Abstract
Mechanisms that regulate the growth and form of the vertebrate skeleton are largely unknown. The zebrafish mutant rapunzel has heterozygous defects in bone development, resulting in skeletal overgrowth, thus identification of the genetic lesion underlying rapunzel might provide insight into the molecular basis of skeletogenesis. In this report, we demonstrate that the rapunzel mutant results from a missense mutation in the previously uncharacterized rpz gene. This conclusion is supported by genetic mapping, identification of a missense mutation in rapunzelc14 in a highly conserved region of the rpz gene, and suppression of the rapunzel homozygous embryonic phenotype with morpholino knockdown of rpz. In addition, rpz transcripts are identified in regions correlating with the homozygous embryonic phenotype (head, pectoral fin buds, somites and fin fold). This report provides the first gene identification for a mutation affecting segment number in the zebrafish fin and development of both the fin ray (dermal) and the axial skeleton.
Keywords: zebrafish, fin, bone, skeleton, growth, overgrowth
Introduction
The molecular mechanisms underpinning growth and the establishment of proper size and form are largely unknown. Vertebrate morphology is inextricably related to the growth and form of the supporting skeletal structures. Identifying mutations that result in defects in bone morphology might provide insights regarding the molecular mechanisms that regulate bone shape and size.
We used the zebrafish fin and axial skeleton as a model to examine genes critical for vertebrate bone development. Fin rays are composed of multiple segments, each segment in turn comprised of two hemirays (lepidotrichia) of dermal bone in apposition, surrounding an intra-ray mesenchyme (Santamaria et al., 1992). In addition to bone, the mesenchymal compartment contains nerves, blood vessels, pigment cells and undifferentiated fibroblasts, all surrounded by a basement membrane and an overlying epithelium. Lepidotrichia are covered on both surfaces by a monolayer of bone-forming osteoblasts (scleroblasts) that form as undifferentiated mesenchymal cells in the distal portion of the intraray proliferate (Goldsmith et al., 2003), condense laterally along actinotrichia, differentiate, and begin secreting bone matrix (Goss, 1978; Haas, 1962). Fin growth occurs via the sequential, distal addition of new segments of bone to each fin ray (Goss and Stagg, 1957; Haas, 1962) with fin length ultimately being determined by the number and size of these individual fin ray segments (Iovine and Johnson, 2000). Zebrafish mutants have been described that affect both the number of segments (rapunzel and long fin) (Goldsmith et al., 2003; Iovine and Johnson, 2000) and the size of individual segments (short fin) ((Iovine and Johnson, 2000) and some molecular details of these events have been elucidated. For example, segment length is regulated in part by the function of the gap junction protein, connexin 43 (cx43) and hypomorphic alleles of the zebrafish cx43 gene result in the zebrafish short fin mutant (Hoptak-Solga et al., 2007; Iovine et al., 2005). To date, the molecular identity of mutations affecting segment number have not been published, nor have mutations been described that affect growth of both the fin ray and the axial skeleton.
We have cloned and characterized the zebrafish mutant rapunzel. Adult zebrafish display a heterozygous overgrowth phenotype of both the fin ray and the axial skeleton. In addition, rapunzel has a distinct, homozygous lethal embryonic phenotype (Goldsmith et al., 2003 and Figure 1). Utilizing this embryonic phenotype, we identified a missense mutation in a previously undescribed gene (rpz) of unknown function. Sequence analysis demonstrated that this mutation is not a common polymorphism and morpholino knockdown of rpz completely suppressed the rapunzel embryonic phenotype, demonstrating that rapunzel is a gain of function allele. In addition, extensive analysis of the rapunzel critical region demonstrates that rpz founds a family of five related genes. Finally, the region of the predicted rapunzel protein containing the missense mutation is highly conserved in teleosts. Therefore rapunzel provides an opportunity to gain new information about the fundamental mechanisms shaping vertebrate morphology.
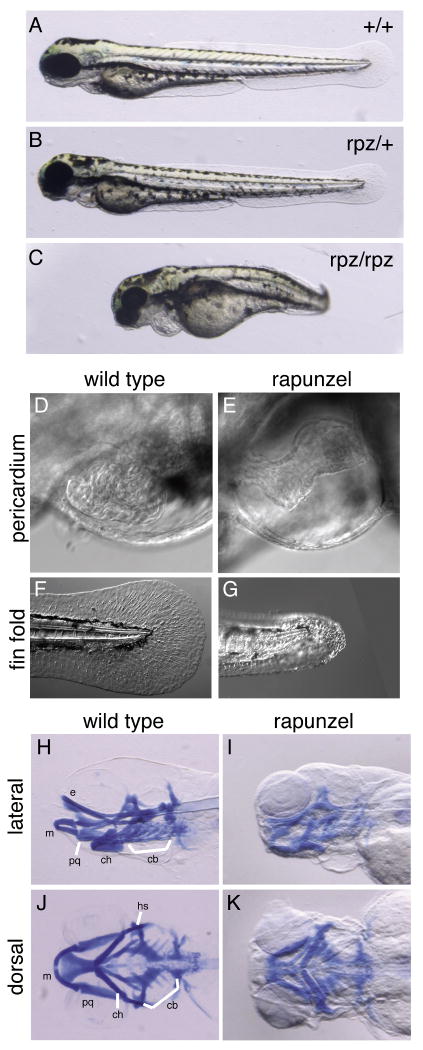
Figure 1.
Phenotype of rapunzel embryos. A-C. Wild type, heterozygous rapunzel and homozygous rapunzel embryos. D-G. Nomarski images of wild type (D, F) and homozygous rapunzel (E, G) embryos at 72hpf demonstrating pericardial edema (E) and fin fold defects (G) in rapunzel mutants. H-K. Lateral (H, I) and ventral (J, K) views of whole mount Alcian blue-stained wild type (H, J) and rapunzel (I, K) homozygous mutants at 96 hpf demonstrating reduced development of the craniofacial cartilages in rapunzel mutants. Craniofacial structures labeled as in Piotrowski et al., 1996; bh: baihyal, cb: ceratobranchial, ch: ceratohyal, e: ethmoid plate, hs: hyosymplectic, m: Meckel's cartilage, and pq: palatoquadrate.
Materials and Methods
Fish husbandry
Wild type fish stocks used for these studies were from the C32 (Rawls et al., 2003) and AB strains. Fish were reared at a constant temperature of 25 °C and maintained on a 14L: 10D photoperiod. Fish were fed three times daily with both micropellets (Hikari, Aquatic Eco-Systems) and brine shrimp (Biomarine, Aquafauna Biomarine).
Alizarin red and Alcian blue staining
Zebrafish were euthanized in Tricaine and fixed in 4% buffered paraformaldehyde. Embryos were then stained for developing cartilage with Alcian blue as described by Schilling et al., 1996. Embryos and larvae were stained with alizarin red as described by Walker and Kimmel, 2007. Following staining, embryos were stored in glycerol. Whole-mount specimens were imaged using a Nikon SMZ1500 stereomicroscope and photographed using Nikon ACT-1 software. Images were processed in Photoshop CS3 (Adobe).
Calcein staining
A 0.2% calcein solution was made by dissolving 2g of calcein in 1L of MilliQ water and adjusting the pH to ∼7.0 with NaOH. Fish were immersed in calcein solution for 5-10 minutes and then washed with fresh system water for 10 minutes to clear away excess dye as previously described (Du et al., 2001). Fish were then anesthetized in 0.03% Tricaine for viewing and imaging as described above.
Sirius red staining
Wild type and rapunzel heterozygous fish (18 months of age) were euthanized in Tricaine, their viscera removed, and fixed over night in 4% buffered paraformaldehyde at 4 degrees. Following fixation the fish were decalcified in 0.35M EDTA, pH 7.8 for several days. Fish were then embedded in paraffin and 5 μM sections were cut on a microtome. Sections were then stained with Sirius red as previously described (Blumer et al., 2004).
MicroCT and DEXA Scanning
rapunzel heterozygotes (2 months, 8 months, 18 months) and their wild type siblings were euthanized in Tricaine and fresh frozen at -20 degrees until scanned. Prior to scanning the fish were embedded in 1% agarose. Fish were scanned on a μ CT40, Scanco Medical (5 kV, 177 A, 200 ms, 16 μ m voxel size). Manufacturer's software (Eval v6.0) was used to calculate bone volume and apparent mineral density (calibrated to the manufacturer's hydroxyapatite [HA] mineral phantom). For DEXA scanning, total body bone mineral density (BMD) was measured by DEXA using a PIXImus scanner GE/Lunar; Madison, WI). Calibration was performed daily with a standard phantom as suggested by the manufacturer. The precision of whole-body BMD, assessed by the root mean square method is 1.34% (coefficient of variation).
Microscopy
General microscopy was performed with a Nikon SMZ1500 stereomicroscope and images captured using Nikon DXM1200F digital camera and Nikon ACT-1 software. Fluorescent images were taken using an Olympus MVX10 microscope and Olympus MicroSuite software. Nomarski images were captured using an Olympus IX71 microscope and MicroSuite Biological Suite software. All images were processed using Photoshop CS2 (Adobe).
Mapping
We localized the rapunzelc14 mutation to linkage group (LG) 16 by centromere linkage analysis (Johnson et al., 1996) using simple sequence repeat (SSR) markers (Shimoda et al., 1999). SSR markers were then used to finely map the rapunzelc14 mutation (780 meioses with zero recombinations). In addition to mapping with SSR markers, we also developed single nucleotide polymorphism (SNP) markers from genes localized to this region (Hukriede et al., 2001). A description of these markers and their respective polymorphisms may be found in supplemental Table 1. 3′ and 5′ rapid amplification of cDNA ends (RACE) was then used to clone two of the genes in the rapunzel critical region (rpz and rpz2). Total RNA was isolated from wild type or homozygous rapunzel embryos at 72 hpf using a standard Trizol (Invitrogen) extraction. Wild type and rapunzel mRNA was then isolated using the Poly (A) Purist Kit (Ambion). Wild type or rapunzel mRNA was used as a template to generate 1st strand, RACE-ready cDNA (SMART RACE, Clontech). 5′ and 3′ RACE PCR primers were designed (PrimerSelect, Lasergene) against coding sequences for the putative transcripts contained in the rapunzel critical region, based upon EST assemblies (http://zfish.wustl.edu). Following PCR amplification, RACE products were separated on 1% agarose gels, excised, purified (Quiagen Gel Purification Kit), and TA cloned into the pCRII vector (Invitrogen). Inserts were sequenced at the Washington University Genome Sequencing Center using the T7 and Sp6 sites in the vector. cDNA contigs were assembled (SeqMan, Lasergene) and designing PCR primers that spanned the gaps and sequencing amplicons resolved gaps. The remaining three paralogues in the rpz family (rpz3, rpz4 and rpz5) were identified by performing tBLASTx and BLAST2 searches against the rpz sequence. To search for additional mutations in these three paralogues, the single exons were sequenced from both rapunzel mutant and wild type DNA. CLUSTALW analysis was then performed to determine relatedness of the rpz family genes. Genbank accession numbers for the rpz family of genes are: FJ539169 (rpz), FJ539170 (rpz2), FJ539171 (rpz3), FJ539172 (rpz4), and FJ539173 (rpz5).
In situ hybridization
Templates for sense and anti-sense probes were cloned by RT-PCR using forward (ACACAATGGCCGACAATGAAT) or reverse (TAGAGGAAAAAGGCAATAAAGAGC) primers targeted against rpz. PCR amplicons were gel-purified (Qiagen) and TA-cloned (pCRII, Invitrogen). Insert-containing plasmids were grown and harvested (Maxi-prep, Qiagen). Plasmids were linearized using BamH1 or EcoRV to serve as template for transcription of sense or anti-sense probes, respectively. Digoxigenin-labeled RNA probes were transcribed from these templates using SP6 (EcoRV) or T7 (BamH1) RNA polymerase. Whole-mount in situ hybridization was performed as described in Jowett and Yan, 1996. Following staining, embryos were washed into MeOH for storage at -20°C. Embryos were cleared in benzyl benzoate: benzyl alcohol (2:1) for microscopy. For ISH on sections the fish were embedded in OCT compound and frozen in an ethanol/dry ice bath, then stored at -80° until sectioned. 40 μm sections were mounted on Superfrost plus slides (Fisher) and ISH was performed in slide boxes.
Quantitative RT-PCR
To quantify rpz transcript in embryos, rpz/+ adults were intercrossed and RNA was prepared from individual embryos at 24 hpf. 1st strand cDNA was then prepared and sequenced to determine the genotype of each embryo. Quantitative RT-PCR (qPCR) was done in triplicate for both the reference (forward CCCTCACCAAACCTAAGATCGT; reverse CTCCAGTTTGCCCTGATCTTG) and rpz (forward GACCATCCAATCGTTTTCACT; reverse GCTTCAGCTTCTCGGGGTCAT) primers. ∼500 ng cDNA was used in each rtPCR reaction. Rpl32, a ribosomal protein, was used as a reference for the reactions. Fold change for each embryo was determined using the delta-delta Ct method. Data were normalized to the wild type embryos. For qPCR on adult fins, RNA was purified from amputated fins using a standard Trizol (Invitrogen) extraction. RNA was used as a template for 1st strand cDNA transcription and cDNA was used for each qPCR reaction as above. Data were normalized to transcript abundance in wild type fins.
Morpholino injections
To inhibit gene translation, a morpholino oligonucleotide (Gene Tools) targeting the start site of rpz (5′-CATCGAATAATTCATTGTCGGCCAT-3′) was injected into fertilized zebrafish embryos at the one-cell stage. A 5 base pair mismatched morpholino (5′-CATGGAATAATTGATTCTCCGCGAT-3′) and a morpholino targeting the start site of rpz2 (a separate gene in the rapunzel critical region) (5′-GCTGATTGTCTGCCATAGATGGATC-3′) were used for control injections. A 3′ fluorescein tag facilitated visualization of injected morpholinos. The morpholino was resuspended in 1× Danieau media (58 nM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca (NO3)2 and 5.0 mM HEPES pH 7.6). Injections were made at a dilution of 1:2 with 1× Danieau media and phenol red (1:10) to visualize injection site.
Isolation and analysis of genomic DNA
At 72 hpf, individual embryos were photographed. Genomic DNA was extracted from each embryo by placing individual embryos in 50μl of DNA extraction buffer (80mM KCl, 10mM Tris pH=8, 1mM EDTA, 0.3% Tween, 0.3% Igepal). The embryos were placed at 98°C for 10 minutes, 4°C for 10 minutes. Proteinase K was then added to a concentration of 1mg/ml. The samples were then incubated at 55°C for 75 minutes, 98°C for 10 minutes and then stored at -20°C for analysis by PCR. Primers (F: 5′-GAGAGTTCGGCAGCGTCAAT-3′ and R: 5′-TCTCCCAGGATGCCGTAGTAGC-3′) surrounding the rapunzelc14 mutation were used in PCR reactions using the genomic DNA of each embryo as template. PCR products were purified (Invitrogen), digested with DdeI, and analyzed by gel electrophoresis. The digestion pattern indicated the genotype (+/+, rpz/+, rpz/rpz) of each embryo.
Results
rapunzel has a homozygous, lethal embryonic phenotype
rapunzel is an ENU-induced mutant with a heterozygous skeletal overgrowth phenotype (Goldsmith et al., 2003). In addition, rapunzel embryos have a homozygous lethal phenotype, dying 4-5 days post fertilization (dpf). At 3 dpf, homozygous rapunzel embryos have a curved, shortened body axis (Figure 1A-C), pericardial edema (Figure 1D, E), a poorly formed fin fold (Figure 1F, G), and a poorly developed jaw (Figure 1H-K). In contrast, heterozygous rapunzel embryos cannot be distinguished from their wild type siblings (Figure 1A, B). Homozygous rapunzel embryos also have generalized defects in vascular development and hematopoiesis (data not shown). We took advantage of the homozygous rapunzel phenotype to clone the rapunzel mutant.
The rapunzel phenotype is caused by a missense mutation in the novel rpz gene
We fine mapped rapunzel to a 46 KB critical region on chromosome 16 (Figure 2A). This critical region contained three novel, paralogous genes as evidenced by mapped (http://www.ensembl.org/Danio_rerio/) EST sequences and tBLASTx pair-wise comparisons. Because none of the three paralogues in the critical region seemed a more likely candidate for rapunzel than the others, we examined the coding sequences of all three genes in wild type zebrafish and in rapunzel mutants.
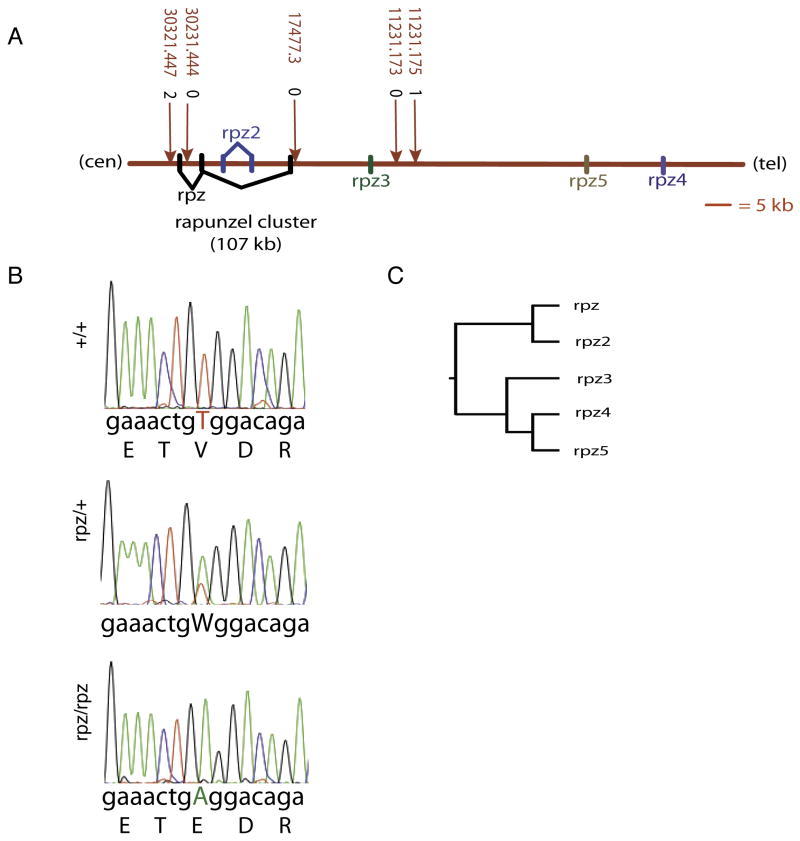
Figure 2.
Cloning of rapunzelc14. A. A meiotic map representing approximately 47 kb of the rapunzelc14 region on chromosome 16. The critical region contains three paralogous genes (rpz, rpz2 and rpz3). Two additional paralogues (rpz4 and rpz5) are also present on chromosome 16, outside of the critical region. All five genes are located on the same strand. Marker positions are shown along with their number of crossovers from rapunzel (see supplemental data for more details regarding marker descriptions). B. Electropherograms showing the rpz T269A mutation in heterozygous and homozygous rapunzel embryos. W indicates the single nucleotide change of thymine to adenine present on chromosomes carrying the rapunzelc14 allele at position 269. The translated amino acid sequence for wild type and rpz homozygotes is shown below each electropherogram. The T269A mutation results in an amino acid change from valine to glutamic acid at amino acid 90 in the encoded protein. C. A dendogram demonstrating that the five rapunzel-like paralogues subdivide into two clades.
We used 3′ and 5′ random amplification of cDNA ends (RACE) to clone two of the three paralogous genes in the rapunzel critical region. A large open reading frame for the third paralogue was obtained by sequencing genomic DNA from rapunzel mutants and wild type zebrafish. One of the three genes, heretofore referred to as rapunzel (rpz), contained a missense mutation (T269A) in the mutant cDNA sequence (Figure 2B). This T269A mutation in rpz would result in a non-conserved amino acid change (V90E) in the predicted protein sequence (Figure 2B). To exclude the possibility T269A might be a common polymorphism, we partially sequenced rpz from 4 genetic strains of zebrafish: AB, WIK, C32 and SJD. rapunzel arose on an AB genetic background and neither our AB laboratory strain nor the other 3 polymorphic strains (WIK, C32, SJD) harbored a T269A polymorphism, although a synonymous polymorphism (G345A) was identified in C32 and SJD. In addition, a BLAST search was performed to identify all EST's demonstrating rpz sequence similarity. Four EST's showed sequence similarity with rpz coding sequence. None of these EST's contained the T269A mutation, although a non-synonymous polymorphism was identified at a different site that results in a neutral amino acid change (H169Q). Finally, to be certain that the T269A mutation was not introduced during transcription of the cDNA RACE template, we partially sequenced rpz from genomic DNA in 20 homozygous mutant rapunzel embryos (scored by phenotype). In every case, genomic DNA from a rapunzel mutant harbored the T269A mutation (data not shown). No differences between mutant and wild type sequence were identified in the remaining two paralogues within the critical region, referred to as rapunzel 2 (rpz2) and rapunzel 3 (rpz3).
rpz founds a family of five paralogous genes in zebrafish
In addition to rpz, rpz2 and rpz3, further tBLASTx searches revealed two additional paralogues on chromosome 16 but outside of the critical region, which we have named rapunzel 4 (rpz4) and rapunzel 5 (rpz5) (Figure 2A). Even though rpz4 and rpz5 were outside of the critical region, we sequenced open reading frames for both genes from mutant and wild type DNA to further characterize the rpz gene family and to ask whether any other paralogues contained additional mutations present in the mutant, but not the wild type allele. No sequence differences were identified. A dendrogram (Figure 2C) reveals that the five members of the rpz family sort into two main clades.
rapunzelc14 is a gain of function allele
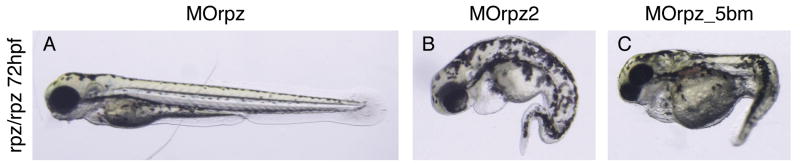
To establish causality between the rapunzelc14 genotype and phenotype, we took advantage of rapunzel's homozygous phenotype. We first injected a morpholino oligonucleotide directed against the putative translation start site of rpz (MOrpz) into wild type embryos. Wild type embryos were injected at the 1-cell stage with 2-5 ng of MOrpz or its 5 base pair mismatched control morpholino (MOrpz_ 5bm). 98% of wild type embryos injected with MOrpz had a normal phenotype at 3 dpf (n=95). A normal phenotype was observed in 100% of control-injected (MOrpz_5bm) embryos (n=45). At higher concentrations, injections of both MOrpz and MOrpz_5bm had non-specific effects of similar magnitude (data not shown). These data raised the possibility that rapunzelc14 is not a loss of function allele and furthermore, that rpz is not essential for early embryonic development. We therefore hypothesized that rapunzelc14 is a gain of function allele. To test this hypothesis, we injected embryos derived from adult rapunzel/+ intercrosses with 2-5 ng of either MOrpz or MOrpz_5bm. Injection of MOrpz suppressed the homozygous rapunzel phenotype, with all of the embryos injected with MOrpz displaying a wild type phenotype at 3 dpf (n=132) (Table 1 and Figure 3). To prove that MOrpz was indeed rescuing homozygous rapunzel embryos, we took advantage of the fact that the T269A mutation introduces a DdeI restriction site into the rpz coding sequence. All injected embryos were individually photographed 3 dpf followed by DNA extraction. Genotype was then established by DdeI digest (not shown). Twenty-four percent (32/132) of the embryos injected with MOrpz had the homozygous rapunzel genotype, but a normal phenotype (Table 1 and Figure 3A). For control (MOrpz_5bm) injected embryos, 21% (16/75) had a rapunzel phenotype (not statistically dissimilar from the 25% Mendelian prediction [Table 1 and Figure 3C]) and of these 16 embryos, 94% (15/16) had a homozygous rapunzel genotype confirmed by DdeI digest. Thus misclassification of an abnormal appearing embryo as a rapunzel mutant was an extremely uncommon event. As an additional control, we used a morpholino to knock down rpz2 (MOrpz2) (the topology of the rapunzel critical region is such that rpz2's two exons lie within the second intron of rpz [Figure 2A]). 28% (n=53) of MOrpz2 injected embryos had a rapunzel phenotype (again not significantly different from the 25% predicted by Mendelian inheritance [Table 1 and Figure 3B]) and 100% (15/15) of the phenotypically abnormal embryos were confirmed to be rapunzel homozygotes by DdeI digest. Together, these data demonstrate that the rapunzel embryonic phenotype results from a gain of function mutation in the rpz gene.
Table 1.
Suppression of the rapunzel embryonic mutant phenotype following injection of a morpholino oligonucleotide targeting the translation start site of rpz.
Suppression of the rapunzel embryonic mutant phenotype following injection of morpholino oligonucleotide to translation start site of rpz. Adult rapunzel heterozygotes were intercrossed and the resulting embryos injected with a rpz morpholino (1:2 dilution in 1× Danieau buffer), a rpz2 morpholino (1:2 dilution in 1× Danieau buffer), and a 5 base pair mismatched control morpholino (1:5 dilution in 1× Danieau buffer) at the 1-cell stage. At 72 hpf, individual embryos were photographed and collected for genomic DNA extraction. The rpz/rpz phenotype indicates the number of embryos resembling the rapunzelc14 embryonic mutant phenotype when photographed at 72 hpf. Primers flanking the rapunzelc14 mutation (T269A) were used in PCR reactions using the genomic DNA of each embryo as template. The PCR products were purified (Invitrogen), digested with DdeI, and analyzed by gel electrophoresis. The digestion pattern indicated the genotype (+/+, rpz/+, rpz/rpz) of each embryo. The # confirmed rpz/rpz indicates the number of embryos with the rpz/rpz digestion pattern after gel electrophoresis. The fraction of confirmed rpz/rpz embryos equals the # confirmed rpz/rpz divided by the total embryos.
| Embryos | Injection | Total Embryos | rpz/rpz phenotype | # confirmed rpz/rpz | Fraction confirmed rpz/rpz |
|---|---|---|---|---|---|
| rpz/+ i.c. | MOrpz | 132 | 0 | 32 | 0.24 |
| rpz/+ i.c. | MOrpz2 | 53 | 15 | 15 | 0.28 |
| rpz/+ i.c. | MOrpz_5bm | 75 | 16 | 15 | 0.20 |
Adult rapunzel heterozygotes were intercrossed and the resulting embryos injected at the 1-cell stage with a morpholino targeting rpz (MOrpz) (1:2 dilution in 1× Danieau buffer). For controls, we injected either a morpholino targeting rpz2 (MOrpz2) (1:2 dilution in 1× Danieau buffer) or a 5 base pair mismatched morpholino against rpz (MOrpz_5bm) (1:5 dilution in 1× Danieau buffer). At 72hpf, individual embryos were photographed and collected for genomic DNA extraction. The rpz/rpz phenotype indicates the number of embryos resembling the rapunzelc14 embryonic mutant phenotype when photographed at 72hpf. For each embryo, the region flanking the rapunzelc14 mutation (T269A) was PCR amplified using the genomic DNA of each injected embryo as template. PCR amplicons were purified (Invitrogen), digested (DdeI) and analyzed by gel electrophoresis. The restriction digest pattern identifies the genotype of each embryo (+/+, rpz/+, rpz/rpz). The # confirmed rpz/rpz indicates the number of embryos with the rpz/rpz genotype. The fraction of confirmed rpz/rpz embryos equals the # confirmed rpz/rpz embryos divided by the total embryos.
Figure 3.
Morpholino knockdown of rpz suppresses the homozygous rapunzel phenotype. Heterozygous rapunzel adult zebrafish were intercrossed and the resulting embryos injected at the 1-2 cell stage with either a morpholino targeting the start site of rpz (MOrpz) (A), a morpholino targeting rpz2 (MOrpz2) (B) or a rpz 5 base pair mismatched control morpholino (MOrpz_5bm) (C). A. Injection of MOrpz suppresses the homozygous embryonic rapunzel phenotype. The rapunzel homozygous phenotype was not suppressed in embryos injected with MOrpz2 (B) or MOrpz_5bm (C). PCR, DdeI digestion and electrophoresis confirmed that the embryos in A, B, and C are all homozygous for the rapunzelc14 allele (not shown).
Expression of rpz in wild type and mutant embryos
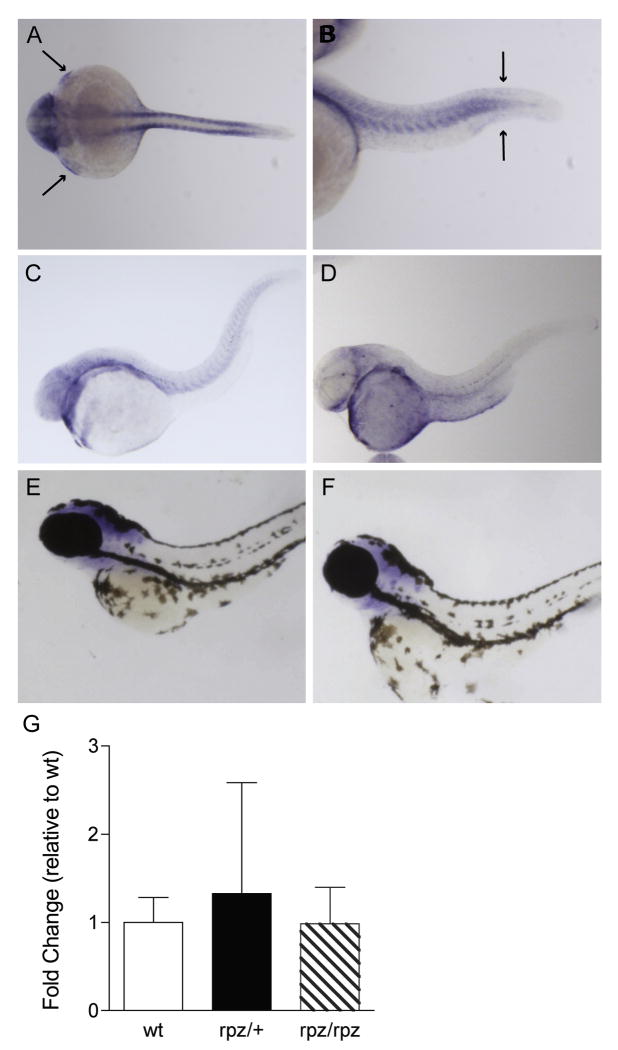
We hypothesize that the homozygous phenotype in rapunzel arises because of the critical alteration the genetic lesion (T269A) brings about in protein structure (V90E) and function. To provide further evidence that the T269A allele does not simply segregate in rapunzel with a separate lesion affecting the expression of an otherwise functionally normal protein product, we first looked at rpz expression in mutant and wild type embryos using in situ hybridization (ISH). At 24 hpf, rpz transcripts are localized in areas consistent with the embryonic phenotype including the somites, tail bud region adjacent to the fin fold, pectoral fin buds and the head (Figure 4 A, B). We then asked whether rpz is expressed later during development, when definitive cartilaginous and bony elements of the skeleton begin to form. At 48 hpf, rpz expression is beginning to decrease, although the general distribution is unchanged. By 120 hpf, rpz transcript is almost completely absent (Figure 4). Importantly, we could define no significant differences in rpz expression between mutant and wild type embryos by ISH (data not shown), consistent with the hypothesis that because T269A is a missense allele, transcript localization and quantity is unlikely to be affected. To further support the notion that rapunzelc14 is not a misexpression allele, we performed qPCR on mutant and wild type embryos. At 24 hpf, no significant differences in levels of rpz transcript are noted between wild type, heterozygous rapunzel and homozygous rapunzel embryos (one way ANOVA, p=0.84) (Figure 4C).
Figure 4.
Expression pattern and qPCR of rpz. A. Dorsal view of an ISH for rpz in a wild type embryo at 24 hpf. Transcript is clearly seen in the somites, pectoral fin buds (small arrows) and head. B. In a lateral view, transcript is seen in the somites and in the tail bud adjacent to the fin fold (large arrows). C-F. Lateral views of wild type embryos at 48 hpf (C), 72 hpf (D), 80 hpf (E), and 120 hpf (F) following ISH for rpz. Expression is beginning to decrease at 48 hpf, and further still at 72 hpf, but the general distribution is unchanged from 24 hpf embryos. By 80 hpf rpz expression is localized exclusively to the head (E) and by 120 hpf expression is almost completely absent (F). G. Quantitative RT-PCR on wild type, heterozygous and homozygous rapunzel embryos reveals no differences in rpz transcript abundance. Data are normalized to the wild type embryo. Data represent means ± SD.
rpz is a highly conserved gene in teleosts
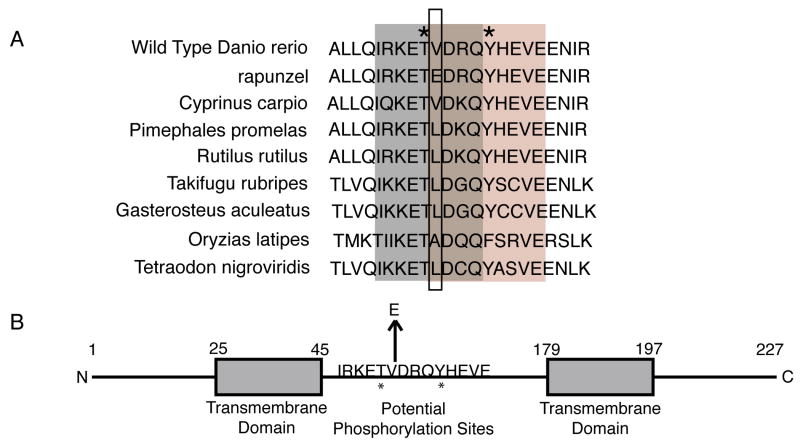
rpz encodes a predicted 227 amino acid protein. A blastp search revealed no significant homologies aside from orthologous genes in other teleosts; moreover a search of the Pfam database of protein families reveals no significant matches. Thus, rpz encodes a novel protein of unknown function. A translated blast search (tblastn) of multiple databases (nr, EST, HTGS, WGS) did reveal rpz orthologues in other teleosts, including cyprinid species closely related to Danio rerio (e.g. Cyprinus carpio) and quite distantly related teleosts (e.g. Takifugu rubripes). Given the dramatic phenotype caused by the V90E mutation and the fact that this lesion is embryonic lethal, we hypothesized that Rpz protein homologs would be highly conserved, particularly around the site of the rapunzel mutation (V90). Consistent with this hypothesis, a partial alignment of Rpz predicted protein homologs (Figure 5A) demonstrates that the V90 amino acid residue is invariably a valine, alanine, or leucine in other teleost species. Also, all rpz paralogs have a nonpolar amino acid at this position (not shown). To date, only the rapunzelc14 allele contains a non-conserved amino acid (glutamic acid) at the position normally occupied by V90.
Figure 5.
Conservation of the amino acid sequence of the rpz gene. A. A translated blast search (tblastx) of the EST database revealed rpz orthologues in cyprinid species closely related to Danio rerio. [Genbank accession numbers for each species: Rutilus rutilus (EG531508), Cyprinus carpio (CF660604), Pimephales promelas (DT229800)] Other related teloests were identified by a tblastx search in EMBL zv.7 (http://www.ensembl.org/Multi/blastview) using the species of interest. Each protein sequence was then aligned in the area flanking the rpz mutation to look for homology. The box outlines the amino acid change in rapunzelc14. Only rapunzelc14 contains a non-neutral amino acid at this position. NetPhos 2.0 identified putative threonine and tyrosine phosphorylation sequences shown in grey and pink, respectively. Asterisks indicate the putative phosphorylation sites. B. A schematic of the predicted Rpz protein structure. Numbers indicate the amino acid position. The arrow highlights the V90E mutation. Asterisks indicate putative phosphorylation sites predicted by NetPhos 2.0. Putative transmembrane domains are shown as boxes at positions predicted by TMpred.
Adult rapunzel heterozygotes display overgrowth of both the fin ray and the axial skeleton
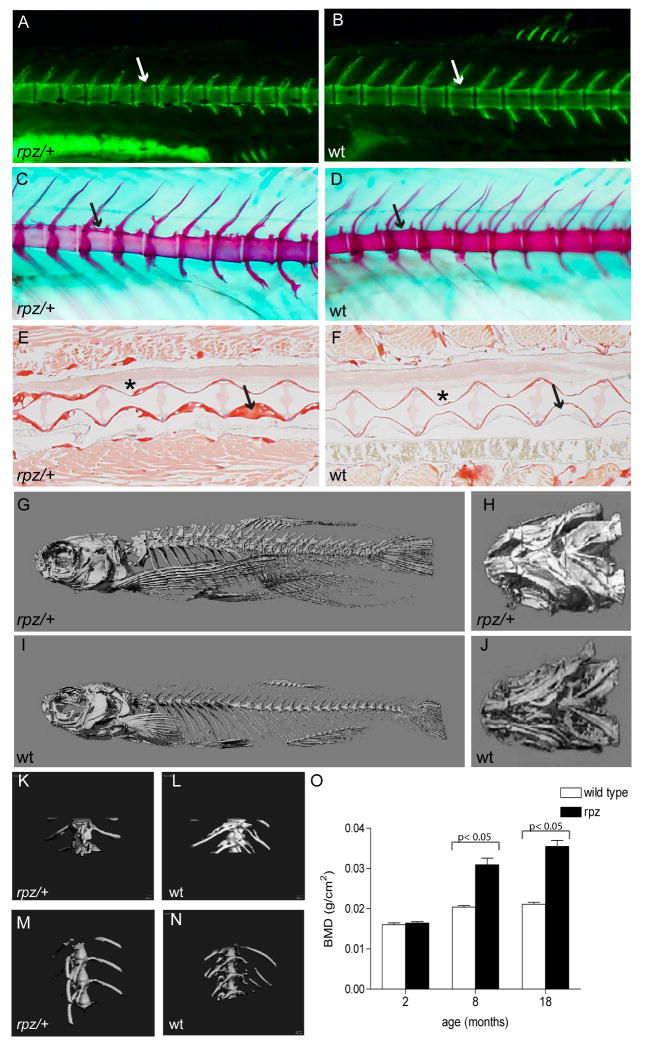
rapunzelc14 is a homozygous, lethal, gain of function allele in zebrafish embryos. In adult zebrafish, rapunzelc14 has a heterozygous fin phenotype consisting of an excess number of fin ray segments (Goldsmith et al., 2003). In addition to overgrowth of the fin ray skeleton, adult rapunzelc14 heterozygotes have pronounced hyperossification of the axial skeleton (Figure 6). We performed bone histomorphometry using Alcian blue, alizarin red, calcein, and Sirius red staining, along with microCT and bone densitometry (DEXA) to more closely define the skeletal overgrowth phenotype of rapunzel heterozygotes. Alcian blue, alizarin red and calcein staining were initially used to ask whether hyperossification develops during early skeletal development (3-10 dpf) in rapunzel heterozygotes. In particular, we focused on the developing craniofacial skeleton for this question, as these are the first skeletal elements to form in developing zebrafish. At these early stages of skeletogenesis, no evidence of hyperossification is noted (Figure 7). Using in vivo calcein staining, we first begin to see differences in bone morphology in rapunzel heterozygotes as compared to their wild type siblings in two week-old larvae (Figure 6 A, B). The vertebrae are less scalloped in shape, although the intervertebral spaces are preserved. A similar phenotype is observed with Alcian blue/alizarin red stained larvae at three weeks of age (Figure 6 C, D). Of note, we do not yet see a hyperossification phenotype in these 2-3 week old heterozygous rapunzel larvae. A loss of scalloping in the vertebral bodies persists into adulthood (18 months), however by this time hyperossification of the axial skeleton is readily seen by Sirius red staining. Sections through the vertebral column show rapunzel heterozygotes have an increased amount of bone compared to their wild type siblings (Figure 6 E, F). We used both microCT (Fig 6 G-N) and bone densitometry (DEXA) (Fig 6 O) to define the time course over which the hyperossification develops in heterozygous rapunzel mutants. Increased bone mineral density in heterozygous rapunzel mutants compared to their wild type siblings is not evident at 2 months of age, but is easily quantifiable at 8 and 18 months of age.
Figure 6.
Adult rapunzel mutants have overgrowth and morphological changes in the axial skeleton. A, B. In two week-old larvae stained with calcein, the vertebrae of rapunzel heterozygotes (A) are less scalloped (arrows) compared to their wild type siblings (B). C, D. Similarly, Alizarin red and Alcian blue staining show a loss of scalloping (arrows) in the vertebral bodies in 3 week-old heterozygous rapunzel larvae (C) in comparison to their wild type siblings (D). No skeletal hyperossification is seen at these early developmental stages (A-D). E, F. The loss of scalloping persists into adulthood (18 months old) and can be seen (asterisk) in Sirius red stained sections from rapunzel heterozygotes (E) compared to their wild type siblings (F). Sirius red-stained sections through the vertebral column also demonstrate that rapunzel heterozygotes have increased bone deposition compared to wild type fish (arrows). G-J. MicroCT's of 18 month-old fish demonstrate marked hyperossification in rapunzel heterozygotes compared to wild type siblings, most easily seen in the vertebral (G, I) and ventral craniofacial (H, J) skeletons. K-L. In higher power views of the mid vertebrae, hyperossification is also readily apparent in 18 month-old rapunzel heterozygotes (K) compared to wild type (L). M-N. Hyperossification is not seen in the vertebrae of 2 month-old rapunzel mutants (M) when compared to age-matched siblings (N). O. This finding is corroborated by measurements of total bone mineral density, where significant differences between rapunzel mutants and their age-matched siblings are seen at 8 and 18 months, but not at 2 months (data represent mean ± SD).
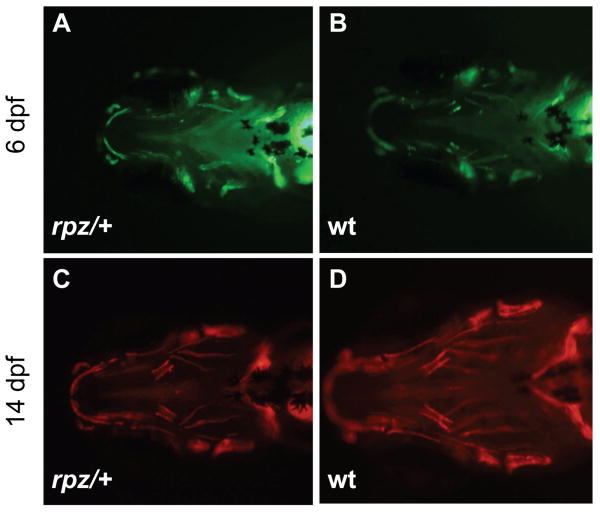
Figure 7.
rapunzel heterozygotes show no evidence of hyperossification during early craniofacial development. A, B. Calcein staining at 6 dpf shows a similar degree of mineralization of the craniofacial skeletons of rapunzel mutants (A) and their wild type siblings (B). C, D. Alizarin red staining at 14 dpf again shows no increased ossification of the craniofacial bones in rapunzel mutants (C) versus their wild type siblings (D). Similar results were seen at other time points between 4-16 dpf (not shown).
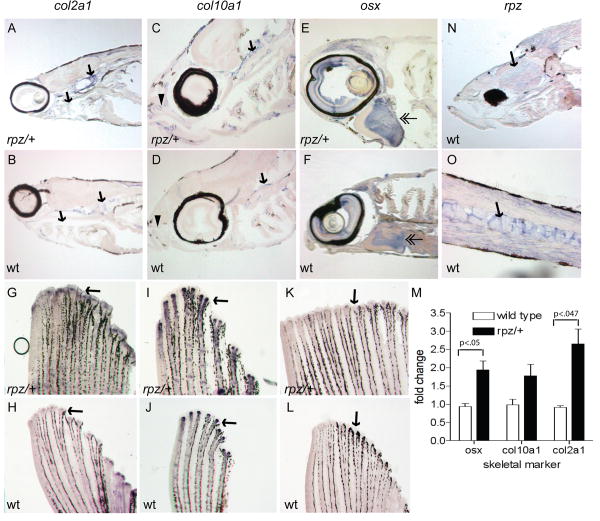
Although we did not observe a skeletal phenotype in rapunzel heterozygotes at pre-larval stages, we used ISH to ask whether the expression of well-defined markers of skeletal development is altered. ISH was performed on 48-120 hpf embryos (heterozygous rapunzel and their wild type siblings) using markers of cartilage (col2a1) and bone (col10a1, osx) development. Consistent with previously published data, we saw strong expression in the pharyngeal arches, otic vesicle, endochondral disc, and notochord of col2a1 (Yan et al., 2005) (Supplemental Figure). Expression in the developing bony skeleton, including the cleithrum, opercle, parasphenoid, and brachiostegal ray was seen with col10a1 and osx (Li et al., 2009) (Supplemental Figure). Importantly, at these pre-larval stages we did not detect differences in expression of any of these genes between heterozygous mutant and wild type embryos. These data are perhaps not surprising in light of that fact that a skeletal phenotype has not developed at these early stages. Therefore, we performed ISH on sections of young larvae (2-4 weeks of age) to ask whether the skeletal phenotype seen in heterozygous rapunzel mutants is linked to increased expression of skeletal genes. At two weeks of age we begin to see overexpression of col2a1 (Figure 8 A, B) in heterozygous rapunzel mutants. By four weeks of age, overexpression of col10a1 and osx are also observed (Figure 8 C-F). Finally, we find overexpression of col2a1, col10a1, and osx in the fins from 3 month-old rapunzel mutants (Figure 8 G-L). This increased expression of col2a1, col10a1, and osx that we observe in the caudal fin was further quantified by qPCR (Figure 8 M). col2a1 and osx are significantly increased (p<.05) in expression (2.65 ±.71 and 1.94±.43, respectively) in the fins from heterozygous rapunzel mutants versus wild type. col10a1 is also increased in expression (1.78±.54), although this fold change did not achieve statistical significance (data represent mean ± SD).
Figure 8.
Adult rapunzel mutants over express skeletal genes during early development. A-F. ISH was performed on saggital sections from rapunzel mutants and their wild type siblings. A, B. Overexpression (arrows) of col2a1 is seen in 2 week old rapunzel mutants (A) compared to wild type (B). C-F. Overexpression of col10a1 (C, D) and osx (E, F) is seen in the rostral (arrowheads) and caudal (arrows) craniofacial skeleton, as well as the opercle (double arrowheads) in 4-week-old rapunzel mutants (C, E) compared to wild type (D, F). G-M. Similar results are observed in the fin ray skeleton. Whole mount ISH reveals overexpression of col2a1 (G, H), col10a1 (I, J), and osx (K, L) in rapunzel fins. These results were confirmed by qPCR (M). N, O. Whole mount ISH reveals that rpz is diffusely expressed in the head (N, arrow) and vertebral column (O, arrow) of 4 week-old wild type larvae, but does not co-localize with skeletal markers. There were no differences in rpz expression between rapunzel heterozygotes (not shown) and wild type siblings.
In order to further characterize rpz expression at time points when a skeletal phenotype is apparent in heterozygous rapunzel mutants, ISH was performed on zebrafish larvae (2-4 weeks of age). At two weeks of age, rpz transcripts are seen diffusely in the vertebral column, muscle, and head (Figure 8 N, O). At three and four weeks of age, levels of rpz transcript decrease slightly, however the distribution remains unchanged (not shown). Interestingly, rpz does not show colocalization with the skeletal genes col2a1, col10a1, or osx.
Our data (see above) indicate that the gain of function phenotype in homozygous rapunzel embryos does not arise from misexpression or overexpression of the rpz gene (Figure 4). Although unlikely, these data do not exclude the possibility that overgrowth of the rapunzel fin ray skeleton arises from misexpression or overexpression of the rpz gene in the fin, as is true in other fin overgrowth mutations such as long fin (Kathy Iovine and Stephen Johnson, not shown). Consistent with our data from rapunzel embryos, qPCR revealed no differences in levels of rpz transcripts between the fins of rapunzel heterozygotes and wild type zebrafish (not shown). We also observed no quantitative differences in rpz transcript in wild type fin regenerates and rapunzel/+ fin regenerates, when expression is normalized to wild type fins (data not shown).
Discussion
We report here the identification of a missense mutation in a novel gene, rpz. rapunzelc14 demonstrates both a homozygous embryonic phenotype (lethal) as well as a heterozygous adult phenotype (overgrowth of the axial and appendicular skeleton). Furthermore, we show that morpholino knockdown of rpz suppresses the homozygous embryonic phenotype, however morpholino knockdown of rpz in wild type embryos produces no demonstrable morphant phenotype. From these data we can derive the following conclusions. First, rpz is not essential for early embryonic development. It remains formally plausible that the morpholino knockdown of rpz translation in wild type embryos was partial, allowing for translation of sufficient protein to prevent a morphant phenotype. This seems less likely since identical doses of morpholino were sufficient to completely suppress the mutant phenotype, however definitive proof will require an antibody (that we are currently generating) that recognizes the endogenous Rpz protein. It also remains possible that the rpz family of genes has redundant functions, thus knockdown of a single paralogue will not yield a morphant phenotype, but knockdown of multiple genes could produce a loss of function phenotype (see below). Second, rapunzelc14 is a gain of function allele. Although it remains possible that the heterozygous mutant phenotype could result from a dominant negative effect, this seems less likely given the absence of a morphant phenotype. RNA-targeting via antisense technology (including morpholinos) has emerged as a promising mechanism for rescuing null phenotypes induced by splice mutations (Wood et al., 2007). This approach has been used successfully in cell culture (Bruno et al., 2004; Du et al., 2007; Gebski et al., 2003; Suwanmanee et al., 2002) and in vivo (Alter et al., 2006; Lu et al., 2005; Madsen et al., 2008; van Deutekom et al., 2007). Our data provide the first example of rescue of a gain of function zebrafish mutant by morpholino injection. This has important ramifications for using zebrafish as a complex vertebrate model system to gain insight into human diseases, as a large number of human diseases are caused by gain of function missense mutations (Fisher et al., 2003). We hypothesize that the gain of function phenotype arises from accumulation of the mutant protein and we find no evidence that rpz is aberrantly expressed in the embryo. Again, more definitive proof must wait the generation of an antibody against the Rpz protein. We did attempt to phenocopy the rapunzel embryonic phenotype by over expressing wild type and mutant rpz in embryos using RNA injections (data not shown). However, we found that injecting either mutant or wild type rpz RNA's produced markedly abnormal embryos. This was likely due to the absence of an endogenous promoter regulating the expression of the injected RNA's.
rpz is a founding member of a novel gene family. We identified five zebrafish paralogues in the rpz family, both within and immediately adjacent to the rapunzelc14 critical region. To date, rpz homologs have only been identified in teleosts. While zebrafish appear to have five members of this novel gene family (see above), the number of paralogues varies in other species. tBlastx comparisons (http://www.ensembl.org) identify six orthologous genes in Orizias latipes and Gasterosteus aculeatus, while Takifugu rubripes contains five. tBlastx results show that all orthologous genes of these species localize to a single chromosome, as is the case in zebrafish. As shown above, both exons for rpz2 are located within the second intron of rpz, although the complete coding sequence for both of these genes is contained within a single exon. Since we did not clone full-length cDNA's for the rpz gene family from other teleosts, we do not know if these species share the same curious genomic arrangement as seen in zebrafish. The presence of multiple rpz orthologues in zebrafish raises the question of whether these genes have overlapping and redundant functions during development. We have performed double morpholino injections (rpz and rpz2) into wild type embryos (not shown) and find no evidence of redundancy to date. Morpholinos targeting the remaining three orthologues are currently being generated.
In addition to a homozygous embryonic phenotype, rapunzel has a heterozygous adult phenotype affecting both the fin ray (Goldsmith et al., 2003) and the axial (see above) skeleton. Interestingly, rpz gene expression appears strongest prior to the formation of definitive skeletal elements and does not show co-localization with well-defined skeletal genes, suggesting that rpz might act in a non cell-autonomous manner. However, heterozygous rapunzel mutants demonstrate increased expression of the skeletal markers col2a1, col10a1, and osx, providing a plausible mechanistic link wherein the mutation results in an increased number or activity of bone forming cells, leading to skeletal overgrowth. These data are consistent with our previously published results (Goldsmith et al., 2003) demonstrating an expanded zone of cell proliferation in the fins of heterozygous rapunzel mutants. It has been shown previously (Goldsmith et al., 2003; Iovine and Johnson, 2000) that fin growth in adult, wild type zebrafish is episodic and isometric. Furthermore, fin overgrowth mutations have been previously described (Goldsmith et al., 2003; Haffter et al., 1996; Iovine and Johnson, 2000; Iovine and Johnson, 2002; van Eeden et al., 1996) wherein isometric and episodic control of fin growth is lost. rapunzel represents the first fin overgrowth mutant affecting segment number to be cloned. Zebrafish mutants (dolphin, stocksteif) causing craniofacial abnormalities and hyperossification have been previously described (Laue et al. 2008, Spoorendonk et al., 2008). These mutants differ from rapunzel in that they show fusion of the vertebrae (not seen in rapunzel mutants) and do not affect the fin ray skeleton. An additional mutant, touchtone, affecting both endochondral and intramembranous ossification (Elizondo et al., 2005) has been described, but again, abnormalities of the fin ray skeleton are not observed. Also, the zebrafish mutant chihuahua displays skeletal abnormalities and a shorter caudal fin, but these are phenotypes not seen in rapunzel mutants (Fisher et al., 2003). Therefore, rapunzel is the first mutant identified causing overgrowth of both the axial and the fin ray skeleton. Interestingly, rapunzel's function is unknown; moreover cloning projects underway suggest that candidate genes for other fin overgrowth mutants do not play currently known roles in growth control (S. Johnson, not shown). Therefore, zebrafish genetics provides an important segue to identify previously uncharacterized growth control pathways in an adult, vertebrate organism.
We hypothesize that the rapunzel mutant phenotypes result from accumulation of the mutant protein (i.e. gain of function). Presumably a single rapunzelc14 allele produces sufficient mutant protein to induce a phenotype in adult fish, however embryos require two copies of the mutant allele to demonstrate a phenotype. In addition, we find no evidence in rapunzel heterozygotes for aberrant expression leading to up regulation of the rpz gene in the fin (not shown), even though this is the mechanistic basis for overgrowth in other zebrafish mutants (S. Johnson and K. Iovine, not shown). We were unable to localize rpz transcript in the fin by ISH (not shown), although ISH is notoriously unreliable for the identification of fin transcripts, even when these transcripts are present in high abundance such as during regeneration (Smith et al., 2008). How might the V90E mutation alter the function of the rapunzel protein? In silico analysis predicts that rapunzel encodes a transmembrane protein (Figure 5B), a finding supported by preliminary, in vitro experiments. Additionally, computational analyses (NetPhos 2.0) (Figure 5B) identify a putative phosphorylation site at T89 (IRKETVDRQ). Interestingly, the amino acid sequence of the mutant, but not of the wild type rapunzel allele predicts a putative phosphorylation site at Y94 (EDRQYHEVE). Perhaps conformational changes induced by the V90E mutation alter phosphorylation and downstream signaling of the rapunzel protein. There exists precedence for this model, as previous studies have shown that substituting glutamic acid for valine can activate the RAS kinase pathway, resulting in cellular proliferation (Turner et al., 2005), moreover malignant melanomas often contain a valine to glutamic acid mutation (Sharma et al., 2005; Turner et al., 2005). We are currently testing whether the rapunzel protein is phosphorylated in vitro. Future studies will elaborate the mechanism by which the mutant rapunzel protein results in overgrowth.
In summary, we have cloned rapunzel and identified a gain of function mutation in a novel gene (rpz) of unknown function. This conclusion is supported by three lines of evidence: the rapunzelc14 mutation maps to the rapunzel gene, expression of the rpz gene in embryos is consistent with the embryonic phenotype, and abrogation of rpz gene activity via morpholino injection suppresses the homozygous rapunzelc14 phenotype. In addition, the rpz gene is highly conserved in teleosts. Indeed, the V90 amino acid residue that is mutated in rapunzelc14 is invariably a conserved, neutral amino acid in all species for which sequence data is available. We also demonstrate that rapunzel heterozygotes have increased expression of cartilage and bone-forming genes providing a mechanism for the skeletal hyperossification phenotype. Although the function of rpz remains unknown and must await further experimentation, the identification of the genetic basis for skeletal overgrowth in rapunzelc14 provides new insight into the mechanisms underpinning vertebrate morphology and growth.
Supplementary Material
Supplemental Figure 1. ISH for markers of cartilage (col2a1) and bone (col10a1, osx) was performed on 120 hpf heterozygous rapunzel embryos and age-matched wild type siblings. Genotype was confirmed by sequencing following ISH (not shown). No differences in expression between rapunzel mutants and their wild type siblings were observed for any of the 3 genes. A, B. col2a1 expression is seen in the pharyngeal arches (pa), otic vesicle (ov), endochondral disc (ed) and fin fold (arrow). Expression of col10a1 (C, D) and osx (E, F) is localized to the cleithrum (cl), opercle (op), parasphenoid (ps), and brachiostegal ray (bs). Similar results were seen at 48, 72, and 96 hpf (not shown).
Supplemental Table 1. Markers used in rapunzelc14 positional cloning. Markers beginning with “z” are from Shimoda et al., 1999. Position of markers is on V7 of whole genome assembly. For sequencing, the F primer was used. Size polymorphisms were assessed on 1.5% agarose gels and SNP's were assessed by sequencing reads.
Acknowledgments
We thank Marnie Halpern for the gift of rapunzelc14, Bendi Gong for help with qPCR analysis, Patricia Jeremias for help with the alizarin red/Alcian blue staining, Kristi Stemler for help on the ISH, Jenny Lynch for help with the microCT, and Marcus Watkins for use of the PIXimus scanner. This work was supported by grants to MIG from the NIH (K08HD046656) and the March of Dimes (6-FY08-253) and to SLJ (GM056988).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alter J, Lou F, Rabinowitz A, Yin H, Rosenfeld J, Wilton SD, Partridge TA, Lu QL. Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nat Med. 2006;12:175–177. doi: 10.1038/nm1345. [DOI] [PubMed] [Google Scholar]
- Blumer MJ, Longato S, Fritsch H. Cartilage canal in the chicken embryo are involved in the process of endochondral bone formation within the epiphyseal growth plate. Anat Rec A Discov Mol Cell Evol Biol. 2004;279:692–700. doi: 10.1002/ar.a.20058. [DOI] [PubMed] [Google Scholar]
- Bruno IG, Jin W, Cote GJ. Correction of aberrant FGFR1 alternative RNA splicing through targeting of intronic regulatory elements. Hum Mol Genet. 2004;13:2409–2420. doi: 10.1093/hmg/ddh272. [DOI] [PubMed] [Google Scholar]
- Du L, Pollard JM, Gatti RA. Correction of prototypic ATM splicing mutations and aberrant ATM function with antisense morpholino oligonucleotides. Proceedings of the National Academy of Sciences. 2007;104:6007–6012. doi: 10.1073/pnas.0608616104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du SJ, Frenkel V, Kindschi G, Zohar Y. Visualizing normal and defective bone development in zebrafish embryos using the fluorescent chromophore calcein. Dev Biol. 2001;238:239–246. doi: 10.1006/dbio.2001.0390. [DOI] [PubMed] [Google Scholar]
- Elizondo MR, Arduini BL, Paulsen J, MacDonald EL, Sabel JL, Henion PD, Cornell RA, Parichy DM. Defective skeletogenesis with kidney stone formation in dwarf zebrafish mutant for trpm7. Current Biology. 2005;15:667–671. doi: 10.1016/j.cub.2005.02.050. [DOI] [PubMed] [Google Scholar]
- Fisher S, Jagadeeswaran P, Halpern ME. Radiographic analysis of zebrafish skeletal defects. Developmental Biology. 2003;264:64–76. doi: 10.1016/s0012-1606(03)00399-3. [DOI] [PubMed] [Google Scholar]
- Gebski BL, Mann CJ, Fletcher S, Wilton SD. Morpholino antisense oligonucleotide induced dystrophin exon 23 skipping in mdx mouse muscle. Hum Mol Genet. 2003;12:1801–1811. doi: 10.1093/hmg/ddg196. [DOI] [PubMed] [Google Scholar]
- Goldsmith MI, Fisher S, Waterman R, Johnson SL. Saltatory control of isometric growth in the zebrafish caudal fin is disrupted in long fin and rapunzel mutants. Dev Biol. 2003;259:303–17. doi: 10.1016/s0012-1606(03)00186-6. [DOI] [PubMed] [Google Scholar]
- Goss RJ. The physiology of growth. Academic Press; New York: 1978. [Google Scholar]
- Goss RJ, Stagg MW. The regeneration of fins and fin rays in Fundulus heteroclitus. Journal of Experimental Zoology. 1957;136:487–508. doi: 10.1002/jez.1401360306. [DOI] [PubMed] [Google Scholar]
- Haas HJ. Studies on mechanisms of joint and bone formation in the skeletal rays of fish fins. Developmental Biology. 1962;5:1–34. doi: 10.1016/0012-1606(62)90002-7. [DOI] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, Kelsh RN, Furutani-Seiki M, Vogelsang E, Beuchle D, Schach U, Fabian C, Nusslein-Volhard C. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Hoptak-Solga AD, Klein KA, DeRosa AM, White TW, Iovine MK. Zebrafish short fin mutations in connexin43 lead to aberrant gap junctional intercellular communication. FEBS Letters. 2007;581:3297–3302. doi: 10.1016/j.febslet.2007.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukriede N, Fisher D, Epstein J, Joly L, Tellis P, Zhou Y, Barbazuk B, Cox K, Fenton-Noriega L, Hersey C, Miles J, Sheng X, Song A, Waterman R, Johnson SL, Dawid IB, Chevrette M, Zon LI, McPherson J, Ekker M. The LN54 radiation hybrid map of zebrafish expressed sequences. Genome Res. 2001;11:2127–32. doi: 10.1101/gr.210601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine MK, Higgins EP, Hindes A, Coblitz B, Johnson SL. Mutations in connexin43 (GJA1) perturb bone growth in zebrafish fins. Developmental Biology. 2005;278:208–219. doi: 10.1016/j.ydbio.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Iovine MK, Johnson SL. Genetic analysis of isometric growth control mechanisms in the zebrafish caudal fin. Genetics. 2000;155:1321–1329. doi: 10.1093/genetics/155.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine MK, Johnson SL. A genetic, deletion, physical, and human homology map of the long fin region on zebrafish linkage group 2. Genomics. 2002;79:756–9. doi: 10.1006/geno.2002.6769. [DOI] [PubMed] [Google Scholar]
- Johnson SL, Gates MA, Johnson M, Talbot WS, Horne S, Baik K, Rude S, Wong JR, Postlethwait JH. Centromere-Linkage Analysis and Consolidation of the Zebrafish Genetic Map. Genetics. 1996;142:1277–1288. doi: 10.1093/genetics/142.4.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue K, Janicke M, Plaster N, Sonntag C, Hammerschmidt M. Restriction of retinoic acid activity by Cyp26b1 is required for proper timing and patterning of osteogenesis during zebrafish development. Development. 2008;135:3775–3787. doi: 10.1242/dev.021238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Felber K, Elks P, Croucher P, Roehl HH. Tracking gene expression during zebrafish osteoblast differentiation. Dev Dynamics. 2009;238:459–466. doi: 10.1002/dvdy.21838. [DOI] [PubMed] [Google Scholar]
- Lu QL, Rabinowitz A, Chen YC, Yokota T, Yin H, Alter J, Jadoon A, Bou-Gharios G, Partridge T. Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles. Proceedings of the National Academy of Sciences. 2005;102:198–203. doi: 10.1073/pnas.0406700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen EC, Morcos PA, Mendelsohn BA, Gitlin JD. In vivo correction of a Menkes disease model using antisense oligonucleotides. PNAS. 2008;105:3909–3914. doi: 10.1073/pnas.0710865105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Frieda MR, McAdow AR, Gross JP, Clayton CM, Heyen CK, Johnson SL. Coupled Mutagenesis Screens and Genetic Mapping in Zebrafish. Genetics. 2003;163:997–1009. doi: 10.1093/genetics/163.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria JA, Mari-Beffa M, Becerra J. Interactions of the lepidotrichial matrix components during tail fin regeneration in teleosts. Differentiation. 1992;49:143–150. doi: 10.1111/j.1432-0436.1992.tb00662.x. [DOI] [PubMed] [Google Scholar]
- Sharma A, Trivedi NR, Zimmerman MA, Tuveson DA, Smith CD, Robertson GP. Mutant V599EB-Raf Regulates Growth and Vascular Development of Malignant Melanoma Tumors. Cancer Res. 2005;65:2412–2421. doi: 10.1158/0008-5472.CAN-04-2423. [DOI] [PubMed] [Google Scholar]
- Shimoda N, Knapik EW, Ziniti J, Sim C, Yamada E, Kaplan S, Jackson D, de Sauvage F, Jacob H, Fishman MC. Zebrafish Genetic Map with 2000 Microsatellite Markers. Genomics. 1999;58:219–232. doi: 10.1006/geno.1999.5824. [DOI] [PubMed] [Google Scholar]
- Smith A, Zhang J, Guay D, Quint E, Johnson A, Akimenko MA. Gene expression analysis on sections of zebrafish regenerating fins reveals limitations in the whole-mount in situ hybridization method. Developmental Dynamics. 2008;237:417–425. doi: 10.1002/dvdy.21417. [DOI] [PubMed] [Google Scholar]
- Spoorendonk KM, Peterson-Maduro J, Renn J, Trowe T, Kranenbarf S, Winkler C, Schulte-Merker S. Retinoic acid and Cyp26b1 are critical regulators of osteogenesis in the axial skeleton. Development. 2008;135:3764–3774. doi: 10.1242/dev.024034. [DOI] [PubMed] [Google Scholar]
- Suwanmanee T, Sierakowska H, Fucharoen S, Kole R. Repair of a splicing defect in erythroid cells from patients with beta-thalassemia/HbE disorder. Mol Ther. 2002;6:718–726. doi: 10.1006/mthe.2002.0805. [DOI] [PubMed] [Google Scholar]
- Turner DJ, Zirvi MA, Barany F, Elenitsas R, Seykora J. Detection of the BRAF V600E mutation in melanocytic lesions using the ligase detection reaction. Journal of Cutaneous Pathology. 2005;32:334–339. doi: 10.1111/j.0303-6987.2005.00338.x. [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, den Dunnen JT, Koop K, van der Kooi AJ, Goemans NM, de Kimpe SJ, Ekhart PF, Venneker EH, Platenburg GJ, Verschuuren JJ, van Ommen GJB. Local Dystrophin Restoration with Antisense Oligonucleotide PRO051. N Engl J Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- van Eeden FJ, Granato M, Schach U, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, Kelsh RN, Mullins MC, Odenthal J, Warga RM, Nusslein-Volhard C. Genetic analysis of fin formation in the zebrafish, Danio rerio. Development. 1996;123:255–262. doi: 10.1242/dev.123.1.255. [DOI] [PubMed] [Google Scholar]
- Walker MB, Kimmel CB. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech Histochem. 2007;82:23–28. doi: 10.1080/10520290701333558. [DOI] [PubMed] [Google Scholar]
- Wood M, Yin H, McClorey G. Modulating the Expression of Disease Genes with RNA-Based Therapy. PLoS Genetics. 2007;3:e109. doi: 10.1371/journal.pgen.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Willoughby J, Liu D, Crump JG, Wilson C, Miller CT, Singer A, Kimmel C, Westerfield M, Postlethwait JH. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. 2005;132:1069–1083. doi: 10.1242/dev.01674. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. ISH for markers of cartilage (col2a1) and bone (col10a1, osx) was performed on 120 hpf heterozygous rapunzel embryos and age-matched wild type siblings. Genotype was confirmed by sequencing following ISH (not shown). No differences in expression between rapunzel mutants and their wild type siblings were observed for any of the 3 genes. A, B. col2a1 expression is seen in the pharyngeal arches (pa), otic vesicle (ov), endochondral disc (ed) and fin fold (arrow). Expression of col10a1 (C, D) and osx (E, F) is localized to the cleithrum (cl), opercle (op), parasphenoid (ps), and brachiostegal ray (bs). Similar results were seen at 48, 72, and 96 hpf (not shown).
Supplemental Table 1. Markers used in rapunzelc14 positional cloning. Markers beginning with “z” are from Shimoda et al., 1999. Position of markers is on V7 of whole genome assembly. For sequencing, the F primer was used. Size polymorphisms were assessed on 1.5% agarose gels and SNP's were assessed by sequencing reads.