Abstract
Electrostatic interactions play an important role in the formation of noncovalent complexes. Our previous work has highlighted the role of certain amino acid residues, such as arginine, glutamate, aspartate, and phosphorylated/sulfated residues in the formation of salt bridges resulting in noncovalent complexes between peptides. Tandem MS studies of these complexes using collision-induced dissociation (CID) have provided information on their relative stability. However, product-ion spectra produced by CID have been unable to assign specifically the site of interaction for the complex. In this work, tandem MS experiments were conducted on noncovalent complexes using both electron capture dissociation (ECD) and electron transfer dissociation (ETD). The resulting spectra were dominated by intramolecular fragments of the complex with the electrostatic interaction site intact. Based upon this data, we were able to affirm the binding site for the peptides forming the noncovalent complex.
Keywords: noncovalent complexes, electrostatic interactions, ECD, ETD
Introduction
Electrostatic interactions are one of the main factors that shapes biomolecular conformation.1–5 Certain amino acid residues and posttranslational modifications play an important role in the formation of noncovalent complexes (NCXs) by electrostatic interactions.6 We have previously observed7–10 that NCXs can form between proteins/peptides if two or more adjacent Arg are present on one epitope and two or more adjacent Glu/Asp or one phosphorylated/sulfated residue are present on the other epitope. Attempts to fragment NCXs formed with phospho/sulfopeptides, using collision-induced dissociation (CID) revealed two dissociation pathways for these complexes: one in which the electrostatic interaction is disrupted and another in which the covalent bond attaching the phosphate/sulfate group to the amino acid residue is fragmented, while the electrostatic interaction is maintained.9,10 While these results confirm the role of phosphate/sulfate in the formation of NCXs, they do not allow for the assignment of the specific residues involved in the electrostatic interactions, if more than one possible binding site is present on an epitope. Furthermore, if only adjacent Glu/Asp are involved in the electrostatic interaction, the NCX dissociates into its parent peptides giving no information about the site of interaction.
CID fragmentation can be complemented by using either electron capture dissociation (ECD) or electron transfer dissociation (ETD).11 Both ECD12 and ETD13 cleave the peptide backbone while side chains and post-translational modifications, such as phosphorylation, are left intact. Furthermore, ECD and ETD are more effective than CID for the sequencing of longer, more highly charged peptides. In one previous study14, ECD was conducted on weakly bound polypeptide complexes and produced some intramolecular fragmentation ions without weak bond dissociation. Electrostatic interactions are known to be greatly strengthened in the gas phase when compared to the solution phase, due to the dielectric constant of the solvent, which weakens electrostatic interactions in solution phase.6,15 Thus, ECD/ETD could prove to be effective tools for the identification of the specific amino acid residues involved in electrostatic interactions between peptides and proteins. In this study, ECD/ETD was conducted on NCXs in order to identify the site of interaction between the two peptides. The results of these experiments are discussed below.
Experimental
Mass Spectrometers
Two ESI mass spectrometers in positive ion mode were employed in this study. ETD experiments were conducted on a LTQ XL ion trap mass spectrometer (Thermo Electron, San Jose, CA) with a modification involving a chemical ionization source at the rear of the instrument. Fluoranthene ions were used as the ETD reagent. The anion target was 2e5 and the activation time for the ETD reaction was 120 milliseconds. ETD mass spectra presented are the sum of 25 scans.
ECD experiments were performed using a linear ion trap FTICR mass spectrometer (Finnigan LTQ FT) equipped with an ECD ion source assembly (Thermo Electron, Bremen, Germany). The resolving power of the FTICR was selected at 50,000 FWHM. The ion count targets for the FTICR experiments were 2 × 106. The ECD parameters were adjusted for each sample to ensure maximum efficiency of fragmentation of the primary amino acid chain. In this experiment, the precursor ions were excited by ECD at 5V for 15 millisecond duration. The ECD spectra are an average of 100 transitions.
Sample Preparation and Analysis
Two basic peptides (BP) YGGFLRR and SFKRRRSSK and two acidic peptides (AP) A3sYA3 and GpSSEDLKKEE were used in this study. Stock solutions were prepared in water at a concentration of 1nmol/μL. Sample mixtures, consisting of one BP at 10 pmol/μL and one AP at 50 pmol/μL in 50% ethanol, were used for mass analysis. Ten microliters of the sample mixture was transferred into the offline static nanospray Pico Tip emitter with a 4 μm i.d. (New Objective Inc. Woburn, MA) using a gel-loader pipette tip. The spray voltage was 1.1–1.2 kV resulting in a flow rate of 20–80 nL/min.
Results and Discussion
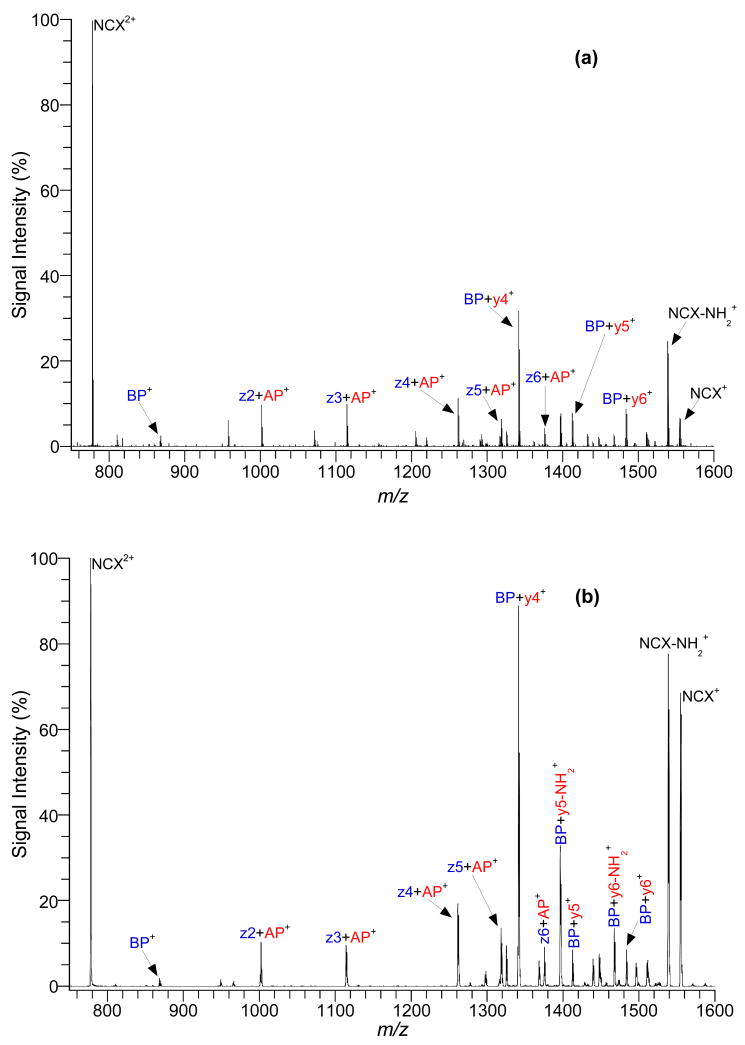
Initial studies focused on two peptides that have only one possible site for electrostatic interactions. The two adjacent arginine residues on the C-terminus of the basic peptide, YGGFLRR, are capable of forming a NCX. In the acidic peptide, A3sYA3, the absence of acidic residues and an amidated carboxyl terminus insured that the only negative charge in the peptide is from the sulfate group. Figure 1 shows product-ion spectra of the NCX2+ between YGGFLRR and A3sYA3 by (a) ECD and (b) ETD. Unlike CID, when either ECD or ETD was conducted, the NCX did not dissociate into the parent peptides, instead it mainly produced intramolecular fragments with the electrostatic bond intact. These diagnostic ions allowed for the identification of the residues involved in the electrostatic interaction resulting in NCX formation. YGGFLRR fragmented from the NCX, were exclusively from its N-terminus, producing intramolecular fragments with z ions down to z2. The fragmentation stopped at z2 due to the presence of the two adjacent arginines. Additionally, no intramolecular fragment ions from the C-terminus of YGGFLRR were recorded due to the involvement of the adjacent arginines in the electrostatic interaction. A3sYA3 yielded intramolecular fragments generated from the NCX, mainly from the N-terminus (y6-y4 and y6-NH2 thru y4-NH2); however some weak intramolecular fragments from the C-terminus (a6-a4) are observed. In both cases, the fragmentation stopped at the sulfated tyrosine residue; the site of interaction on the acidic peptide. Furthermore, both ECD and ETD gave similar number and type of fragment ions for the NCX.
Figure 1.
Product-ion spectra of NCX2+ from a peptide mixture of YGGFLRR (BP) and A3sYA3 (AP) with (a) ECD and (b) ETD.
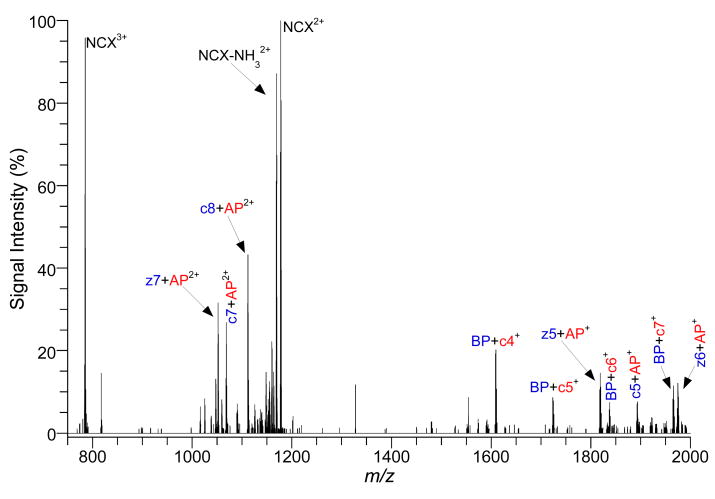
In order to test the utility of ECD/ETD in locating the electrostatic interaction for NCXs, more complex peptides were studied, in which two or more sites of interaction are possible. Figure 2 shows product-ion spectrum of the NCX3+ between SFKRRRSSK and GpSSEDLKKEE by ECD. The three adjacent arginines (residues 4-6) in SFKRRRSSK represent two possible interaction sites while GpSSEDLKKEE has three possible sites of interaction (pS, ED, EE). ECD fragmentation of the NCX3+ produced both a doubly- and singly-charged series of intramolecular fragment ions with the electrostatic interaction remaining intact. Table 1 lists the ECD product-ions that were observed in Figure 2. SFKRRRSSK fragmented off the NCX from the N-terminus producing intramolecular fragments with z ions down to z5 and from the C-terminus producing intramolecular fragments with c ions down to c5. In both directions on the basic peptide, the fragmentation stopped to preserve two adjacent arginines. The presence of z5+ GpSSEDLKKEE and c5+ GpSSEDLKKEE confirm that both possible interaction sites on SFKRRRSSK form noncovalent bonds. However, the z5+ GpSSEDLKKEE peak is more than twice as abundant as the c5+ GpSSEDLKKEE, suggesting that residues 5&6 are favored as an interaction site with the acidic peptide, compared to residues 4&5. GpSSEDLKKEE yielded intramolecular fragments of the NCX from the N-terminus (z9 and y9) and from the C-terminus (c8-c3 and a9, a8). Of the three possible binding sites for GpSSEDLKKEE, only the phosphorylated serine was conserved suggesting that it is the most likely and the favored binding site for the basic peptide, confirming previous findings.8
Figure 2.
Product-ion spectrum of NCX3+ from a peptide mixture of SFKRRRSSK (BP) and GpSSEDLKKEE (AP) with ECD.
Table 1.
Assignments of ECD product-ions from NCX3+ between SFKRRRSSK (BP) and GpSSEDLKKEE (AP).
| Peak Assignment | m/zobs (mono) | m/zcalc (mono) | Relative Intensity (%) |
|---|---|---|---|
| z6+AP+ | 1974.96 | 1974.96 | 12.1 |
| BP+ c7+ | 1965.00 | 1965.00 | 8.30 |
| c5+AP+ | 1892.92 | 1892.92 | 7.03 |
| BP+ c6+ | 1836.91 | 1836.91 | 5.71 |
| z5+AP+ | 1818.86 | 1818.86 | 14.6 |
| BP+ c5+ | 1723.83 | 1723.82 | 8.59 |
| BP+ c4+ | 1608.80 | 1608.80 | 19.2 |
| BP+ c3+ | 1479.75 | 1479.76 | 2.66 |
| NCX2+ | 1176.59 | 1176.59 | 100 |
| NCX-NH32+ | 1168.09 | 1168.08 | 87.1 |
| NCX-2NH32+ | 1159.58 | 1159.57 | 22.2 |
| BP+ y92+ | 1148.07 | 1148.07 | 14.8 |
| BP+ z92+ | 1140.07 | 1140.06 | 5.12 |
| z8+AP2+ | 1125.57 | 1125.57 | 6.87 |
| c8+AP2+ | 1112.08 | 1112.05 | 43.3 |
| BP+ a92+ | 1089.57 | 1089.56 | 4.90 |
| c7+AP2+ | 1068.53 | 1068.53 | 26.8 |
| z7+AP2+ | 1052.03 | 1052.03 | 31.5 |
| BP+ c82+ | 1047.06 | 1047.05 | 13.1 |
| c6+AP2+/BP+ a82+ | 1025.02 | 1025.01/1025.04 | 8.34 |
| NCX3+ (precursor ion) | 784.73 | 784.73 | 95.8 |
Conclusion
This work demonstrates the utility of ECD/ETD for studying interactions leading to the formation of NCX. Both ECD and ETD conserve the electrostatic site of interaction in NCXs and produced similar number and type of intramolecular fragments. In the case of multiple binding sites; sufficient information regarding the peptide backbone was obtained to decipher the most favorable site for the electrostatic interaction in the gas phase. Future studies will include using ECD/ETD to probe for the interaction sites in more complex NCXs involving proteins.
References
- 1.O’Neil KT, DeGrado WF. A Predicted Structure of Calmodulin Suggests an Electrostatic Basis for its Function. Proc Natl Acad Sci USA. 1985;82:4954–4958. doi: 10.1073/pnas.82.15.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woods AS, Ciruela F, Fuxe K, Agnati LF, Lluis C, Franco R, Ferre S. Role of Electrostatic Interaction in Receptor-Receptor Heteromerization. J Mol Neurosci. 2005;26:125–132. doi: 10.1385/JMN:26:2-3:125. [DOI] [PubMed] [Google Scholar]
- 3.Kovacic P, Draskovich CD, Pozos RS. Unifying Electrostatic Mechanism for Phosphates and Sulfates in Cell Signaling. J Recept Signal Transduct. 2007;27:433–443. doi: 10.1080/10799890701699702. [DOI] [PubMed] [Google Scholar]
- 4.Langner M, Kubica K. The Electrostatics of Lipid Surfaces. Chem Phys Lipids. 1999;101:3–35. doi: 10.1016/s0009-3084(99)00052-3. [DOI] [PubMed] [Google Scholar]
- 5.Johnson LN, Lewis RJ. Structural Basis for Control by Phosphorylation. Chem Rev. 2001;101:2209–2242. doi: 10.1021/cr000225s. [DOI] [PubMed] [Google Scholar]
- 6.Schug KA, Lindner W. Noncovalent Binding between Guanidnium and Anionic Groups: Focus on Biological- and Synthetic-Based Arginine/Guanidinium and Interactions with Phosph[on]ate and Sulf[on]ate Residues. Chem Rev. 2005;105:67–114. doi: 10.1021/cr040603j. [DOI] [PubMed] [Google Scholar]
- 7.Woods AS, Huestis MA. A Study of Peptide–Peptide Interaction by Matrix-Assisted Laser Desorption/Ionization. J Am Soc Mass Spectrom. 2001;12:88–96. doi: 10.1016/S1044-0305(00)00197-5. [DOI] [PubMed] [Google Scholar]
- 8.Jackson SN, Wang HYJ, Yergey A, Woods AS. Phosphate Stabilization of Intermolecular Interactions. J Proteome Res. 2006;5:122–126. doi: 10.1021/pr0503578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson SN, Wang HYJ, Woods AS. A Study of the Fragmentation Patterns of the Phosphate-Arginine Noncovalent Bond. J Proteome Res. 2005;4:2360–2363. doi: 10.1021/pr050261d. [DOI] [PubMed] [Google Scholar]
- 10.Woods AS, Wang HYJ, Jackson SN. Sulfation and the up-and-coming post-translational modification: its role and mechanism in protein-protein interaction. J Proteome Res. 2007;6:1176–1182. doi: 10.1021/pr060529g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zubarev RA, Zubarev AR, Savitski MM. Electron Capture/Transfer versus Collisionally Activated/Induced Dissociations: Solo or Duet? J Am Soc Mass Spectrom. 2008;19:753–761. doi: 10.1016/j.jasms.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Zubarev RA. Electron-Capture Dissociation Tandem Mass Spectrometry. Curr Opin Biotech. 2004;15:12–16. doi: 10.1016/j.copbio.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Coon JJ, Syka JEP, Shabanowitz J, Hunt DF. Tandem Mass Spectrometry for Peptides and Protein Sequence Analysis. BioTechniques. 2005;38:519–523. doi: 10.2144/05384TE01. [DOI] [PubMed] [Google Scholar]
- 14.Haselmann KF, Jorgensen TJD, Budnik BA, Jensen F, Zubarev RA. Electron Capture Dissociation of Weakly Bound Polypeptide Polycationic Complexes. Rapid Commun Mass Spectrom. 2002;16:2260–2265. doi: 10.1002/rcm.853. [DOI] [PubMed] [Google Scholar]
- 15.Loo JA. Electrospray Ionization Mass Spectrometry: A Technology for Studying Noncovalent Macromolecular Complexes. Int J Mass Spectrom. 2000;200:175–186. [Google Scholar]