they provide impinges on key regulatory systems that control gene activity in early development. Although modern animals have genetically determined systems that appear in many cases to have superceded the influence of mitochondria, these are built on (and hence retain components of) a foundation of metabolic signaling systems that probably played a dominant role in the epigenesis of our primordial ancestors.
Acknowledgments
The research in my laboratory reviewed in this article is supported by a grant from the NIH (ES016722). I thank Drs. Neil Blackstone and Remi Dumollard for helpful comments and suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Coffman JA, Denegre JM. Mitochondria, redox signaling and axis specification in metazoan embryos. Dev Biol. 2007;308:266–80. doi: 10.1016/j.ydbio.2007.05.042. [DOI] [PubMed] [Google Scholar]
- 2.Child CM. Patterns and Problems of Development. Chicago, IL: University of Chicago Press; 1941. [Google Scholar]
- 3.Covarrubias L, et al. Function of reactive oxygen species during animal development: passive or active? Dev Biol. 2008;320:1–11. doi: 10.1016/j.ydbio.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 4.Finkel T. Oxidant signals and oxidative stress. Curr Opin Cell Biol. 2003;15:247–54. doi: 10.1016/s0955-0674(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 5.Connor KM, et al. Mitochondrial H2O2 regulates the angiogenic phenotype via PTEN oxidation. J Biol Chem. 2005;280:16916–24. doi: 10.1074/jbc.M410690200. [DOI] [PubMed] [Google Scholar]
- 6.Nelson KK, et al. Redox-dependent matrix metalloproteinase-1 expression is regulated by JNK through Ets and AP-1 promoter motifs. J Biol Chem. 2006;281:14100–10. doi: 10.1074/jbc.M601820200. [DOI] [PubMed] [Google Scholar]
- 7.Gunter TE, et al. Calcium and mitochondria. FEBS Lett. 2004;567:96–102. doi: 10.1016/j.febslet.2004.03.071. [DOI] [PubMed] [Google Scholar]
- 8.Rutter GA, Rizzuto R. Regulation of mitochondrial metabolism by ER Ca2+ release: an intimate connection. Trends Biochem Sci. 2000;25:215–21. doi: 10.1016/s0968-0004(00)01585-1. [DOI] [PubMed] [Google Scholar]
- 9.Campbell K, Swann K. Ca2+ oscillations stimulate an ATP increase during fertilization of mouse eggs. Dev Biol. 2006;298:225–33. doi: 10.1016/j.ydbio.2006.06.032. [DOI] [PubMed] [Google Scholar]
- 10.Jouaville LS, et al. Synchronization of calcium waves by mitochondrial substrates in Xenopus laevis oocytes. Nature. 1995;377:438–41. doi: 10.1038/377438a0. [DOI] [PubMed] [Google Scholar]
- 11.Hajnoczky G, Hager R, Thomas AP. Mitochondria suppress local feedback activation of inositol 1,4, 5-trisphosphate receptors by Ca2+ J Biol Chem. 1999;274:14157–62. doi: 10.1074/jbc.274.20.14157. [DOI] [PubMed] [Google Scholar]
- 12.Pacher P, et al. Uncoupling of ER-Mitochondrial Calcium Communication by Transforming Growth factor-{beta} Am J Physiol Renal Physiol. 2008 doi: 10.1152/ajprenal.90343.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. San Diego: Academic Press/Elsevier; 2006. [Google Scholar]
- 14.Davidson EH, McClay DR, Hood L. Regulatory gene networks and the properties of the developmental process. Proc Natl Acad Sci U S A. 2003;100:1475–80. doi: 10.1073/pnas.0437746100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Losick R, Desplan C. Stochasticity and cell fate. Science. 2008;320:65–8. doi: 10.1126/science.1147888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pires-daSilva A, Sommer RJ. The evolution of signalling pathways in animal development. Nat Rev Genet. 2003;4:39–49. doi: 10.1038/nrg977. [DOI] [PubMed] [Google Scholar]
- 17.Dumollard R, et al. Regulation of redox metabolism in the mouse oocyte and embryo. Development. 2007;134:455–65. doi: 10.1242/dev.02744. [DOI] [PubMed] [Google Scholar]
- 18.Sun Y, Oberley LW. Redox regulation of transcriptional activators. Free Radic Biol Med. 1996;21:335–48. doi: 10.1016/0891-5849(96)00109-8. [DOI] [PubMed] [Google Scholar]
- 19.Clive DR, Greene JJ. Cooperation of protein disulfide isomerase and redox environment in the regulation of NF-kappaB and AP1 binding to DNA. Cell Biochem Funct. 1996;14:49–55. doi: 10.1002/cbf.638. [DOI] [PubMed] [Google Scholar]
- 20.Nishi T, et al. Spatial redox regulation of a critical cysteine residue of NF-kappa B in vivo. J Biol Chem. 2002;277:44548–56. doi: 10.1074/jbc.M202970200. [DOI] [PubMed] [Google Scholar]
- 21.Rutter J, et al. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–4. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 22.McKnight S. Gene switching by metabolic enzymes--how did you get on the invitation list? Cell. 2003;114:150–2. doi: 10.1016/s0092-8674(03)00563-4. [DOI] [PubMed] [Google Scholar]
- 23.Zheng L, Roeder RG, Luo Y. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell. 2003;114:255–66. doi: 10.1016/s0092-8674(03)00552-x. [DOI] [PubMed] [Google Scholar]
- 24.Dai RP, et al. Histone 2B (H2B) expression is confined to a proper NAD+/NADH redox status. J Biol Chem. 2008 doi: 10.1074/jbc.M804307200. [DOI] [PubMed] [Google Scholar]
- 25.Tu BP, et al. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–8. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 26.Tu BP, et al. Cyclic changes in metabolic state during the life of a yeast cell. Proc Natl Acad Sci U S A. 2007;104:16886–91. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fulco M, et al. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Q, Piston DW, Goodman RH. Regulation of corepressor function by nuclear NADH. Science. 2002;295:1895–7. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- 29.Coffman JA, Davidson EH. Oral-aboral axis specification in the sea urchin embryo. I. Axis entrainment by respiratory asymmetry. Dev Biol. 2001;230:18–28. doi: 10.1006/dbio.2000.9996. [DOI] [PubMed] [Google Scholar]
- 30.Hansen JM, Harris C. A novel hypothesis for thalidomide-induced limb teratogenesis: redox misregulation of the NF-kappaB pathway. Antioxid Redox Signal. 2004;6:1–14. doi: 10.1089/152308604771978291. [DOI] [PubMed] [Google Scholar]
- 31.Davidson SM, Duchen MR. Calcium microdomains and oxidative stress. Cell Calcium. 2006;40:561–74. doi: 10.1016/j.ceca.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen A, Chen P, Cai H. Role of CaMKII in hydrogen peroxide activation of ERK1/2, p38 MAPK, HSP27 and actin reorganization in endothelial cells. FEBS Lett. 2004;572:307–13. doi: 10.1016/j.febslet.2004.06.061. [DOI] [PubMed] [Google Scholar]
- 33.Howe CJ, et al. Redox regulation of the calcium/calmodulin-dependent protein kinases. J Biol Chem. 2004;279:44573–81. doi: 10.1074/jbc.M404175200. [DOI] [PubMed] [Google Scholar]
- 34.Zima AV, Blatter LA. Redox regulation of cardiac calcium channels and transporters. Cardiovasc Res. 2006;71:310–21. doi: 10.1016/j.cardiores.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 35.Camello-Almaraz MC, et al. Mitochondrial production of oxidants is necessary for physiological calcium oscillations. J Cell Physiol. 2006;206:487–94. doi: 10.1002/jcp.20498. [DOI] [PubMed] [Google Scholar]
- 36.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 37.Funato Y, et al. The thioredoxin-related redox-regulating protein nucleoredoxin inhibits Wnt-beta-catenin signalling through dishevelled. Nat Cell Biol. 2006;8:501–8. doi: 10.1038/ncb1405. [DOI] [PubMed] [Google Scholar]
- 38.Shibata Y, et al. Redox regulation of germline and vulval development in Caenorhabditis elegans. Science. 2003;302:1779–82. doi: 10.1126/science.1087167. [DOI] [PubMed] [Google Scholar]
- 39.Finkel T. Intracellular redox regulation by the family of small GTPases. Antioxid Redox Signal. 2006;8:1857–63. doi: 10.1089/ars.2006.8.1857. [DOI] [PubMed] [Google Scholar]
- 40.Joseph SK, Nakao SK, Sukumvanich S. Reactivity of free thiol groups in type-I inositol trisphosphate receptors. Biochem J. 2006;393:575–82. doi: 10.1042/BJ20050889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dumollard R, Duchen M, Sardet C. Calcium signals and mitochondria at fertilisation. Semin Cell Dev Biol. 2006;17:314–23. doi: 10.1016/j.semcdb.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 42.Bowerman B, et al. The maternal gene skn-1 encodes a protein that is distributed unequally in early C. elegans embryos. Cell. 1993;74:443–52. doi: 10.1016/0092-8674(93)80046-h. [DOI] [PubMed] [Google Scholar]
- 43.Maduro MF, et al. Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3beta homolog is mediated by MED-1 and -2 in C. elegans. Mol Cell. 2001;7:475–85. doi: 10.1016/s1097-2765(01)00195-2. [DOI] [PubMed] [Google Scholar]
- 44.Bowerman B, Eaton BA, Priess JR. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;68:1061–75. doi: 10.1016/0092-8674(92)90078-q. [DOI] [PubMed] [Google Scholar]
- 45.Lee JM, Johnson JA. An important role of Nrf2-ARE pathway in the cellular defense mechanism. J Biochem Mol Biol. 2004;37:139–43. doi: 10.5483/bmbrep.2004.37.2.139. [DOI] [PubMed] [Google Scholar]
- 46.Numazawa S, Yoshida T. Nrf2-dependent gene expressions: a molecular toxicological aspect. J Toxicol Sci. 2004;29:81–9. doi: 10.2131/jts.29.81. [DOI] [PubMed] [Google Scholar]
- 47.An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–93. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.An JH, et al. Regulation of the Caenorhabditis elegans oxidative stress defense protein SKN-1 by glycogen synthase kinase-3. Proc Natl Acad Sci U S A. 2005;102:16275–80. doi: 10.1073/pnas.0508105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salazar M, et al. Glycogen synthase kinase-3beta inhibits the xenobiotic and antioxidant cell response by direct phosphorylation and nuclear exclusion of the transcription factor Nrf2. J Biol Chem. 2006;281:14841–51. doi: 10.1074/jbc.M513737200. [DOI] [PubMed] [Google Scholar]
- 50.Inoue H, et al. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 2005;19:2278–83. doi: 10.1101/gad.1324805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thornton TM, et al. Phosphorylation by p38 MAPK as an alternative pathway for GSK3beta inactivation. Science. 2008;320:667–70. doi: 10.1126/science.1156037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alonso M, et al. Mitochondrial extracellular signal-regulated kinases 1/2 (ERK1/2) are modulated during brain development. J Neurochem. 2004;89:248–56. doi: 10.1111/j.1471-4159.2004.02323.x. [DOI] [PubMed] [Google Scholar]
- 53.Torres M, Forman HJ. Redox signaling and the MAP kinase pathways. Biofactors. 2003;17:287–96. doi: 10.1002/biof.5520170128. [DOI] [PubMed] [Google Scholar]
- 54.Nemoto S, et al. Role for mitochondrial oxidants as regulators of cellular metabolism. Mol Cell Biol. 2000;20:7311–8. doi: 10.1128/mcb.20.19.7311-7318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jaakkola P, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–72. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 56.Ivan M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–8. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 57.Brunelle JK, et al. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 2005;1:409–14. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 58.Guzy RD, et al. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1:401–8. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 59.Mansfield KD, et al. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-alpha activation. Cell Metab. 2005;1:393–9. doi: 10.1016/j.cmet.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin X, et al. A chemical genomics screen highlights the essential role of mitochondria in HIF-1 regulation. Proc Natl Acad Sci U S A. 2008;105:174–9. doi: 10.1073/pnas.0706585104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maxwell PH, Ratcliffe PJ. Oxygen sensors and angiogenesis. Semin Cell Dev Biol. 2002;13:29–37. doi: 10.1006/scdb.2001.0287. [DOI] [PubMed] [Google Scholar]
- 62.Nagao K, et al. HIF-1alpha signaling upstream of NKX2.5 is required for cardiac development in Xenopus. J Biol Chem. 2008;283:11841–9. doi: 10.1074/jbc.M702563200. [DOI] [PubMed] [Google Scholar]
- 63.Iyer NV, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–62. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Provot S, et al. Hif-1alpha regulates differentiation of limb bud mesenchyme and joint development. J Cell Biol. 2007;177:451–64. doi: 10.1083/jcb.200612023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ryan HE, Lo J, Johnson RS. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. Embo J. 1998;17:3005–15. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang MH, Wu KJ. TWIST activation by hypoxia inducible factor-1 (HIF-1): implications in metastasis and development. Cell Cycle. 2008;7:2090–6. doi: 10.4161/cc.7.14.6324. [DOI] [PubMed] [Google Scholar]
- 67.Yang MH, et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 68.Zheng X, et al. Interaction with factor inhibiting HIF-1 defines an additional mode of cross-coupling between the Notch and hypoxia signaling pathways. Proc Natl Acad Sci U S A. 2008;105:3368–73. doi: 10.1073/pnas.0711591105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gustafsson MV, et al. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell. 2005;9:617–28. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 70.Yamasaki S, et al. Zinc is a novel intracellular second messenger. J Cell Biol. 2007;177:637–45. doi: 10.1083/jcb.200702081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maret W. Zinc coordination environments in proteins as redox sensors and signal transducers. Antioxid Redox Signal. 2006;8:1419–41. doi: 10.1089/ars.2006.8.1419. [DOI] [PubMed] [Google Scholar]
- 72.Krezel A, Hao Q, Maret W. The zinc/thiolate redox biochemistry of metallothionein and the control of zinc ion fluctuations in cell signaling. Arch Biochem Biophys. 2007 doi: 10.1016/j.abb.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 73.Haase H, Maret W. Fluctuations of cellular, available zinc modulate insulin signaling via inhibition of protein tyrosine phosphatases. J Trace Elem Med Biol. 2005;19:37–42. doi: 10.1016/j.jtemb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 74.Haase H, Maret W. Intracellular zinc fluctuations modulate protein tyrosine phosphatase activity in insulin/insulin-like growth factor-1 signaling. Exp Cell Res. 2003;291:289–98. doi: 10.1016/s0014-4827(03)00406-3. [DOI] [PubMed] [Google Scholar]
- 75.Sensi SL, et al. Modulation of mitochondrial function by endogenous Zn2+ pools. Proc Natl Acad Sci U S A. 2003;100:6157–62. doi: 10.1073/pnas.1031598100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Falchuk KH, Montorzi M. Zinc physiology and biochemistry in oocytes and embryos. Biometals. 2001;14:385–95. doi: 10.1023/a:1012994427351. [DOI] [PubMed] [Google Scholar]
- 77.Kambe T, Weaver BP, Andrews GK. The genetics of essential metal homeostasis during development. Genesis. 2008;46:214–28. doi: 10.1002/dvg.20382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kobayashi N, Okamura H. Effects of heavy metals on sea urchin embryo development. Part 2. Interactive toxic effects of heavy metals in synthetic mine effluents. Chemosphere. 2005;61:1198–203. doi: 10.1016/j.chemosphere.2005.02.071. [DOI] [PubMed] [Google Scholar]
- 79.Kobayashi N, Okamura H. Effects of heavy metals on sea urchin embryo development. 1. Tracing the cause by the effects. Chemosphere. 2004;55:1403–12. doi: 10.1016/j.chemosphere.2003.11.052. [DOI] [PubMed] [Google Scholar]
- 80.Driesch H. Entwicklungsmechanisme Studien. I. Der Werth der beiden ersten Furchungszellen in der Echinodermentwicklung. Experimentelle Erzeugen von Theil und Doppelbildung. Zeitschrift für wissenschaftliche Zoologie. 1892;53:160–78. 83–84. [Google Scholar]
- 81.Duboc V, et al. Nodal and BMP2/4 signaling organizes the oral-aboral axis of the sea urchin embryo. Dev Cell. 2004;6:397–410. doi: 10.1016/s1534-5807(04)00056-5. [DOI] [PubMed] [Google Scholar]
- 82.Flowers VL, et al. Nodal/activin signaling establishes oral-aboral polarity in the early sea urchin embryo. Dev Dyn. 2004;231:727–40. doi: 10.1002/dvdy.20194. [DOI] [PubMed] [Google Scholar]
- 83.Nam J, et al. Cis-regulatory control of the nodal gene, initiator of the sea urchin oral ectoderm gene network. Dev Biol. 2007;306:860–9. doi: 10.1016/j.ydbio.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Range R, et al. Cis-regulatory analysis of nodal and maternal control of dorsal-ventral axis formation by Univin, a TGF-{beta} related to Vg1. Development. 2007;134:3649–64. doi: 10.1242/dev.007799. [DOI] [PubMed] [Google Scholar]
- 85.Duboc V, et al. Lefty acts as an essential modulator of Nodal activity during sea urchin oral-aboral axis formation. Dev Biol. 2008;320:49–59. doi: 10.1016/j.ydbio.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 86.Sakuma R, et al. Inhibition of Nodal signalling by Lefty mediated through interaction with common receptors and efficient diffusion. Genes Cells. 2002;7:401–12. doi: 10.1046/j.1365-2443.2002.00528.x. [DOI] [PubMed] [Google Scholar]
- 87.Chen Y, Schier AF. Lefty proteins are long-range inhibitors of squint-mediated nodal signaling. Curr Biol. 2002;12:2124–8. doi: 10.1016/s0960-9822(02)01362-3. [DOI] [PubMed] [Google Scholar]
- 88.Turing AM. The chemical basis of morphogenesis. Bull Math Biol. 1952;52:153–97. doi: 10.1007/BF02459572. [DOI] [PubMed] [Google Scholar]
- 89.Gierer A, Meinhardt H. A theory of biological pattern formation. Kybernetik. 1972;12:30–9. doi: 10.1007/BF00289234. [DOI] [PubMed] [Google Scholar]
- 90.Child CM. Formation and reduction of indophenol blue in development of an echinoderm. Proc Natl Acad Sci U S A. 1941;27:523–9. doi: 10.1073/pnas.27.11.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Czihak G. Entwicklungsphysiologische untersuchungen an echiniden (Verteilung und bedeutung der cytochromoxydase) Roux’Archiv fur Entwicklungsmechanik. 1963;154:272–92. doi: 10.1007/BF00582031. [DOI] [PubMed] [Google Scholar]
- 92.Pease DC. Echinoderm bilateral determination in chemical concentration gradients. I. The effects of cyanide, ferricyanide, iodoacetate, picrate, dinitrophenol, urethane, iodine, malonate, etc. J Exp zool. 1941;86:381–404. [Google Scholar]
- 93.Pease DC. Echinoderm bilateral determination in chemical concentration gradients. II. The effects of azide, pilocarpine, pyocyanine, diamine, cysteine, glutathione, and lithium. J Exp zool. 1942;89:329–45. [Google Scholar]
- 94.Pease DC. Echinoderm bilateral determination in chemical concentration gradients. III. The effects of carbon monoxide and other gases. J Exp zool. 1942;89:347–56. [Google Scholar]
- 95.Coffman JA, et al. Oral-aboral axis specification in the sea urchin embryo II. Mitochondrial distribution and redox state contribute to establishing polarity in Strongylocentrotus purpuratus. Dev Biol. 2004;273:160–71. doi: 10.1016/j.ydbio.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 96.Bradham CA, McClay DR. p38 MAPK is essential for secondary axis specification and patterning in sea urchin embryos. Development. 2005;133:21–32. doi: 10.1242/dev.02160. [DOI] [PubMed] [Google Scholar]
- 97.Nemoto Y, De Camilli P. Recruitment of an alternatively spliced form of synaptojanin 2 to mitochondria by the interaction with the PDZ domain of a mitochondrial outer membrane protein. Embo J. 1999;18:2991–3006. doi: 10.1093/emboj/18.11.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Coffman JA, et al. Oral-aboral axis specification in the sea urchin embryo III. Role of mitochondrial redox signaling via H2O2. Dev Biol. 2009;330:123–30. doi: 10.1016/j.ydbio.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lopez-Maury L, Marguerat S, Bahler J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet. 2008;9:583–93. doi: 10.1038/nrg2398. [DOI] [PubMed] [Google Scholar]
- 100.Coffman JA. Developmental ascendency: from bottom-up to top-down control. Biol Theor. 2006;1:165–78. [Google Scholar]
- 101.Rea SL, Ventura N, Johnson TE. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Morrow G, Tanguay RM. Mitochondria and ageing in Drosophila. Biotechnol J. 2008;3:728–39. doi: 10.1002/biot.200800015. [DOI] [PubMed] [Google Scholar]
- 103.Navarro A, Boveris A. The mitochondrial energy transduction system and the aging process. Am J Physiol Cell Physiol. 2007;292:C670–86. doi: 10.1152/ajpcell.00213.2006. [DOI] [PubMed] [Google Scholar]
- 104.Newman SA, Muller GB. Epigenetic mechanisms of character origination. J Exp Zool. 2000;288:304–17. doi: 10.1002/1097-010X(20001215)288:4<304::AID-JEZ3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 105.Newman SA. The pre-Mendelian, pre-Darwinian world: Shifting relations between genetic and epigenetic mechanisms in early multicellular evolution. J Biosci. 2005;30:75–85. doi: 10.1007/BF02705152. [DOI] [PubMed] [Google Scholar]
- 106.Westfall SD, et al. Identification of oxygen-sensitive transcriptional programs in human embryonic stem cells. Stem Cells Dev. 2008;17:869–81. doi: 10.1089/scd.2007.0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Blackstone NW. Charles Manning Child (1869–1954): the past, present, and future of metabolic signaling. J Exp Zoolog B Mol Dev Evol. 2006;306:1–7. doi: 10.1002/jez.b.21085. [DOI] [PubMed] [Google Scholar]
- 108.Werner E, Werb Z. Integrins engage mitochondrial function for signal transduction by a mechanism dependent on Rho GTPases. J Cell Biol. 2002;158:357–68. doi: 10.1083/jcb.200111028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kamata H, Hirata H. Redox regulation of cellular signalling. Cell Signal. 1999;11:1–14. doi: 10.1016/s0898-6568(98)00037-0. [DOI] [PubMed] [Google Scholar]
- 110.Simon AR, et al. Activation of the JAK-STAT pathway by reactive oxygen species. Am J Physiol. 1998;275:C1640–52. doi: 10.1152/ajpcell.1998.275.6.C1640. [DOI] [PubMed] [Google Scholar]
- 111.Madamanchi NR, et al. Reactive oxygen species regulate heat-shock protein 70 via the JAK/STAT pathway. Arterioscler Thromb Vasc Biol. 2001;21:321–6. doi: 10.1161/01.atv.21.3.321. [DOI] [PubMed] [Google Scholar]