Abstract
Precise regulation of eukaryotic gene expression requires interactions between distal cis-acting regulatory sequences with the looping out of the intervening DNA, but how trans-acting regulatory proteins work to establish and maintain DNA loops during gene activation remains largely unexplored. LPS-induced transcription of the mouse Igκ gene in B lymphocytes utilizes three distal enhancers and requires the transcription factor NF-κB, whose family members include RelA and c-Rel. Using chromosome conformation capture technology in combination with chromatin immunoprecipitation, here we demonstrate that LPS-induced Igκ gene activation creates chromosomal loops by bridging together all three pair-wise interactions between the distal enhancers and RNA polymerase II, the apparent molecular tie for the bases of these loops. RelA and actin polymerization are essential for triggering these processes, which do not require new transcription, protein synthesis or c-Rel. We have thus identified both essential and non-essential events that establish higher-order chromatin reorganization during Igκ gene activation.
Keywords: chromosome conformation capture, mechanism of enhancer action, NF-κB, chromosomal loops, Igκ, gene regulation, B cells
Introduction
It is becoming increasingly evident that precise regulation of eukaryotic gene expression requires communication between cis-acting elements through alterations in higher-order chromatin structure (for recent reviews, see references 1 and 2). During gene activation or silencing, distal regulatory elements, such as enhancers, locus control regions, differentially methylated regions and insulators, adopt a close proximity to their target genes with the looping out of the intervening DNA (3, 4, 5). In addition to intrachromosomal associations, interchromosomal interactions also occur between the interferon-γ (IFNγ) and interleukin-4 (IL-4) genes (6), the imprinted Igf2/H19 and Wsb1/Nf1 genes (7), and the olfactory receptor gene loci (8). Thus, long-range physical interactions between distal cis-acting elements and their target genes is emerging as a wide spread mechanism for gene regulation, yet much information is lacking regarding the requirements to establish and maintain such DNA loops.
The mouse Igκ light chain gene locus represents an ideal system to elucidate a paradigm for stimulus-induced transcriptional activation, gene rearrangement and nuclear reorganization during cellular differentiation (for reviews, see references 9 and 10). During B cell development the mouse IgH locus rearranges first, by sequential D-J and then V-DJ joining, leading to the pro- and pre-B cell stages of development, respectively (for a review on recombination, see reference 11). The Igκ locus rearranges next, which results in its transcriptional activation because it positions a Vκ gene carrying its own promoter into a chromatin domain containing three powerful downstream enhancers: an intronic enhancer (Ei) within the transcription unit (12), and two enhancers downstream of the transcription termination region, termed E3′ and Ed (13, 14). Ei and E3′ play roles in gene rearrangement (15, 16, 17), while E3′ and Ed function in rearranged gene transcription (15, 16, 18), with Ed maximally acting during the latest stages of B cell differentiation (18). Upon Ig gene rearrangement, the germ line, unrearranged IgH and Igκ alleles become deposited into pericentromeric heterochromatin and silenced (9, 19).
In an earlier study we used chromosome conformation capture technology (3C) (20; reviewed in references 1 and 21) - a method that reveals the in vivo relative proximity between specific DNA sequences in chromatin - to investigate the higher-order organization of transcriptionally active, rearranged Igκ loci in mouse B cells relative to inactive gene controls (22). We found that the three transcriptional enhancers and rearranged V gene promoters exhibited B-cell-specific proximal interactions over 22 kb, with the intervening DNA looping out. These results represented the first example a looping mechanism for enhancer action in an Ig gene locus, whose physical organization and developmental regulation differs dramatically from that of the well-studied β-globin locus as well as several other gene systems (1, 5, 21, 23). However, the mechanisms underlying loop formation as well as the identity of trans-acting factors that initiate and maintain complex formation between the Igκ gene enhancers remained to be elucidated, which is the subject of the present investigation.
Transcription factors of the NF-κB family are widespread regulators of inflammatory responses, immune system development and apoptosis (for review, see reference 24). Vertebrates have five members: NF-κB1 (p50 and its precursor, p105), NF-κB2 (p52 and its precursor, p100), RelA (p65), RelB and c-Rel. Most NF-κB family members can form homo- or heterodimers, which in turn bind to κB DNA sequence motifs and can act as transactivators depending on dimer compositions. Notably, each of the three Igκ gene enhancers contain single κB sites and these sites each become occupied by NF-κB species upon LPS-induction of Igκ gene transcription in B lymphocytes (vide infra). LPS engagement of toll like receptor 4, as well as ligand engagement of certain other B-cell surface receptors, results in the activation of cytoplasmically localized p50-RelA and p50-c-Rel heterodimers, which are the major NF-κB family members in B lymphocytes (25). The process is mediated by the so called “canonical pathway” through IKK kinase phosphorylation of associated inhibitory IκB proteins in response to the various stimuli, leading to IκB degradation and the release of DNA-binding competent NF-κB species, which translocate to the nucleus to activate target gene transcription. By contrast, activation of p52-RelB heterodimers occurs by a so called “noncanonical pathway”, independent of IκB protein phosphorylation and degradation, but dependent on IKK kinase phosphoryating p100, which triggers its proteolytic processing to p52 allowing its nuclear translocation in association with RelB.
In spite of the fact that NF-κB was initially discovered as a regulator of Igκ gene transcription more than twenty years ago (26, 27), more recent detailed studies on mouse B cells derived from knockouts of RelA, c-Rel and p50 NF-κB family members reveal that Igκ gene rearrangement and transcription still occurs if B cell survival can be maintained, but many aspects of B cell development become altered (for review, see reference 28). NF-κB family members are not essential for triggering Igκ gene expression because each of the three Igκ gene enhancers has multiple binding sites for numerous other transcription factors (13, 10). In fact, the transcription factors Pax5, E2A, and IRF4/IRF8 are crucial for proper Igκ gene rearrangement and expression (29, 30, 31). Nevertheless, in the present investigation we take advantage of the fact that in response to LPS, transcriptional induction of the Igκ gene in B lymphocytes is totally dependent on the canonical NF-κB pathway (vide infra).
Here we investigate interactions between distal Igκ enhancers and promoters before and after gene rearrangement, as well as before and after LPS-induced transcriptional activation, allowing us to deduce the step-wise events that establish the active state of the locus. Prior to transcriptional induction, we find that rearranged V gene promoters are complexed with either the enhancer Ei or E3′ (but not with both nor with Ed), forming the “poised” conformation, but that pair-wise interactions between the three enhancers occur only after LPS-activation and the transient and essential recruitment of RelA. ChIP-3C experiments reveal that these enhancer complexes are in close proximity to RNA polymerase II, providing evidence that transcription factories are the molecular ties for the bases of these DNA loops. These processes are actin-filament-dependent but independent of new protein synthesis, transcription or c-Rel. We have thus identified both essential and non-essential events that precede and temporally occur during the process of gene activation at the level of protein involvement and chromosomal looping of a specific Ig gene locus.
Materials and Methods
Cell culture, fractionation and mouse strains
70Z/3 and P815 cells were cultured in RPMI1640 medium containing 10% FBS, 50 μM β-mercaptoethanol, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. Where indicated, cells were treated with 10 μg/ml LPS (Sigma), 100 IU/ml IFNγ ( BD Bioscience), 10 μg/ml α-amanitin (Sigma), 10 μg/ml cyclohexamide (Sigma), 20 mM 2-3-Butanedione monoxime (Sigma), and 4 μM Latrunculin A (Sigma). 70Z/3 cells containing a stably integrated dominant negative IκB expression vector were kindly provided by Eugene Oltz and have been described (32). For mouse experiments, mature B and T cells were purified from 6-8 weeks old Vκ8-J5 knock-in mice (33) or c-Rel-/- mice (described in reference 37; kindly provided by Stephen Smale) as described previously except that the B cells were stimulated by culturing with 40 μg/ml LPS for three to four days (19). All mice were used in accordance with protocols approved by the UT Southwestern Medical Center Institutional Animal Care and Use Committee (IACUC).
Northern and RT-PCR Assays
Total cellular RNA was prepared using Tri-Zol (Invitrogen). For RT-PCR experiments, 4 μg of RNA was reverse transcribed with SuperScriptII reverse transcriptase (Invitrogen) in 20 μl at 42°C for 1 h with 300 ng oligo(dT) (Invitrogen) according to the manufacturer’s recommendations. Subsequent PCR or quantitative real time PCR was performed with the primers indicated in Table I and the quantity of assayed transcripts were referenced to those of the porphobilinogen deaminase (PBGD) housekeeping gene, with the level of uninduced Igκ, Vκ4 or Vκ8 transcripts assigned a value of 1.0. For Northern experiments, 10 μg of RNA was run on 1.5% formaldehyde-agarose gels (34). After electrophoresis, gels were stained with ethidium bromide for 10 min, destained and 28S rRNA was used as a RNA loading control. RNA was blotted onto Zeta-Probe GT filters (Bio-Rad) and hybridized with a 200 bp cDNA fragment located in the rearranged Vκ4 transcript according to standard methods. Quantitation was based on the signal ratios of Vκ4 transcripts relative to those of ribosomal RNA.
TABLE I.
Primers and Taqman probes that were used in 3C, ChIP, ChIP-3C and RT-PCR assays
| Primers for 3C Assays | 1 | 5′ cac gaa tac tct ctg gta att ga 3′ |
| 1′ | 5′ gca tgg aga caa gat ttc 3′ | |
| 2 | 5′ cta cag cca gca aaa gtc atg g 3′ | |
| 3 | 5′ ggc aac tat tta agg acc c 3′ | |
| 4 | 5′ acc aaa atg tcc acc aac agt 3′ | |
| 5 | 5′ tcc tac ctc ttg aga atg ta 3′ | |
| 6 | 5′ acc atg ttc ctg tgg ctg at 3′ | |
| 7 | 5′ aga agc aga act gtc tag aga ct 3′ | |
| 8 | 5′ cca tcc tat ctt ccc ttc taa gg 3′ | |
| 9 | 5′ cat caa agg tga ggc cat 3′ | |
| 10 | 5′ gga ggg tgg aga aag aat 3′ | |
| 11 | 5′ act gag gca gga agt cct 3′ | |
| 12 | 5′ cct ttg tct tta gtt ctc tgt cta t 3′ | |
| 13 | 5′ cca tca att gtt cat ttc ct 3′ | |
| 14 | 5′ tga ctc tac ttt gtt ccc c 3′ | |
| 15 | 5′ act caa gcc ctg ggt agg ag 3′ | |
| 16 | 5′ ttg taa gcc tct gga gga 3′ | |
| 17 | 5′ ggc acc aac att cat gtg 3′ | |
| AseI PBGD1 | 5′ cct acc ctg tct aac cta gt 3′ | |
| AseI PBGD2 | 5′ ttt act att gga gcc atc tg 3′ | |
| BglII PBGD1 | 5′ gcc taa cta cct ggg gaa tc 3′ | |
| BglII PBGD2 | 5′ tgt cct gga act ctc tct gt 3′ | |
| Control AseIVκ8f | 5′ tct atg cat gga gac aag at 3′ | |
| Control AseIVκ8r | 5′ aga gct ttc tta atc cac aa 3′ | |
| ControlAseIVκ4f | 5′ gag ata tga tac atg ttc c 3′ | |
| Control AseIVκ4r | 5′ cct aca gct tct caa cct gc 3′ | |
| NspI1 | 5′ gtg aac cat tat gat tgc a 3′ | |
| NspI2 | 5′ ccc aat gct ttt gca cag tc 3′ | |
| NspI3 | 5′ gtg gag tag taa ccc acg ga 3′ | |
| 18 | 5′ atg atg atg atg atg atg tt 3′ | |
| 25 | 5′ cag aaa taa gaa act gcc cac gg 3′ | |
| 26 | 5′ cat tat cag ttg acg tgg c 3′ | |
| 27 | 5′ tct gtt gag atg cca act c 3′ | |
| 28 | 5′ gtt tcc atc ttg cta cct c 3′ | |
| 29 | 5′ aaa gaa tac agc atg tga c 3′ | |
| 30 | 5′ caa aat ttg agg tca ttg gg 3′ | |
| 31 | 5′ gga gtc tgc tgt cct tac agg 3′ | |
| 32 | 5′ atg tgc cag agt ggg ttg gta tga 3′ | |
| Taqman Probes | AseI Ei | 6FAMtga cat gtt cac aat tga gct att act taa TAMRA |
| AseI E3′ | 6FAMcag gcc ttc tgt gca cac aat caa MGB | |
| AseI Ed | 6FAMcac tct gtg tgt att ttt ctt MGB | |
| BglII Ei | 6FAMaag cga cct acc act gtt gcg gtg c TAMRA | |
| BglII E3′ | 6FAMatt aaa gca ata cct aca TAMRA | |
| AseI PBGD | 6FAMtcc ctc cc a tgc atg tcc tcc ccg ct TAMRA | |
| BglII PBGD | 6FAMcat gaa ttc aag gct aaa tct ggt TAMRA | |
| Primers for ChIP | 1f | 5′ cag ctt cct gct aat cag tgc t 3′ |
| 1r | 5′ ata cca ccc aac att cag at 3′ | |
| 1′f | 5′ ctt gct cca cct gcc ttt cag at 3′ | |
| 1′r | 5′ cac att gtc cag gca ttt gct a 3′ | |
| 2f (Eif) | 5′ cag agg gga ctt tcc gag agg cc 3′ | |
| 2r (Eir) | 5′ acc ctg gtc taa tgg ttt gta ac 3′ | |
| 3f | 5′ cag tcc ttt cta agg ttc acg 3′ | |
| 3r | 5′ ggt tgc ctt tgc ttc ctg tg 3′ | |
| 4f (E3′f) | 5′ ata gca act gtc ata gctacc gt 3′ | |
| 4r (E3′r) | 5′ gca ggt gta tga ggc ttt gga aa 3′ | |
| 5f | 5′ cta ata gtc cca atg ctc tcc 3′ | |
| 5r | 5′ tcc ttc ctt tgg tgg ctt c 3′ | |
| 6f | 5′ ggg tag ttt tcc caa cga aca gt 3′ | |
| 6r | 5′ gga tgg ttt tct taa aga gtg tga g 3′ | |
| 7f (Edf) | 5′ ggg aca gat cac cat ttc gtt aa 3′ | |
| 7r (Edr) | 5′ ctt cag gcc agc tgg gca gtg ag 3′ | |
| β-actinf | 5′ cgc cat gga tga cga tat cg 3′ | |
| β-actinr | 5′ cga agc cgg ctt tgc aca tg 3′ | |
| α-actinf | 5′ acg gac gta agc ctc act tc 3′ | |
| α-actinr | 5′ tac ctg ctg ctc tga ctc tg 3′ | |
| Primers for RT-PCR | Vκ8f | 5′ ggt acc tgt ggg gac att gtg 3′ |
| Cκr | 5′ gag gca cct cca gat gtt aac 3′ | |
| Vκ4f | 5′ aca atg act tgc agg gcc 3′ | |
| 5′ germline transf | 5′ cca cat gcc ttt ctt cag gga caa gtg gga 3′ | |
| 5′ germline transr | 5′ gtt atg tcg ttc ata ctc gtc ctt ggt caa c 3′ | |
| PBGDf | 5′ tgt ccc ggt aac ggc ggc gcg gcc aca ac 3′ | |
| PBGDr | 5′ gcc acc aca gtc tcg gtc tgt atg cga gc 3′ |
Gel mobility super shift
Cell extracts were prepared using NucBuster protein extraction kit (Novagen) according to the manufacturer’s instructions. Double-stranded oligonucleotides corresponding to the κB motif 5′AGTTGAGGGGACTTTCCCAGGC3′ (Promega) were end-labelled with [γ-32P]-ATP using T4 polynucleotide kinase. After purification of the radiolabeled oligonucleotides with Sephadex-G25, 0.1 pM oligonucleotides were incubated with 2 μg of nuclear protein extracts at room temperature for 30 min in binding buffer (50 mM KCl, 0.5 mM EDTA, 10 mM Tris-HCl (pH 7.9), 1 mM DTT, 0.04% Nonidet P-40, 5% glycerol and 1 μg poly(dI-dC). For super shift assays, 2 μg of nuclear protein extracts were incubated with RelA antibody for 10 min on ice before incubating with κB oligonucleotide. The reaction mixtures were analyzed on 5% non-denaturing polyacrylamide gels followed by phosphorimaging (Molecular Dynamics).
Reporter gene assays
A luciferase reporter gene containing plasmid whose expression was driven by κB sites (a gift from Dr. Zhijian Chen at UT Southwestern Medical Center) was transiently transfected in triplicate into cell lines using LipofectAMINE 2000 (Invitrogen). Typically, 106-107 cells and 1-2 μg of DNA were used per transfection, along with 20-50 ng of pRL-CMV Renilla luciferase reporter (Promega). Cell extracts were assayed for luciferase activity using the Dual-Luciferase™ reporter assay system (Promega) following the manufacturers’ instructions. The Renilla luciferase activity was used for normalization of transfection efficiencies.
3C assay and controls
3C assays were performed as described previously with minor modifications (22). These previous studies established the validity of the 3C technique in our hands by performing many controls, including the demonstration by genomic Southern assays that restriction enzyme digestion goes to completion at efficiencies ranging from 65 to 89% within cross-linked chromatin DNA for both transcriptionally active and inactive Igκ loci. 107 cells were cross-linked, lysed and nuclei were digested with 1000 IU of BglII or AseI at 37°C for 16 h. After ligation and subsequent DNA purification, the cross-linking frequencies between the anchor and test fragments, as measured by the amount of corresponding ligation product, were estimated by duplicate Taqman real time PCR reactions relative to standards. The standards were used to normalize for the PCR primer efficiencies. Yeast DNA prepared from a strain bearing the YAC FAW.A3 (22), which spans from Vκ19 to all downstream regulatory elements, was mixed at equal molar ratios with a PCR product spanning the Vκ4-72 or Vκ8-19 promoter region AseI cutting site and the PCR product spanning the house keeping gene, porphobilinogen deaminase (PBGD) BglII or AseI cutting site. The mixture was digested with AseI or BglII to completion and ligated at high concentration to generate eqimolar mixtures of all possible ligation products. The cross-linking and ligation efficiencies between different samples and different experiments were also normalized to the cross-linking frequencies of two adjacent BglII or AseI fragments of the PBGD gene. The relative cross-linking frequency was then calculated using the following formula: (PCR signal of a given Igκ gene ligation product from chromatin/PCR signal of that Igκ gene ligation product from naked DNA standard)/(PCR signal of a given PBGD gene ligation product from chromatin/PCR signal of that PBGD gene ligation product from naked DNA standard). The highest cross-linking frequency for each experimental series was then normalized to 1.0 (23). The error bars presenting 3C results represent the standard deviations from the means from 2-5 independent experiments as indicated in the Figure Legends. Primers used in this study are listed in Table I.
ChIP assays
Chip assays were performed as described previously (22). Antibodies used were purchased from Santa Cruz Biotechnology, Inc. Anti-Pol II used in ChIP and ChIP-3C experiments was a rabbit polyclonal against the N-terminal domain of the large subunit of Pol II (sc-899). Results were quantitated by real-time PCR with SYBR Green dye using the ABI Prism 7300 system. All PCR signals from IP samples were referenced to their respective inputs to normalize for differences in primer efficiencies. α-actin was treated as a negative control and its enrichment fold was considered as 1. The enrichment of every test fragment was referenced to α-actin. Primers used in this study are listed in Table I.
ChIP-3C assays
108 cells were cross-linked, harvested, lysed and digested with restriction enzymes as described in 3C assays. After digestion, samples were centrifuged at maximum speed for 5 min in an Eppendorf centrifuge and the supernatants were immunoprecipitated with the indicated Abs. After washing five times with 50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, pH 8, two times with 10 mM Tris-HCl, 250 mM LiCl2, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA, pH 8, and three times in Tris-EDTA buffer, the precipitated complexes were eluted from the Ab-bound Sepharose beads with 10 mM DTT and ligated at 16°C for 16 h. Proteinase K was added and samples were incubated at 65°C for 16 h to reverse cross-links. DNA was purified as described previously (22). The fragment cross-linking frequencies were evaluated by touch down PCR at 40 cycles. Primers used in this study are listed in Table I.
shRNA knockdown of RelA
Short hairpin RNA (shRNA) sequences for RNA interference of RelA were designed with the help of Stephen Smale (personal communication). The effective sequence proved to be: 5′AGCTTGAAGACAGCCTTTACTGAAATTCAA-GAGATTTCAGTAAAGGCTGTCTTTTTTTTG3′. The lentiviral transfer construct pCCL.PPT.hPGK.GFP.Wpre, and helper viruses pMD2.VSVG, pMDLg/pRRE, and pRSV-REV were the kind gifts of Antonia Follenzi. The lentivirus was packaged in Fx293 cells and infected into target cells 70Z/3 as previously described (35).
Results
B-Cell-specific constitutive and inducible interactions between distal cis-acting regulatory sequences precede and accompany transcriptional activation of Igκ loci
We have previously shown that in actively transcribed mouse Igκ loci, the three enhancers, Ei, E3′ and Ed, and rearranged V gene promoter regions, exhibit mutual interactions with the looping out of the intervening DNA (22). Here we investigate which of these interactions might precede transcriptional activation, and subsequently dissect in detail the requirements for the temporal establishment of these interactions during transcriptional activation. We address these questions using both a pre-B-lymphoma cell line (70Z/3) and an animal model in which we can induce germline and rearranged gene transcription upon LPS treatment 9- to 100-fold (Fig. 1A and B), thus providing the opportunity to investigate the relationship between the activation of transcription and higher-order chromatin conformations. As non-B cell negative controls with inactive Igκ loci, we used P815 mastocytoma cells and primary mouse T cells from the corresponding knock-in mice (22).
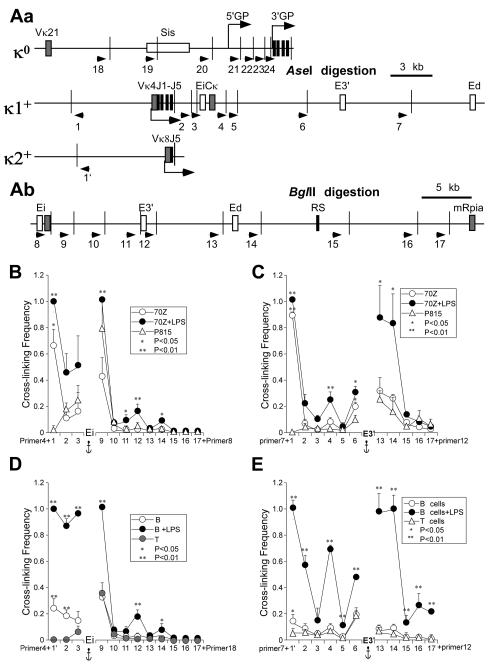
FIGURE 1.
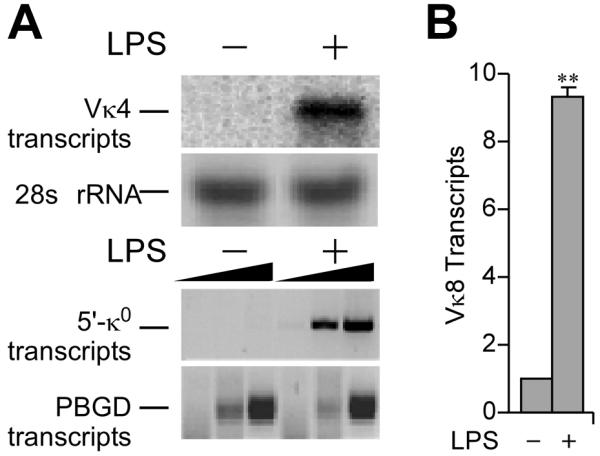
LPS-induction of Igκ gene transcription. A, RNA samples from 70Z/3 cells before and after 20 h of LPS treatment as indicated. (Upper) Northern blot of rearranged Vκ4 gene transcripts with ethidium bromide staining of rRNA as a loading control. (Lower) RT-PCR analysis germline (κo) Igk gene transcripts with the housekeeping gene porphobilinogen deaminase (PBGD) transcripts used as internal controls, with a 3-fold serial dilution series of the templates. Representative results from several independent RNA preparations are indicated. B, Real time RT-PCR analysis of rearranged Igκ gene transcript levels in splenic B cells before and after 3 days of LPS treatment in culture. The value of 1.0 was assigned for the level of uninduced transcripts, and data were referenced to PBGD transcript levels. Standard deviations of at least five independent RNA preparations are indicated. Here and throughout, double and single asterisks indicate P<0.01 and P<0.05, respectively, resulting from statistical analysis of standard deviations from several independent experiments.
We utilized the 3C technique to investigate whether interactions between distal cis-acting sequences in Igκ loci were constitutive and/or inducible (20; reviewed in references 1 and 21). Using either Ei or E3′ as fixed primers (anchors) for their successive pairing with other primers in 3C samples from 70Z/3 cells, before or after LPS treatment, and in P815 cells as a control, we observed B-cell-specific strong interactions between Ei or E3′ with the rearranged Vκ4 gene promoter region prior to transcriptional induction (Fig. 2B and C). Furthermore, using primers specific for either germline or rearranged Vκ4 alleles reveals that these enhancer-promoter interactions occur exclusively on the κ+ allele as expected (Fig. S1). Interestingly, because Ei and E3′ do not exhibit significant interactions prior to induction, we infer that either Ei or E3′, but not both, interacts with the Vκ4 gene promoter region in uninduced single cells, indicating cellular heterogeneity. Upon induction of transcription in 70Z/3 cells, each of the three enhancers now exhibit significant mutual interactions (Fig. 2B and C). Additional conformational changes can be detected upon transcriptional activation of the locus, such as closer interactions of Ei with its adjacent downstream fragments 9 and 11 (Fig. 2B), and related increases in interactions of E3′ with flanking fragments 6 and 13 (Fig. 2C). We will comment on these increased adjacent fragment interactions later below. We also used Ed as an anchor in 3C experiments with 70Z/3 cells, and found that the Vκ4 gene promoter region did not interact with Ed even after LPS-induction of these pre-B cells (Fig. S2), a result consistent with Ed’s known function later in B cell development (18). In summary, constitutive interactions exist between the Vκ4 gene promoter region and either Ei or E3′, but not with Ed, whereas induction gives rise to all-possible pair-wise interactions between each of the three enhancers, but not Vκ4-Ed interactions.
FIGURE 2.
3C analysis of germline and rearranged Igκ loci before and after induction of transcription. A, Maps of 3′ regions of mouse Igκ loci. Cell line 70Z/3 possesses one germline (κo) and one Vκ4 rearranged Igκ allele (κ1+) (55), while isolated splenic B cells from the animal model possess one rearranged Vκ8 gene (κ2+) (33). Exons and cis-acting sequences are indicated by closed and open rectangles, respectively. Arrows indicate the primers used in 3C assays. Narrow vertical lines depict restriction enzyme cut sites. Bent arrows indicate sites of initiation and the direction of transcription. Aa, Igκ loci were digested with AseI restriction enzyme. κo depicts the germline Igκ alleles in both 70Z/3 and splenic B cells. κ1+ indicates the rearranged Igκ allele in 70Z/3 cells and κ2+ denotes the rearranged Igκ allele in splenic B cells purified from targeted heterozygous mice. Ab, Igκ loci were digested with BglII restriction enzyme. The downstream region shown is common to germline and rearranged alleles. B, 3C assays in 70Z/3 cells before and after LPS treatment with Ei as the anchor fragment (primer 4 for AseI digests, or primer 8 for BglII digests). The amounts of 3C ligation products were measured by real time PCR using Taqman probes. Standard deviations of at least five independent chromatin preparations are indicated. Significant differences are in comparison to P815 mastocytoma cells. (Although the Vκ4 gene is far removed from the locus in the germline alleles of the negative control P815 cells, we can extrapolate the baseline cross-linking frequency expected for an adjacent fragment for purposes of statistical estimate, which predicts a significant interaction between Vκ4 and Ei or E3′ in 70Z/3 cells, either before or after induction of transcription. In addition, using a primer next to the BglII cutting site that is adjacent to the common J5 region in both germline and rearranged alleles paired with a E3′ primer, we obtain a significantly higher cross-linking frequency in 70Z/3 cells than in P815 cells (data not shown). C, 3C assays in 70Z/3 cells before and after LPS treatment with E3′ as the anchor fragment fragment (primer 7 for AseI digests, or primer 12 for BglII digests). Standard deviations of at least five independent chromatin preparations are indicated. Significant differences are in comparison to P815 mastocytoma cells. D, 3C assays in splenic B cells before and after LPS treatment with Ei as the anchor fragment (paired as in panel B). Standard deviations of at least two independent chromatin preparations are indicated. Significant differences are in comparison to T cells. E, 3C assays in splenic B cells before and after LPS treatment with E3′ as the anchor fragment (paired as in panel C). Standard deviations of at least three independent chromatin preparations are indicated. Significant differences are in comparison to T cells.
Related 3C experiments with splenic B cells isolated from mice harboring a rearranged Vκ8 gene revealed no significant interactions between each of the three enhancers before transcriptional induction, and induced mutual interactions between the enhancers after transcriptional induction (Fig. 2D and E), just as in 70Z/3 cells. In fact, the increased flanking fragment interactions noted upon transcriptional induction of 70Z/3 are much more marked in stimulated splenic B cells in which the level of Igκ gene transcription is much higher, as every fragment studied exhibited a highly significant increase in cross-linking frequency (Figure 2D and E), suggesting that upon transcriptional induction the active Igκ allele assumes a more compacted conformation. We imagine that in highly transcribing Igκ genes, the constant physical interactions between Vκ promoter regions and Ei or E3′ bring the flanking fragments much closer together in comparison to the more linear chromosome conformation of inactive Igκ loci. Furthermore, very similar significant increases in flanking fragment cross-linking frequencies have been seen upon transcriptional activation of the β-globin locus (23). In addition, before LPS treatment, we also observed statistically significant interactions of the Vκ8 gene promoter region with Ei and E3′ (Fig. 2D and E), but much less dramatic than those seen between the Vκ4 gene promoter region and either Ei or E3′ in uninduced 70Z/3 cells. We also used Ed as an anchor in 3C experiments with these splenic cells, and found that the Vκ8 gene promoter region did exhibit significant induced-interactions with Ed but not with further upstream sequences (Fig. S2), a result again consistent with our previous findings that Ed’s primary function is at the terminal stages of B cell development (22). In summary, because both primary and immortalized B cells exhibit all three possible LPS-inducible pair-wise interactions between the three enhancers, we will focus on these induced pair-wise enhancer interactions for the remainder of the studies to be presented below.
LPS-inducible interactions between the three Igκ gene enhancers require activation of NF-κB
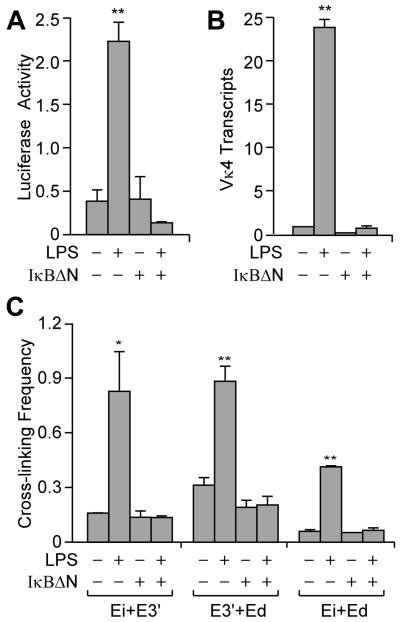
LPS treatment of B cells triggers the rapid degradation of IκBα followed by the nuclear translocation of cytoplasmic p50-RelA and p50-c-Rel heterodimers (reviewed in reference 24), both in 70Z/3 and primary splenic B cells (25). Because each Igκ enhancer possesses κB sites, and our ChIP analysis revealed that RelA and c-Rel associate with each of the three enhancers upon LPS treatment (Fig. S3), we wanted to determine whether NF-κB activation is required for inducing pair-wise interactions between these enhancers. For this purpose we used a 70Z/3 clone that expresses a dominant negative IκB (IκBΔN), which in previous studies was shown to block LPS-inducible Igκ gene transcription (32). As shown in Figure 3A and B, 70Z/3-IκBΔN cells were neither LPS-inducible for a κB site-driven luciferase reporter nor for Vκ4 gene transcription. Furthermore, 3C experiments revealed that formation of each of the three pair-wise enhancer complexes were blocked after LPS treatment of the cells expressing the dominant negative IκB (Fig. 3C). We conclude that activation of RelA or c-Rel (or both) via the canonical pathway is required to induce mutual interactions between the enhancers.
FIGURE 3.
Role of NF-κB activation on LPS-inducible interactions between the Igκ gene enhancers. Where indicated, 70Z/3 ± IκBΔN were treated with LPS for 20 h. A, The luciferase activity of a reporter plasmid whose expression was driven by κB sites was measured after transient transfection of the indicated cell lines. The level of activity is expressed as percent relative to the activity of a co-transfected pRL-CMV Renilla luciferase reporter gene. Standard deviations of three independent experiments are indicated. B, Real time RT-PCR assay of rearranged Vκ4 gene transcript levels. C, 3C assays of enhancer interactions. Standard deviations of three independent chromatin preparations are indicated.
LPS-inducible interactions between the three Igκ gene enhancers still occur in the absence of new protein synthesis and transcription
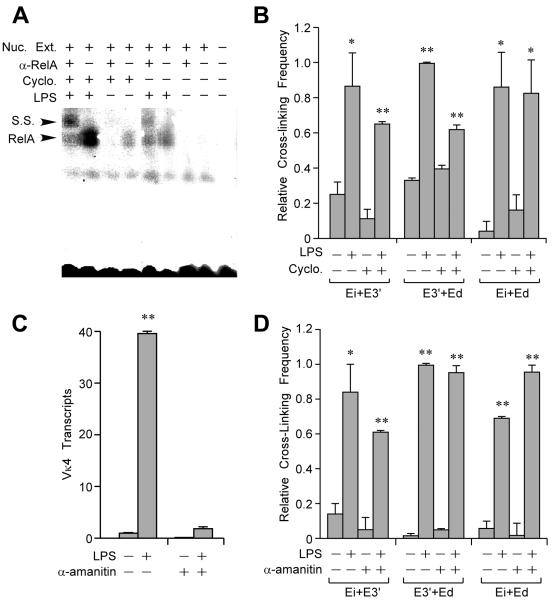
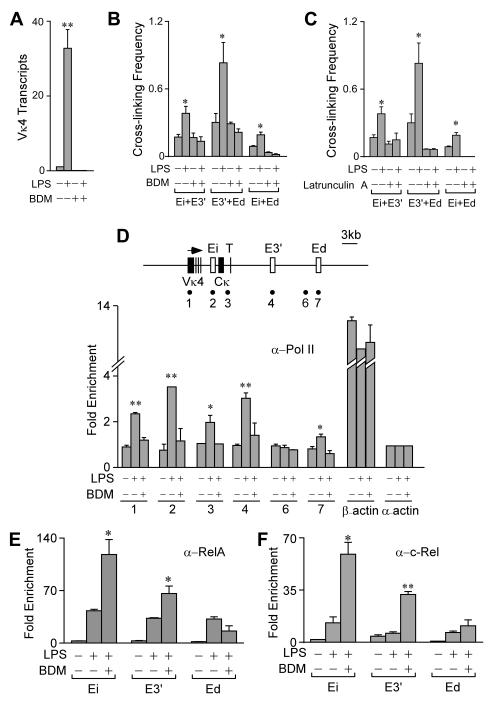
The formation of pair-wise Igκ enhancer interactions might require NF-κB target gene transcription and translation, or might exploit the pre-existing protein complement of B cells poised for transcription. We therefore determined the effect of inhibiting protein synthesis on LPS-induced enhancer interactions. In agreement with Sen and Baltimore (27), cycloheximide treatment did not inhibit LPS mediated activation of RelA as measured by gel mobility super shift, but rather led to superinduction in its DNA binding activity (Fig. 4A). Furthermore, 3C experiments revealed that LPS-induced-interactions between the three enhancers still occurred in the presence of the drug, but at somewhat reduced levels (Fig. 4B). To examine the effect of transcription on LPS-induced enhancer interactions, we pre-treated cells with α-amanitin, an inhibitor of the DNA translocation step Pol II (36), so as to block LPS-inducible Vκ4 gene transcription (Fig. 4C). Significantly, 3C experiments revealed that LPS-inducible enhancer interactions still occurred (Fig. 4D). Hence, neither new protein synthesis nor transcription are essential for NF-κB to trigger pair-wise enhancer interactions.
FIGURE 4.
Effect of cycloheximide and α-amanitin on LPS-inducible interactions between the Igκ gene enhancers. 70Z/3 cells were treated with cycloheximide and LPS for 5 h. Cycloheximide was added 30 min before LPS treatment. (A) Gel mobility super shift (S.S.) assays reveal that RelA is activated by LPS treatment in the presence or absence of cycloheximide. (B) 3C assays reveal that LPS-inducible enhancer interactions occur in the presence or absence of cycloheximide. Standard deviations of three independent chromatin preparations are indicated. (C) Real-time RT-PCR assay of rearranged Vκ4 gene transcript levels in 70Z/3 cells treated with α-amanitin as indicated. α-Amanitin was added to 70Z/3 cells 10 h before 5 h of LPS treatment. (D) 3C assays reveal that LPS-inducible enhancer interactions occur in the presence or absence of α-amanitin. Standard deviations of three independent chromatin preparations are indicated.
RelA is essential and c-Rel is dispensable for mediating LPS-induced Igκ gene transcription and pair-wise enhancer interactions
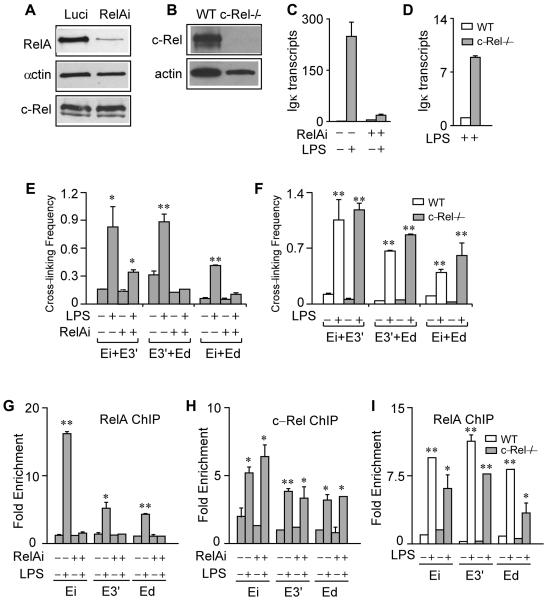
To investigate the individual roles of RelA and c-Rel in fostering Igκ gene transcription and enhancer complex formation, we performed experiments utilizing 70Z/3 cells in which RelA expression was knocked-down by a shRNA-expressing lentivirus (RelAi). Performing the corresponding RelA knock-down experiments with splenic cells, however, is problematic because their survival in culture requires agents that trigger Igκ gene transcription, and RelA-/- knockout mice die during embryogenesis (reviewed in reference 28). By contrast, c-Rel-/- knockout mice are viable and develop splenic B cells (37), thus allowing us to address directly the role of this NF-κB family member in these primary cells. As shown by the Western blots in Figures 5A and B, these cell line and animal systems successfully deplete the corresponding proteins. Significantly, LPS-induced Igκ gene transcription was blocked in RelA knockdown cells (Fig. 5C), whereas IFNγ-stimulated Igκ gene transcription was not affected (data not shown), which occurs by a NF-κB-independent pathway (5). Importantly, LPS-inducible pair-wise enhancer interactions between E3′ and Ed, and between Ei and Ed, no longer occurred in the RelA knock-down cell line, and interactions between Ei and E3′ were dampened (Fig. 5E). ChIP analysis reveals that LPS-induced c-Rel enhancer occupancy was not changed in RelA knock-down cells (Fig. 5G and H), indicating that c-Rel recruitment is independent of RelA. These results prove that RelA is essential for LPS-induced Igκ gene transcription and the formation of major pair-wise enhancer interactions, and that c-Rel is incapable of activating transcription or mediating major enhancer interactions in the absence of RelA. To determine further the role of c-Rel on these functions, we took advantage of c-Rel knockout mice. Surprisingly, in c-Rel knockout mice splenic B cells, LPS-induced transcription of the Igκ gene was increased some 8-fold over that exhibited by wild type cells (Fig. 5D), indicating that c-Rel plays an apparent negative role in LPS-induced Igκ gene transcription. Significantly, LPS-induced pair-wise interactions between the enhancers and RelA occupancy were not significantly altered in B cells from these knockout mice compared to wild type samples (Fig. 5F and I). We also found that in response to LPS, unlike splenic B cells from wild type mice, the corresponding cells from c-Rel knockout mice did not proliferate during cell culturing, in agreement with a previous report (37). We conclude that c-Rel is dispensable in mediating LPS-induced Igκ gene transcription and pair-wise enhancer interactions, and actually may play a negative role in attenuating Igκ gene transcription levels.
FIGURE 5.
Individual roles for RelA and c-Rel on LPS-induced Igκ gene transcription and pair-wise enhancer interactions. A, Western blot of RelA, actin and c-Rel in 70Z/3 cells infected with Luci or RelAi shRNA-expressing lentiviruses. B, Western blot of c-Rel and actin in splenic B cells from wild type and c-Rel knockout mice. C, Real-time RT-PCR analysis of Igκ gene transcript levels before and after LPS treatment for 20 h in control or RelA knock-down 70Z/3 cells. Standard deviations of two independent RNA purifications are indicated. D, Real time RT-PCR analysis of Igκ gene transcript levels in 48 h LPS-stimulated splenic B cells from wild type and c-Rel knockout mice. Standard deviations of two independent RNA purifications are indicated. E, 3C assays of control and RelA knock-down 70Z/3 cells treated with or without LPS as above. F, 3C assays of splenic B cells from wild type or c-Rel knockout mice treated with or without LPS as above. Standard deviations of two independent chromatin preparations are indicated. G, Real time PCR ChIP assays of RelA in control or RelA knock-down 70Z/3 cells treated with or without LPS as above. Fold enrichment refers to the sequence abundance in the immunoprecipitated sample divided by the corresponding sequence abundance in input DNA. Standard deviations of two independent chromatin preparations are indicated. H, Real time PCR ChIP assays of c-Rel in control or RelA knock-down 70Z/3 cells treated with out without LPS as above. Fold enrichment is defined in Figure 5G. Standard deviations of two independent chromatin preparations are indicated. I, Real time PCR ChIP assays of RelA in wild type or c-Rel knockout splenic B cells treated with or without LPS as above. Fold enrichment is defined in Figure 5G. Standard deviations of two independent chromatin preparations are indicated.
Although complexes between the three Igκ gene enhancers have different recipes of NF-κB species, each are associated with RNA polymerase II
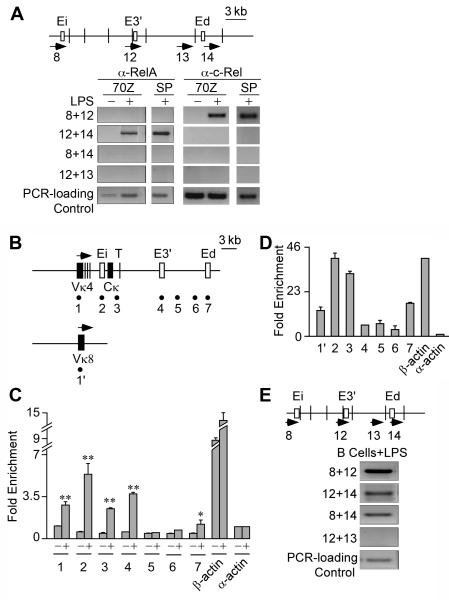
To determine whether NF-κB family members or RNA polymerase II (Pol II) might act as potential molecular ties in the three different pair-wise enhancer complexes we performed ChIP-3C experiments. In both LPS treated 70Z/3 cells and splenic B cells, RelA was not present in Ei-E3′ or Ei-Ed complexes, but was present in E3′-Ed complexes, whereas c-Rel was not present in Ei-Ed or E3′-Ed complexes, but was present in Ei-E3′ complexes (Fig. 6A). Furthermore, neither protein was present in Ei-Ed complexes (Fig. 6A). The fact that identical ChIP-3C results are obtained from the cell line and primary B cells suggests that both systems may share common mechanisms for regulating the Igκ gene through signaling by the same specific NF-κB family members. These results indicate that the three different pair-wise enhancer complexes each have a unique protein composition with respect to NF-κB species, and that neither RelA nor c-Rel play direct roles in maintaining interactions between Ei-Ed complexes. These results demonstrate further that tri-molecular complexes between the three enhancers do not exist with either associated RelA or c-Rel proteins per se.
FIGURE 6.
Potential RelA, c-Rel and RNA polymerase II interactions bridging the Igκ gene enhancers. A, ChIP-3C assays of RelA’s and c-Rel’s presence in Ei-E3′, E3′-Ed, and Ei-Ed complexes in 20 h LPS treated 70Z/3 cells and 3 day LPS stimulated splenic B cells (SP). Pre-immune IgG immunoprecipitation controls reveal that no amplification products are detected. B, Map of the Igκ locus with the sites assayed for by PCR amplification in ChIP samples indicated by the underlying numbered solid circles. C, Real time PCR ChIP assays of Pol II occupancy at the Igκ locus in 70Z/3 cells before (-) and after 20 h of LPS treatment (+). Fold enrichment is defined in Figure 5G. Standard deviations of three independent chromatin preparations are indicated. D Real time PCR ChIP assays of Pol II occupancy at the Igκ locus in splenic B cells after 3 days of LPS treatment. Fold enrichment is defined in Figure 5G. Standard deviations of three independent chromatin preparations are indicated. E, ChIP-3C assay for presence of Pol II in the Ei-E3′, E3′-Ed, Ei-Ed complexes in splenic B cells stimulated with LPS for 3 days. Pre-immune IgG immunoprecipitation controls reveal that no amplification products are detected.
Recently it has been proposed that transcription factories might be the platforms for interactions between cis-acting elements with the looping out of the intervening DNA (38). To investigate this possibility, first we performed ChIP assays to study Pol II association with selected regions of the Igκ locus (Fig. 6B), and with the expressed β-actin gene and the inactive α-actin gene as positive and negative controls, respectively. After LPS induction of both 70Z/3 and splenic B cells and immunoprecipitation with antibodies against Pol II, the Igκ gene’s rearranged gene promoters, coding regions, and Ei, E3′ and Ed were significantly enriched relative to their corresponding sequence abundances in input chromatin DNA samples (Fig. 6C and D). It is noteworthy that E3′ and Ed reside about 8 kb and 18 kb, respectively, downstream of the previously mapped transcription termination region (Fig. 6B, T symbol) (39), so their sequence enrichment in association of Pol II was unexpected. Furthermore, enrichment of fragment 5 and 6 control sequences was not observed, which reside in between these two downstream enhancers (Fig. 6B-D), suggesting that the proximity of the enhancers to Pol II is not due to tracking of the enzyme during gene transcription. After LPS-induction, we also observed similar enrichments of Ei and E3′ sequences in α-amanitin pre-treated cells, but not in IκBΔN expressing 70Z/3 cells (Fig. S4). Therefore, the association of Pol II with these enhancers strictly correlates with the formation of LPS-inducible complexes between these enhancers, consistent with transcription factories being the molecular ties for such interactions. Next, to investigate directly whether Pol II was present in complexes between these enhancers we performed ChIP-3C experiments. Consistent with our expectation, Pol II was indeed present in complexes between Ei-E3′, Ei-Ed, and E3′-Ed in primary splenic B cells (Fig. 6E). In conclusion, these results prove that each of the three pair-wise enhancer complexes is associated with Pol II.
Actin polymerization is required for the formation of LPS-inducible complexes between the Igκ gene enhancers
Our results so far have demonstrated RelA-dependent event(s) following LPS treatment that eventually lead to the association of the Igκ genes’ pair-wise enhancer complexes with Pol II, either by stationary assembly (40), or by gene movement to preassembled transcription factories (41). Previous studies have shown that inhibitors of actin dynamics, like BDM (42), block the long-range directional movement of chromosomal sites targeted by acidic activators like RelA or VP16 during gene activation (43, 44). Therefore, we determined whether actin polymerization might be involved in the loading of Pol II. Significantly, treatment of 70Z/3 cells with BDM or another actin polymerization inhibitor, latrunculin A, inhibited LPS-inducible transcription and interactions between the enhancers (Fig. 7A-C). To determine whether BDM might disrupt interactions between the three enhancers once formed, we treated 70Z/3 cells with LPS first for 5 h and then added BDM for another 2 h, and found no significant change in pre-induced enhancer interactions (data not shown). We conclude that actin polymerization is required for the initiation but not the maintenance of pair-wise enhancer complexes. As expected, BDM treatment also blocked LPS-induced Pol II occupancy on the Igκ gene enhancers (Fig. 7D). BDM treatment, however, did not block LPS-induced RelA and c-Rel occupancy on these enhancers. By contrast, significantly more RelA and c-Rel were associated with Ei and E3′ in cells treated with BDM plus LPS (Fig. 7E). These results prove that this drug’s inhibitory effects are downstream of NF-κB activation pathways. In conclusion, these results demonstrate that actin filaments are required, either indirectly or directly, for the Igκ gene enhancers to generate loop complexes with themselves and with Pol II.
FIGURE 7.
Effect of BDM and latrunculin A on interactions between the Igκ gene enhancers. A, Real time RT-PCR assay of rearranged Vκ4 gene transcript levels in 70Z/3 cells before and after treatment with BDM and/or LPS for 5 h. B, 3C assays of 70Z/3 cells treated as above. Standard deviations of three independent chromatin preparations are indicated. C, 3C assays of 70Z/3 cells before and after treatment with latrunculin A and/or LPS for 5 h. Standard deviations of three independent chromatin preparations are indicated. D, Real time PCR ChIP assays of Pol II occupancy at the Igκ enhancers in 70Z/3 cells treated as in Panel A. Map of the Igκ locus with the sites assayed for by PCR amplification in ChIP samples indicated by the underlying numbered solid circles. Fold enrichment is defined in Figure 5G. Standard deviations of three independent chromatin preparations are indicated. (E and F) Real time PCR ChIP assays of RelA and c-Rel occupancy on the Igκ gene enhancers in 70Z/3 cells treated as above. Fold enrichment is defined in Figure 5G. Standard deviations of three independent chromatin preparations are indicated.
Discussion
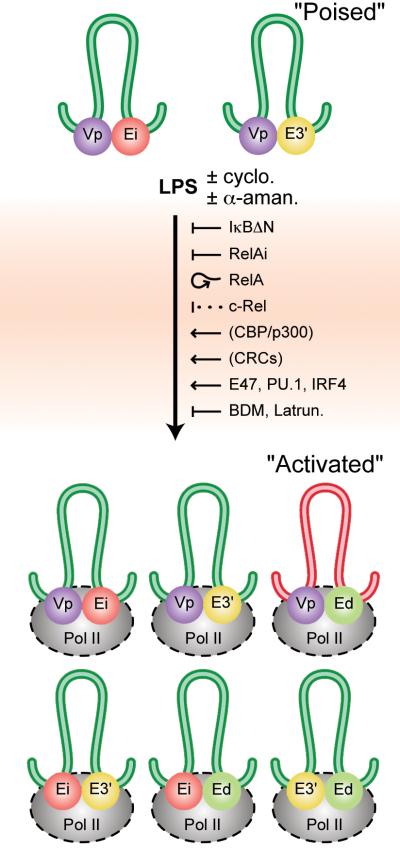
The formation of complexes between distal regulatory elements with the looping out of the intervening DNA has been observed in several model gene systems (reviewed in references 1 and 2), but little is known about the proteins that generate these DNA loops and nor about how they are maintained. For this reason we have taken advantage of LPS inducible systems thus allowing us to determine the events that precede and accompany transcriptional activation of a locus. Figure 8 schematically summarizes our results. We have observed interactions between rearranged gene promoters (Vp) and either Ei or E3′, but not both nor with Ed, in B cells before NF-κB activation and transcriptional induction, thus representing a transcriptionally poised state. We conclude that different interactions must exist in single cells within the population. It is also possible that such interactions “flip-flop” between these enhancers in single cells, like that seen in switching between promoters in the β-globin locus as revealed by in situ hybridization experiments (45).
FIGURE 8.
Schematic summary of the results of this investigation. The diagram depicts interactions between the three Igκ gene enhancers, and the rearranged Vκ4 or Vκ8 gene promoters (Vp), in 70Z/3 and splenic cells, before and after LPS induction, with the looping out of the intervening DNA. Loops are color coded to represent structures common to 70Z/3 and splenic cells (green), those specific to 70Z/3 (blue), and those specific to splenic cells (red). The colored spheres represent protein complexes associated with the indicated cis-acting elements. Innocuous drugs, positive and negative regulators or inhibitors of the LPS activation process are also shown (cyclo., cycloheximide; α-aman., α-amanitin; Latrun., latrunculin A. A proposed negative role for c-Rel is shown with a dotted line and proposed positive roles for CBP/p300 and chromatin remodeling complexes (CRCs) are shown in parentheses. ChIP-3C experiments have demonstrated the presence of Pol II in all three pair-wise enhancer complexes in stimulated splenic cells (Fig. 6E) and also in complexes between the Vκ4 gene promoter region and Ei or E3′ in stimulated 70Z/3 cells (Fig. S7). The presence of Pol II in the other complexes shown in this Figure are generalizations depicted by dashes surrounding the enzyme complexes.
Upon LPS-induced Igκ gene activation we have elucidated an essential role for RelA and a dispensable role for c-Rel in initiating all three pair-wise distal enhancer interactions. Interestingly, although RelA is required for the formation of each of the three pair-wise enhancer complexes, once formed RelA is no longer present in two of them (Ei-E3′ and Ei-Ed). Therefore, RelA is required for the formation but not the maintenance to these complexes. Our results reveal further an actin filament requirement downstream of RelA and that Pol II is a potential molecular tie for the bases of these chromosomal loops (Fig. 8). We have shown further that LPS-induced pair-wise enhancer interactions must largely utilize pre-existing proteins and do not require transcription, as they still can occur in the presence of cycloheximide or α-amanitin. We propose that the initial binding of RelA to the enhancers recruits the histone acetyl transferase CBP/p300 based on previous studies (46), which presumably triggers the binding of chromatin remodeling complexes (CRCs) and several other transcription factors. Once RelA has performed these tasks its presence is apparently no longer required. Indeed, we have found that LPS-treatment of 70Z/3 cells not only leads to the increased occupancy of NF-κB species on each of the three Igκ gene enhancers, but also to increases in the corresponding associations of the transcription factors E47, PU.1 and IRF4 (Fig. S5). In contrast to activated splenic cells or plasmacytomas (22), after transcriptional activation in 70Z/3 cells we did not observe interactions between Ed and the rearranged Vκ4 gene promoter (Fig. S2), yet the Igκ gene enhancers exhibit all three pair-wise interactions. This observation indicates that when Ed interacts with either Ei or E3′, these enhancers are not interacting with rearranged Vκ4 gene promoter regions (Fig. 8). In addition, the lack of an interaction between the Vκ4 gene promoter region and Ed under induced conditions in the 70Z/3 pre-B cell line further supports our earlier observations that Ed’s primary function is in the later stages of B cell development (18). Furthermore, our ChIP-3C experiments revealed that tri-molecular complexes between the enhancers did not occur when the enhancers were associated with either RelA or c-Rel in 70Z/3 or splenic B cells. In this respect it is of interest that although the results of 3C experiments are often summarized in models depicting tri-, tetra-, or even penta-molecular complexes between interacting cis-elements (5, 23), no direct support for these multi-molecular complexes exist, because these models represent a composite of data collected from several independent, single anchor PCR assays. Nevertheless, in the case of the Igκ loci, we do believe that at a minimum tri-molecular complexes between enhancer pairs and rearranged Vκ gene promoters should exist in activated states, because previous studies have demonstrated synergistic transcriptional activation between Ei and E3′ (47), and between Ei and Ed (13). Even if only bi-molecular complexes exist between promoters and enhancers in active Igκ loci, pair-wise enhancer complexes could serve an activating function for a single enhancer’s subsequent participation in a bimoleculat complex with a promoter. However, it is still possible that pair-wise enhancer complexes could serve either neutral or even negative functions, as in the case of Ei-Ed and E3′-Ed complexes in LPS-induced 70Z/3 cells. Clearly, 3C experiments only provide a snapshot of the population average, where only the strongest interactions are readily detected and less frequent or weaker ones might be missed, and cannot distinguish between the dynamics of, or the heterogeneity in, interactions within or between single cells.
Complex formation between the enhancers peaks with the association of Pol II (Fig. 8). We propose that this process occurs by long-range chromosomal movement of the locus to pre-assembled transcription factories using an actin/myosin motor, by integrating our actin-filament inhibitor and ChIP-3C results with those of other investigators in related systems (41, 43, 44). It should also be noted that Igκ gene transcription can be induced by IFNγ by an NF-κB-independent pathway (48), and that we have found that this pathway of induction also leads to all three pair-wise interactions between the enhancers in 70Z/3-IκBΔN cells (Fig. S6). This observation speaks to the significance of these three pair-wise enhancer interactions because they occur by two independent pathways through different sets of initiating proteins. IFNγ is also known to lead to the recruitment of several transcription factors to the Igκ locus and other target genes (48, 49), including the co-activator CBP/p300, which may be a common link between LPS and IFNγ inducible pathways at the level of initiating chromatin modifications. In addition, our results should be generally applicable to more physiological inducers associated with B cell signaling and NK-κB activation, because like LPS, BCR or TNFR engagement also lead to NF-κB activation through the canonical pathway, and CD40R signaling results in NF-κB activation by both the canonical and noncanonical pathways (24, 28). By contrast, BAFF, an important signaling molecule for B cell development and survival, leads to NF-κB activation principally through the noncanonical pathway, resulting in nuclear accumulation of p52-RelB heterodimers (24). Thus, an interesting question remaining to be addressed is whether these NF-κB species will also lead to similar higher order chromatin alterations in the Igκ locus to those described here.
Knockout and knockin experiments have shown the importance of the germline promoters (GP) and the adjacent KI-II sequences, as well as Ei and E3′, in fostering the ability of the Igκ locus to rearrange in pre-B cells (15, 17, 50, 51). However, we did not detect any 3C interactions between any of the three enhancers with the GP or with any other sequence between Vκ21 and Jκ1 in κo alleles of primary pre-B cells from the bone marrow of RAG1oxμ mice (52) (data not shown). We believe that there should be loop complexes between GP and Ei/E3′ during the process of germline transcription, but apparently these loops are dynamic and present in only a few percent of the cells at any one time, because the 3C technique is not sensitive enough to detect such interactions in these cells. In addition, in pro-B cells from the bone marrow of Rag1-/- mice carrying the Vκ8 knock-in allele, no interactions between the rearranged V gene promoter region and the enhancers are seen before or after LPS treatment. However, LPS inducible interactions do exist in these cells between each of the three Igκ gene enhancers (data not shown).
RelA and c-Rel are the only two NF-κB family members activated by the canonical pathway that possess transactivation domains. Generally, these two members regulate different target genes. For example, c-Rel but not RelA is critical for IL12b gene activation (53). In addition, over-expression of RelA or c-Rel has opposite effects in regulating pro- and anti-apoptotic genes, such as DR4, DR5, cIAP1 and c-IAP2 (54). Our results are novel in that we demonstrate for the first time that RelA and c-Rel each initially associate with the Igκ gene enhancers, but only RelA, or c-Rel, or neither eventually ends up in the three pair-wise enhancer complexes. RelA in the absence of c-Rel is sufficient for the generation of all three pair-wise enhancer complexes during LPS-induced Igκ gene transcription. By contrast, the c-Rel that is present in Ei-E3′ complexes, is incapable of activating Igκ gene transcription in RelA knock-down cells. Furthermore, although c-Rel is present in the Ei-E3′ complex by ChIP-3C experiments, its presence is not required for the initiation or maintenance of this complex, which forms in c-Rel KO mice. Interestingly, splenic B cells from c-Rel knockout mice are hyper-LPS-inducible for Igκ gene transcription. While c-Rel deficiency in these mice may have pleiotropic effects on B-cell function and hence might indirectly result in hyper-LPS-inducibility for Igκ gene transcription, it is also possible that c-Rel may have a direct negative effect on Igκ gene function. Hypothetically, c-Rel could be inhibitory because it may compete with p50 and RelA monomers, thereby limiting the availability of crucial p50-RelA heterodimers. Its presence on the Igκ gene enhancers may serve no function, because although RelA and c-Rel recognize the same single κB sites in each of the three enhancers, the different factors do not appear to compete for these sites since the chromatin occupancy of one protein was not affected by the abundance of the other. Alternatively, hypothetically c-Rel could recruit co-repressors, thereby acting to fine tune and govern the level of Igκ gene induced transcription to prevent its over-expression.
In conclusion, using 3C and ChIP-3C technology along with inducible biological systems, dominant negative, knock-down or knockout of NF-κB family members, and chemical inhibitors, we have been able to determine step-wise many of the requirements and events that lead to the establishment of the fully active state of the Igκ locus. We show for the first time that different NF-κB family members have divergent roles in activating transcription of the same gene by establishing unique interactions between different pair-wise enhancer complexes. In addition, both NF-κB-dependent and -independent pathways establish the same three pair-wise enhancer complexes upon gene activation.
Supplementary Material
Acknowledgements
We thank Eugene Oltz, Mark Schlissel, Stephen Smale and Martin Weigert for providing cell lines and mice animal models, and Jose Cabrera for graphic illustrations. We are indebted to Keith Wharton for his insightful comments and suggestions. None of the authors have any financial interest related to this work.
Footnotes
This investigation was supported by Grants GM29935 and AI067906 from NIH and Grant I-823 from the Robert A. Welch Foundation to WTG, and by Grants HL067256 and HL61897 from NIH to LST.
References
- 1.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 2.Sexton T, Schober H, Fraser P, Gasser SM. Gene regulation through nuclear organization. Nature Struc. Mol. Biol. 2007;14:1049–1055. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- 3.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nature Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 4.Spiliankis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nature Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 5.Tolhuis B, Palstra R-J, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 6.Spiliankis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 7.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 8.Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 9.Fuxa M, Skok JA. Transcriptional regulation in early B cell development. Curr. Opin. Immunol. 2007;19:1–8. doi: 10.1016/j.coi.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Schlissel MS. Regulation of activation and recombination of the murine Igκ locus. Immunol. Rev. 2004;200:215–223. doi: 10.1111/j.0105-2896.2004.00157.x. [DOI] [PubMed] [Google Scholar]
- 11.Gellert M. Recombination: RAG proteins, repair factors, and regulation. Ann. Rev. Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 12.Queen C, Baltimore D. Immunoglobulin gene transcription is activated by downstream sequence elements. Cell. 1983;33:741–748. doi: 10.1016/0092-8674(83)90016-8. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z-M, George-Raizen JB, Li S, Chang MY, Meyers KC, Garrard WT. Chromatin structural analyses of the mouse Igκ gene locus reveal new hypersensitive sites specifying a transcriptional silencer and enhancer. J. Biol. Chem. 2002;277:32640–32649. doi: 10.1074/jbc.M204065200. [DOI] [PubMed] [Google Scholar]
- 14.Meyer KB, Neuberger MS. The immunoglobulin κ locus contains a second, stronger B-cell-specific enhancer which is located downstream of the constant region. EMBO J. 1989;8:1959–1964. doi: 10.1002/j.1460-2075.1989.tb03601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorman JR, van der Stoep N, Monroe R, Cogne M, Davidson L, Alt FW. The Igκ 3′ enhancer influences the ratio of Igκ versus Igλ B lymphocytes. Immunity. 1996;5:241–252. doi: 10.1016/s1074-7613(00)80319-2. [DOI] [PubMed] [Google Scholar]
- 16.Inlay M, Alt FW, Baltimore D, Xu Y. Essential roles of the κ light chain intronic enhancer and 3′ enhancer in κ rearrangement and demethylation. Nat. Immunol. 2002;3:463–468. doi: 10.1038/ni790. [DOI] [PubMed] [Google Scholar]
- 17.Xu Y, Davidson L, Alt FW, Baltimore D. Deletion of the lgκ light chain intronic enhancer/matrix attachment region impairs but does not abolish Vκ-Jκ rearrangement. Immunity. 1996;4:377–385. doi: 10.1016/s1074-7613(00)80251-4. [DOI] [PubMed] [Google Scholar]
- 18.Xiang Y, Garrard WT. The downstream transcriptional enhancer, Ed, positively regulates mouse Igκ gene expression and somatic hypermutation. J. Immunol. 2008;180:6725–6732. doi: 10.4049/jimmunol.180.10.6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z, Widlak P, Zou Y, Xiao F, Oh M, Li S, Chang MY, Shay JW, Garrard WT. A recombination silencer that specifies heterochromatin positioning and Ikaros association in the immunoglobulin κ locus. Immunity. 2006;24:405–415. doi: 10.1016/j.immuni.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 21.Simonis M, Kooren J, de Laat W. Nature Methods. 2007;4:895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- 22.Liu Z, Garrard WT. Long-range interactions between three transcriptional enhancers, active Vκ gene promoters and a 3′ boundary sequence spanning 46 kb. Mol. Cell. Biol. 2005;25:3220–3231. doi: 10.1128/MCB.25.8.3220-3231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palstra R-J, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The β-globin nuclear compartment in development and erythroid differentiation. Nature Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 24.Hayden MS, West AP, Ghosh S. NF-κB in the immune response. Oncogene. 2006;25:6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- 25.Liou H-C, Sha WC, Scott ML, Baltimore D. Sequential induction of NF-κB/Rel family proteins during B-cell terminal differentiation. Mol. Cell. Biol. 1994;14:5349–5359. doi: 10.1128/mcb.14.8.5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer. Cell. 1986;46:705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- 27.Sen R, Baltimore D. Inducibility of kappa immunoglobulin enhancer-binding protein NF-kappa B by a posttranslational mechanism. Cell. 1986;47:921–928. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann A, Baltimore D. Circuitry of nuclear factor κB signaling. Immunol. Rev. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 29.Johnson K, Hashimshony T, Sawal CM, Pongubala JMR, Skok JA, Aifantis I, Singh H. Regulation of immunoglobulin light-chain recombination by transcription factor IRF-4 and attenuation of interleukin-7 signaling. Immunity. 2008;28:335–345. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 30.Lazorchak AS, Schlissel MS, Zhang Y. E2A and IRF-4/Pip promote chromatin modification and transcription of the immunoglobulin kappa locus in pre-B cells. Mol. Cell. Biol. 2006;26:810–821. doi: 10.1128/MCB.26.3.810-821.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato H, Saito-Ohara F, Inazawa J, Kudo A. Pax-5 is essential for kappa sterile transcription during Ig kappa gene rearrangement. J. Immunol. 2004;172:4858–4865. doi: 10.4049/jimmunol.172.8.4858. [DOI] [PubMed] [Google Scholar]
- 32.Scherer DC, Brockman A, Bendall HH, Zhang GM, Ballard DW, Oltz EM. Corepression of RelA and c-Rel inhibits immunoglobulin κ gene transcription and rearrangement in precursor B lymphocytes. Immunity. 1996;5:563–574. doi: 10.1016/s1074-7613(00)80271-x. [DOI] [PubMed] [Google Scholar]
- 33.Prak EL, Weigert M. Light chain replacement: A new model for antibody gene rearrangement. J. Exp. Med. 1995;182:541–548. doi: 10.1084/jem.182.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, A Laboratory Manual. 2nd Ed. Cold Spring Harbor Laboratory; Cold Spring Harbor, N. Y.: 1989. [Google Scholar]
- 35.Wang Z, Schraw TD, Kim J, Khan T, Rajala MW, Follenzi A, Scherer PE. Secretion of the adipocyte-specific secretory protein adiponectin critically depends on thiol-mediated protein retention. Mol. Cell. Biol. 2007;27:3716–3731. doi: 10.1128/MCB.00931-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bushnell DA, Cramer P, Kornberg RD. Structural basis of transcription: α-amanitin-RNA polymerase II cocrystal at 2.8 Å resolution. Proc. Natl. Acad. Sci. USA. 2002;99:1218–1222. doi: 10.1073/pnas.251664698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Köntgen F, Grumont RJ, Strasser A, Metcalf D, Li R, Tarlinton D, Gerondakis S. Mice lacking the c-Rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 38.Marenduzzo D, Faro-Trindade I, Cook PR. What are the molecular ties that maintain genomic loops? Trends Genet. 2007;23:126–133. doi: 10.1016/j.tig.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 39.Xu M, Barnard MB, Rose SM, Cockerill PN, Huang S-Y, Garrard WT. Transcription termination and chromatin structure of the active immunoglobulin κ gene locus. J. Biol Chem. 1986;261:3838–3845. [PubMed] [Google Scholar]
- 40.Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT. Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol. Cell. 2007;28:978–990. doi: 10.1016/j.molcel.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 41.Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, Fraser P. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PloS Biol. 2007;5:1763–1772. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yarrow JC, Lechler T, Li R, Mitchison TJ. Rapid de-localization of actin leading edge components with BDM treatment. BMC Cell Biol. 2003;4:5. doi: 10.1186/1471-2121-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle B, Belmont AS. Long-range directional movement of an interphase chromosomal site. Curr. Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 44.Dundr M, Ospina JK, Sung M-H, John S, Upenderr M, Ried T, Hager GL, Matera AG. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J. Cell Biol. 2007;179:1095–1103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gribnau J, de Boer E, Trimborn T, Wijgerde M, Milot E, Grosveld F, Fraser P. Chromatin interaction mechanism of transcriptional control in vivo. EMBO J. 1998;17:6020–6027. doi: 10.1093/emboj/17.20.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong H, Voll RE, Ghosh S. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
- 47.Blasquez VC, Hale MA, Trevorrow W, Garrard WT. Immunoglobulin κ gene enhancers synergistically activate gene expression but independently determine chromatin structure. J. Biol. Chem. 1992;267:23888–23893. [PubMed] [Google Scholar]
- 48.Damore MA, Omori SA, Wall R. IFN-g induced the κ intron enhancer via an IFN-stimulated response element. J. Immunol. 1996;156:2451–2457. [PubMed] [Google Scholar]
- 49.Horvai AE, Xu L, Korzus E, Brard G, Kalafus D, Mullen T-M, Rose DW, Rosenfeld MG, Glass CK. Nuclear integration of JAK/STAT and Ras/AP-1 signaling by CBP and p300. Proc. Natl. Acad. Sci. USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferradini L, Gu H, Smet AD, Rajewsky K, Reynaud C-A, Weill J-C. Rearrangement-enhancing element upstream of the mouse immunoglobulin kappa chain J cluster. Science. 1996;271:1416–1420. doi: 10.1126/science.271.5254.1416. [DOI] [PubMed] [Google Scholar]
- 51.Liang H-E, Hsu L-Y, Cado D, Schlissel MS. Variegated transcriptional activation of the immunoglobulin κ locus in pre-B cells contributes to the allelic exclusion of light-chain expression. Cell. 2004;118:19–29. doi: 10.1016/j.cell.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 52.Spanopoulou E, Roman CA, Corcoran LM, Schlissel MS, Silver DP, Nemazee D, Nussenzweig MC, Shinton SA, Hardy RR, Baltimore D. Functional immunoglobulin transgenes guide ordered B-cell differentiation in Rag-1-deficient mice. Genes Dev. 1994;8:1030–1042. doi: 10.1101/gad.8.9.1030. [DOI] [PubMed] [Google Scholar]
- 53.Sanjabi S, Williams KJ, Saccani S, Zhou L, Hoffmann A, Ghosh G, Gerondakis S, Natoli G, Smale ST. A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation. Genes Dev. 2005;19:2138–2151. doi: 10.1101/gad.1329805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen X, Kandasamy K, Srivastava RK. Differential roles of RelA (p65) and c-Rel subunits of nuclear factor κB in tumor necrosis factor-related apoptosis-inducing ligand signaling. Cancer Res. 2003;63:1059–1066. [PubMed] [Google Scholar]
- 55.Nelson KJ, Mather EL, Perry RP. Lipopolysaccharide-induced transcription of the kappa immunoglobulin locus occurs on both alleles and is independent of methylation status. Nucl. Acids Res. 1984;12:1911–1923. doi: 10.1093/nar/12.4.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.