Summary
In homeostasis of adult vertebrate tissues, stem cells are thought to self-renew by infrequent and asymmetric divisions that generate another stem cell daughter and a progenitor daughter cell committed to differentiate. This model is based largely on in vivo invertebrate or in vitro mammal studies. Here we examine the dynamic behaviour of adult hair follicle stem cells in their normal setting by employing mice with repressible H2B-GFP expression to track cell divisions and Cre inducible mice to perform long-term single cell lineage tracing. We provide direct evidence for the infrequent stem cell division model in intact tissue. Moreover, we find that differentiation of progenitor cells occurs at different times and tissue locations than self-renewal of stem cells. Distinct fates of differentiation or self-renewal are assigned to individual cells in a temporal-spatial manner. We propose that large clusters of tissue stem cells behave as populations, whose maintenance involves unidirectional daughtercell fate decisions.
Introduction
Stem cells (SCs) have the ability to self-renew and differentiate for extended periods of time. Their dynamic behaviour within tissues is key in regulating normal tissue homeostasis and injury repair (Morrison and Kimble, 2006). Based on in vivo work in invertebrates, and supported by in vitro transplantation (functional) assays in mammalian systems adult SCs are thought to have several characteristics: reside in a protective environment or niche important for their long-term maintenance; divide infrequently during life; generate at division two daughter cells that acquire asymmetric fates (one daughter remains in the niche and becomes another SC, and the other leaves the niche and becomes a short-lived rapidly dividing transit-amplifying or progenitor cell); be multipotent or capable to generate all cell types in their own lineage (Lansdorp, 2007; Morrison and Kimble, 2006; Fuchs, 2009). However, due to the complexity of vertebrate tissues, the lack of specific markers, and the overall scarcity of adult SCs, there are few studies demonstrating these features for mammalian tissues in normal settings, without transplantation injury (Morrison and Kimble, 2006; Fuchs, 2009). Recently, long-term lineage tracing in intact tissue (referred to here as “in vivo”) in mouse epidermis, hair follicle (HF), and intestine (Barker et al., 2007; Clayton et al., 2007; Jaks et al., 2008), challenged the infrequently dividing/ “slow-cycling” SC model, previously best supported by in vitro data from blood and HF single-cell transplantation assays (Cotsarelis, 2006; Fuchs, 2009). Here we employ the mouse HF system to reexamine the dynamic behaviour of long-lived adult SCs in their normal setting.
HFs form beneath the skin surface during mouse embryonic life and emerge in early post-natal life as fully differentiated organelles with a cylindrical structure, made of an upper permanent region (bulge), containing epithelial SCs, and a temporary region (bulb) containing differentiated cells (Fig. 1A). Concentric layers of cells surround the centrally located hair shaft, and the SCs are localized in the outer most layer, called the outer root sheath (ORS). At the bulb base there is a pocket of progenitor cells known as matrix (Mx), which divide rapidly and generate terminally differentiated cells forming the inner root sheath (IRS) and the hair shaft. The matrix encloses a mesenchymal pocket of cells called dermal papillae (DP), a signalling centre with fate instructive properties (Fig. 1A). Around 3 weeks into postnatal life HFs begin a cyclic process of destruction and regeneration known as the hair cycle (Cotsarelis, 2006), which consists of recognizable and synchronous phases: catagen, telogen and anagen (Fig. 1B). Catagen is the destructive phase in which bulge cells cease proliferation, bulb cells undergo massive apoptosis, and DP cells are lifted upwards towards the bulge. Telogen is the resting quiescent phase, in which HFs consist of bulge and hair germ (HG). The HG is a small epithelial structure generated at least in part by cells at the bulge base (Cotsarelis, 2006; Jaks et al., 2008; Fig. 1B and Fig. 3A). Anagen is a phase of intense growth, when rapidly dividing progenitor Mx cells emerge underneath the bulge at the HG location and produce a new hair bulb (Cotsarelis, 2006; Greco et al., 2009; Legue and Nicolas, 2005). In later life these phases occur repeatedly, albeit less synchronously (Plikus et al., 2008), and are marked by long periods of rest. Bulge cells can be identified in the skin by their infrequent divisions and specific surface expression of CD34 and α6-integrin (Cotsarelis, 2006; Fig. S1B). Single bulge-cell transplantation assays showed contribution to HF regeneration in transplantation assays, which deemed the bulge as the SC compartment. In addition, data in intact skin revealed that at least some infrequently dividing bulge cells contributed progeny cells to the bulb in anagen in short-term assays. Finally, both K15 or LGR5 promoters marked collectively both bulge and HG cells in lineage tracing experiments, and confirmed the long-term contribution of bulge/germ cell progeny to all hair compartments (Cotsarelis, 2006; Fuchs, 2009). In this paper we marked for the first time the single bulge cells apart from the HG cells and test their long-term fate in intact tissue, in the absence of injury induced by cell isolation, transplantation, or dermabrasion. Specifically, we ask whether long-lived functional bulge HFSCs divide infrequently in unperturbed tissue homeostasis and whether their daughter cells undergo asymmetric fate decision throughout life.
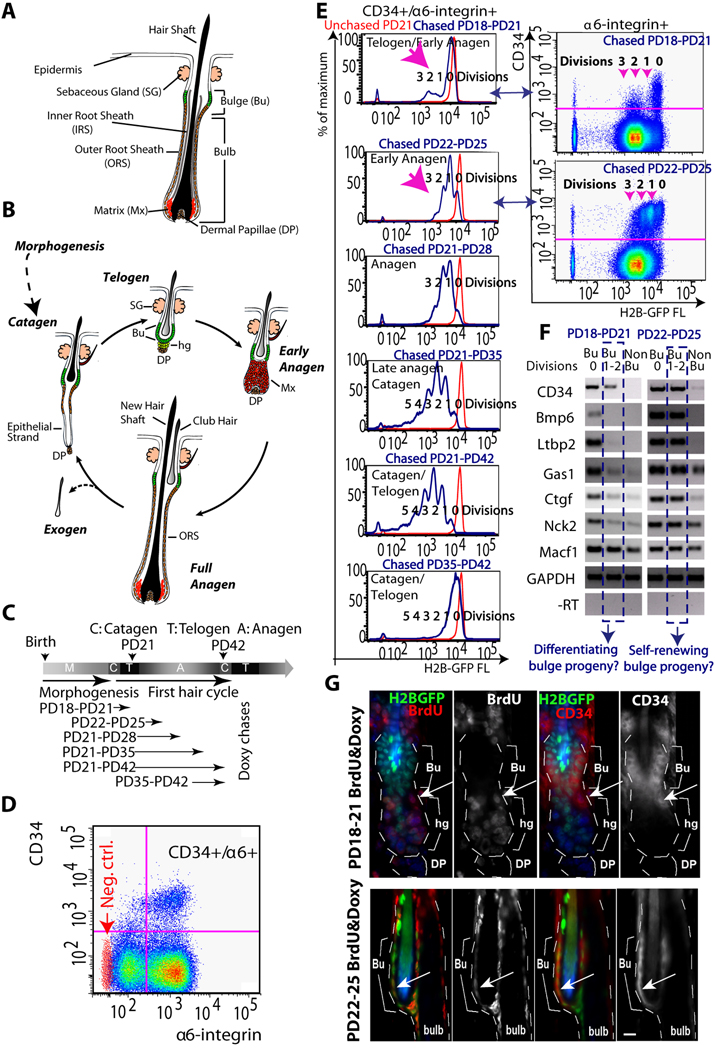
Figure 1.
Distinct characteristics of bulge cells newly generated at different hair cycle stages. (A) The hair follicle structure. (B) Hair cycle phases of growth (anagen), destruction (catagen) and quiescence (telogen) (see text). (C) Doxycycline (doxy) chases in 1st (synchronous) hair cycle to document cell proliferation quantitatively. PD, postnatal day. (D) FACS dot plot of live (PI negative) skin cells stained for CD34 and α6-integrin surface expression (PD21-PD28 chase). Red dots, secondary antibody alone for CD34 staining. (E) FACS histograms of CD34+/α6+ bulge cells from individual mice at stages indicated. Note distinct proliferation profiles at different stages (pink arrows). (F) Cells were sorted and analyzed by RT-PCR for expression of bulge-preferred factors (Tumbar et al, 2004). (G) Skin images from 3-day BrdU labeled and doxy treated mice showed BrdU staining in hair germ (hg) at PD18-PD21 and in bulge (Bu) at PD22-PD25. Arrows indicate CD34+/BrdU+ proliferating cells. Note that lower CD34 levels at the bulge/germ transition zone at PD18- PD21. All experiments were repeated in ≥ 3 mice per stage. DP, dermal papillae. Scale bar in G, 10µm.
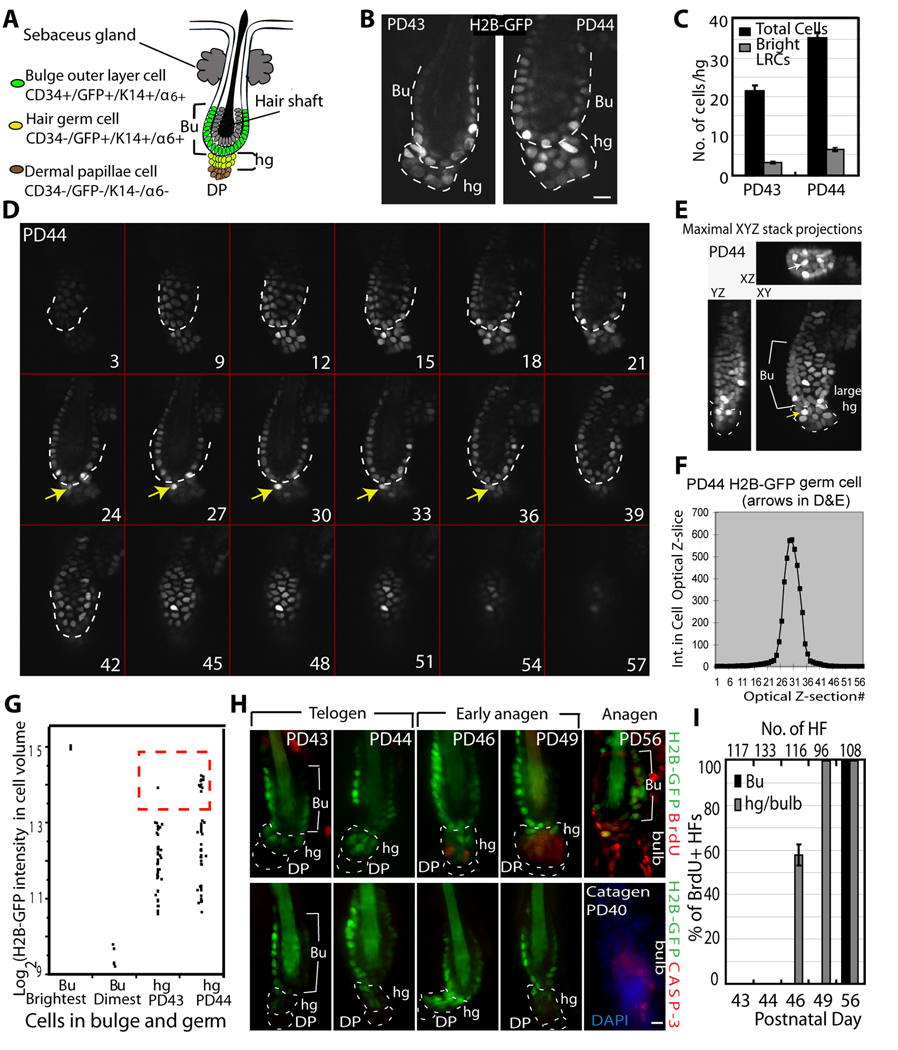
Figure 3.
Bulge-cell departure from the niche during the quiescent phase. (A) Schematic of telogen HF structure, showing 2 layers of cells surrounding the shaft that make the bulge (Bu), and the hair germ (hg) underneath. DP, dermal papillae. (B) Confocal image from whole HFs in 100µm skin sections of pTRE H2B-GFP/K5tTA mice with doxy chase from PD21. Note increased number of hg cells by PD44, quantified in (C) and shown with SEM (N=39 HFs). (D) Confocal stack images of whole HF collected side by side from skin sections of pTRE-H2B-GFP/K5tTA mice at PD43 and PD44 (3-wks doxy chase; PD43 is shown in Fig. S3). Optical z-stacks are shown as tiled images. Numbers indicate actual optical slice and arrows point to a bright H2B-GFP cell throughout the stack located in hg at PD44, normally found in bulge at PD43. TOPRO-3 was used as DNA counterstain for revealing the hair follicle structures (not shown). (E) Stacks in (D) are shown as maximal projection through the slices on XY, ZY, and XZ plane. Arrow points to the same cells as in (D). (F) Total intensity after background subtraction (Int) in each optical z-section for cell indicated by arrow in (D&E) used to obtain total 3D intensity, which was then used to generate the plot in G. (G) H2B-GFP intensity/hg cell volume shows more bright cells at PD44 relative to PD43 (see Fig. S3); brightest (0 divisions) and dimmest bulge cells are shown for comparison. There are only 1–2 cells with 0 division, and they were localized in the bulge at PD43&PD44. (H) Skin section for mice fed doxy and BrdU for 3 days at stages indicated, and stained for BrdU (top) or caspase (bottom). (I) Frequencies of BrdU+ HFs (N at top) averaged in 3 mice/stage, with SEM. Scale bars, 10 µm.
Results
Distinct characteristics of newly generated CD34+/α6-integrin+ cells at two hair cycle stages
We began by analyzing the proliferation and differentiation status of newly generated bulge cells during the first adult hair cycle, when HF remodelling phases are synchronous (Fig. 1B). We employed double transgenic mice, K5tTA x pTRE-H2B-GFP in which an epithelial Keratin 5 (K5) promoter drove repressible histone H2B-GFP expression ( Tumbar et al., 2004) . Repression is achieved by feeding the mice doxycycline (doxy) for a period of time (chase), when the H2B-GFP dilutes in cells by 2-fold at division. This allowed us to quantify precise proliferation history in vivo, from the amount of H2B-GFP fluorescence retained in cells after chase (Waghmare et al., 2008). After various chase periods (Fig. 1C) we sacrificed mice at different ages corresponding to distinct phases of hair cycle, as confirmed by microscopy (Fig. S1A). We isolated skin cell suspensions from these mice and stained them for surface expression of bulge markers CD34 and α6-integrin. Fluorescence-activated cell sorting (FACS) revealed histograms with distinct peaks of 2-fold median H2B-GFP intensity corresponding to distinct divisions (Fig. 1D,E). These data quantified the bulge proliferation during the first hair cycle, and provided information on kinetics and fraction of divided bulge cells. The data showed in Fig. 1E indicated that CD34+/α6+ cells began to divide at the telogen-anagen transition (Postnatal day (PD)18-PD21 chase) and continued on in anagen (PD22-PD25&PD21-PD28 chases). The cells slowed-down divisions by late anagen (PD21-PD35 chase) and became quiescent in catagen (PD21-PD42&PD35-PD42 chases). Most CD34+/α6+ cells divided infrequently (1–3x) during the PD21-PD42 chase, in line with our previous data from a full hair cycle chase (PD21-PD49; Waghmare et al., 2008).
Since single CD34+/α6+ sorted cells gave rise to rapidly dividing progenitor Mx cells upon transplantation (Fuchs, 2009), we examined FACS profiles for emergence of a more rapidly dividing cell population of cells. Histograms from PD18-PD21 doxy chases indicated 2–3 divisions, which occurred in <15% of CD34+/α6+ cells (Fig. 1E, top). In contrast, the histograms from PD22-PD25 chases showed on average only ~1 division in >80% CD34+/α6+ cells. In addition, the CD34 surface levels appeared largely reduced in PD18-PD21 divided cells (Fig. 1E right panels & Fig. S1C). To probe this observation further, we sorted skin cells subpopulations as CD34+/α6+ bulge (Bu) cells, with H2B-GFP signals indicating they were un-divided (Bu, 0 Division) or divided (Bu, 1–2 Divisions) and CD34-/α6+ non-bulge (Non-Bu) cells (Fig. 1E, right panels). We analyzed these cells by RT-PCR for mRNA expression of CD34 and other bulge-enriched factors (Tumbar et al., 2004), which appeared largely down-regulated in divided cells from PD18-PD21 but not from PD22-PD25 chases (Table S1; Fig. 1F). To find the divided CD34+/α6+ cells in situ we administered Bromodeoxyuridine (BrdU) together with doxy for 3 days (PD18-PD21 and PD22-PD25). In the PD18-PD21 experiment CD34+/BrdU+ cells localized exclusively at the bulge/germ transition zone, near the known region where Mx cells emerge and cells begin to proliferate first (Cotsarelis, 2006; Fuchs, 2009) (Fig. 1G, top). In contrast, in the PD22-PD25 experiment CD34+/BrdU+ cells were present inside the bulge (Fig. 1G, bottom). In conclusion, newly generated bulge cells, analyzed as CD34+/α6+ divided cells, seemed to divide at higher rates and show lower expression of bulge-enriched markers at PD18-PD21 relative to PD22-PD25.
Stage-dependent genomic expression signatures of newly generated CD34+/α6+ cells
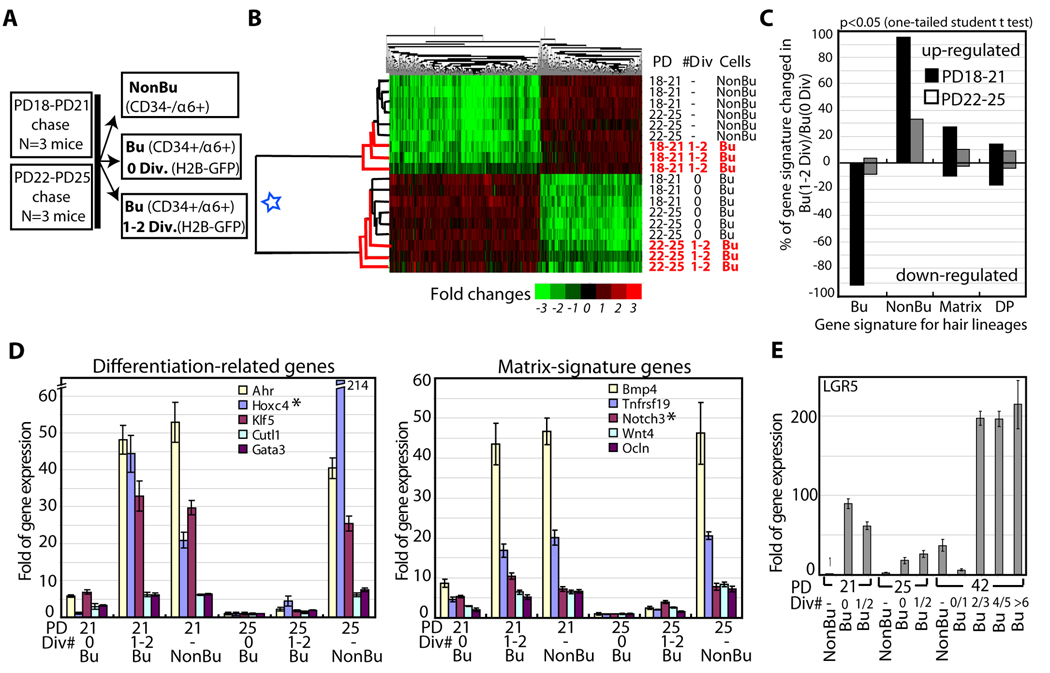
To extract information about differentiation status of newly generated bulge cells at distinct hair cycle phases, we determined the gene expression signatures in subpopulations of HFs isolated from mice treated with doxy from PD18-PD21 and PD22-PD25 stages: telogen-anagen transition and anagen, respectively. We sorted CD34+/α6+ bulge cells as divided (Bu, 1–2 Div) and undivided (Bu, 0 Div) subpopulations and the CD34-/α6+ (non-bulge) subpopulation (N=3 mice per stage; Fig. 2A). We analyzed a total of 18 RNA samples by Affymetrix expression microarrays (Experimental Procedures). First, we extracted a set of 767 probes changed by ≥2 fold (p≤0.05; one-tailed student t test) in all 6 undivided CD34+/α6+ (bulge) populations relative to 6 CD34-/α6+ (non-bulge) populations (Table S2). This set constitutes the bulge and non-bulge signatures of genes that remain expressed preferentially in each subpopulation at both PD18-PD21 and PD22-PD25 stages. Log2 of signal/average values from this set (and an inclusive 9450 probe set) yielded expression dendograms with two major clusters: one for the 6 undivided CD34+/α6+ bulge subpopulations (triplicate per each of the two stages) and another for the 6 CD34-/α6+ non-bulge subpopulations (Fig. 2B; Fig. S2A). Strikingly, the divided CD34+/α6+ cells (Bu, 1–2 Div) from the PD18-PD21 chase, but not from the PD22-PD25 chase, clustered together with the CD34-/α6+ non-bulge subpopulation (Fig. 2B). At PD22-PD25, the divided CD34+/α6+ cells showed little changes in gene expression relative to undivided CD34+/α6+ cells (Fig. 2C), and these changes were likely related to the differences in their proliferative status. In contrast, at PD18-PD21 the divided CD34+/α6+ showed down-regulation of most (~95%) bulge-enriched mRNAs or “bulge signature genes” relative to undivided CD34+/α6+ cells and up-regulation of some previously reported Mx signature genes (Rendl et al., 2005; Fig. 2C). QRT-PCR for 5 transcription factors previously implicated in keratinocyte or HF differentiation and for 5 reported Mx signature genes (Tumbar, 2006; Jones and Reiners, 1997; Luong et al., 2002; Rendl et al., 2005; Rieger et al., 1994; Sur et al., 2006) confirmed the microarray expressions. These mRNAs were high in divided CD34+/α6+ cells from PD18-PD21 chases, but not from PD22-PD25 chases (Fig. 2D). This high expression was not found in undivided α6+/CD34+ cells at PD18-PD21, even when we sorted them into CD34-medium and CD34-high cells (Fig. S1D). The PD18-PD21 divided CD34+/α6+ cells, although clustered with CD34-/α6+ cells in the expression dendograms, displayed their own subset of enriched mRNAs attesting to their distinct identity (Table S3). LGR5, a recently reported lower bulge/HG marker (Jaks et al., 2008), was higher in all PD18-PD21 CD34+/α6+ cell fractions relative to CD34-/α6+ non-bulge cells or relative to PD22-PD25 CD34+/α6+ bulge fractions (Fig. 2E and Fig. S2B,C), as expected (Jaks et al., 2008). Unlike the previous report (Jaks et al., 2008), in catagen (PD21-PD42 chase) we found consistently high levels of LGR5 mRNA expressed in most CD34+/α6+ cell fractions, including those that divided only 2–3x in one hair cycle (see Discussion). LGR5 expression was variable in cells with 0–1 divisions, consistent with previously reported rare association of LGR5+ cells with BrdU label retaining cells (LRCs; Jaks et al., 2008). In summary, our gene expression data demonstrated divergence in expression profiles of divided CD34+/α6+ cells at the two hair cycles stages analyzed (PD18-PD21 and PD22-PD25).
Figure 2.
Expression profiling at 2-hair cycle stages. (A) Cell subpopulations sorted and profiled by expression microarrays. (B) Gene (top), and cell population (left) dendogram heat map shows 2 main population clusters (blue star). Newly divided CD34+/α6+ cells (Bu 1–2 Div) cluster with a different group at each stage. (C) Genes changed in Bu (1–2 Div) cell populations relative to Bu (0 Div) are shown as percent of 4 hair lineages gene signatures: CD34+/α6+ bulge (Bu). CD34−/α6+ non-bulge (NonBu), matrix (Mx), dermal papillae (DP) (Table S2). Note down-regulation of bulge signature genes and higher up-regulation of Mx genes at PD18-PD21. DP was a negative control as an unrelated lineage and showed low and similar representation in both populations. (D). QRT-PCR (relative to GAPDH) of several Non-Bu or Matrix signature genes found up-regulated in our microarray in Bu (1–2 Div) relative to Bu (0 Div) shown with SEM. N=2 mice per stage (triplicate wells). Regular RT-PCR performed for 4 additional mice per stage is not shown. (E) Same QRT-PCR analysis for LGR5; a second experiment with another set of mice at each stage is shown in Fig. S2.
The data thus far suggested that newly divided (1–2 divisions) CD34+/α6+ cells, which may represent newly generated bulge cells, were distinct in molecular makeup, proliferation rates, and localization at PD18-PD21 (telogen-anagen) with respect to PD22-PD25 (anagen). The CD34+/α6+ cells generated at the telogen-anagen stage displayed characteristics consistent with progenitor cell differentiation: more vigorous divisions; localization right outside the SC niche (bulge) in the tissue zone where Mx is first generated; high expression of differentiation and progenitor Mx markers; and lower expression levels of most bulge markers. In contrast, later in anagen, newly generated CD34+/α6+ divided less frequently, remained inside the SC niche (bulge), and maintained the bulge-enriched gene expression signature. Although it is unclear if the divided CD34+/α6+ cells isolated here at telogen-anagen transition were directly bulge-derived, this possibility is likely, given the earlier tracking of downward cellular displacements from bulge into germ (Cotsarelis, 2006; Fuchs, 2009). These data would then imply that newly generated bulge progeny differentiate near the niche exit during an early hair cycle stage while expanding their pool of bulge-like cells (self-renewing) inside the niche at the latter stage. To directly address this hypothesis we explored further the lineage relationship between bulge, HG, and Mx/bulb cells in hair cycle.
Bulge cells leave their niche prior to division
In vivo evidence based on short-term tracking of bulge LRCs, indicated that bulge progeny contribute differentiated cells to the HG/Mx formation (Cotsarelis, 2006; Fuchs, 2009). It remains unclear, however, if the cells divide/self-renew prior to their departure from the bulge. A model derived from transplantation of rat whisker cells suggested that bulge SCs migrate away from the niche during quiescence (Oshima et al., 2001). Here we recognized bulge cells due to higher levels of H2B-GFP retention after PD21-PD42 doxy chase, and subsequently tracked their location in HF compartments in the second telogen. Telogen morphology is defined by 3 molecularly distinct compartments: bulge, made of 2 concentric epithelial cell layers surrounding the club hair, with SCs in the outer CD34+ layer; hair germ (HG), an epithelial cell cluster underneath with little if any CD34; and the mesenchymal dermal papillae (DP) structure (Fig. 3A). In over 25 mice sacrificed between PD42-PD56, HF morphology indicated end of catagen at PD42, telogen with small HGs at PD43, somewhat larger HGs at or after ~PD47, and mostly anagen at PD56 (not shown). To examine if bulge cells directly contributed to HG size increase by first dividing in the niche, we counted number of HG cells at two telogen time points, and correlated it with H2B–GFP levels and BrdU incorporation over time. We removed dermal cells by collagenase treatment to release full HFs at PD43 and PD44 and imaged them by confocal optical sectioning (Rendl et al., 2005; Waghmare et al., 2008). Analysis of image stacks, maximal projections and rotations showed that HGs contained on average ~20 cells at PD43 and 35 cells at PD44 (Fig. 3C). Moreover the number of apparent bright H2B–GFP cells in the HGs also increased at PD44 relative to PD43 (Fig. 3B–E; Fig. S3). This semiquantitative analysis was confirmed by quantitatively integrating the H2B–GFP intensity in a 3D cell volume for a smaller set of HFs (Fig. 3F,G). These data demonstrated that bulge cells, including some cells with H2B–GFP levels consistent with only one division, contributed to an increase in HG size from PD43 to PD44. Moreover, this happened in the absence of divisions because we detected no proliferation in HFs from mice sacrificed 6 hrs after BrdU injection (Fig. 3H,I) or after continuous administration of BrdU for 3 days (PD41-PD44, N=3 mice, data not shown). At later stages BrdU appeared first in HGs and then in bulges (Fig. 3H,I), as expected (Greco, V. et al, 2009), and as seen before in the first telogen (Fig. 1G). Catagen specific caspase staining to detect cell death was absent in all stages examined (Fig. 3H); PD40 was used as positive control for this staining. These data indicated that during telogen bulge cells, including some that divided rarely (~once in 3 weeks), leave their normal niche and go into the HG without a simultaneous bulge-replenishing division.
Retrospective view of single bulge cell fate: differentiation or self-renewal phases
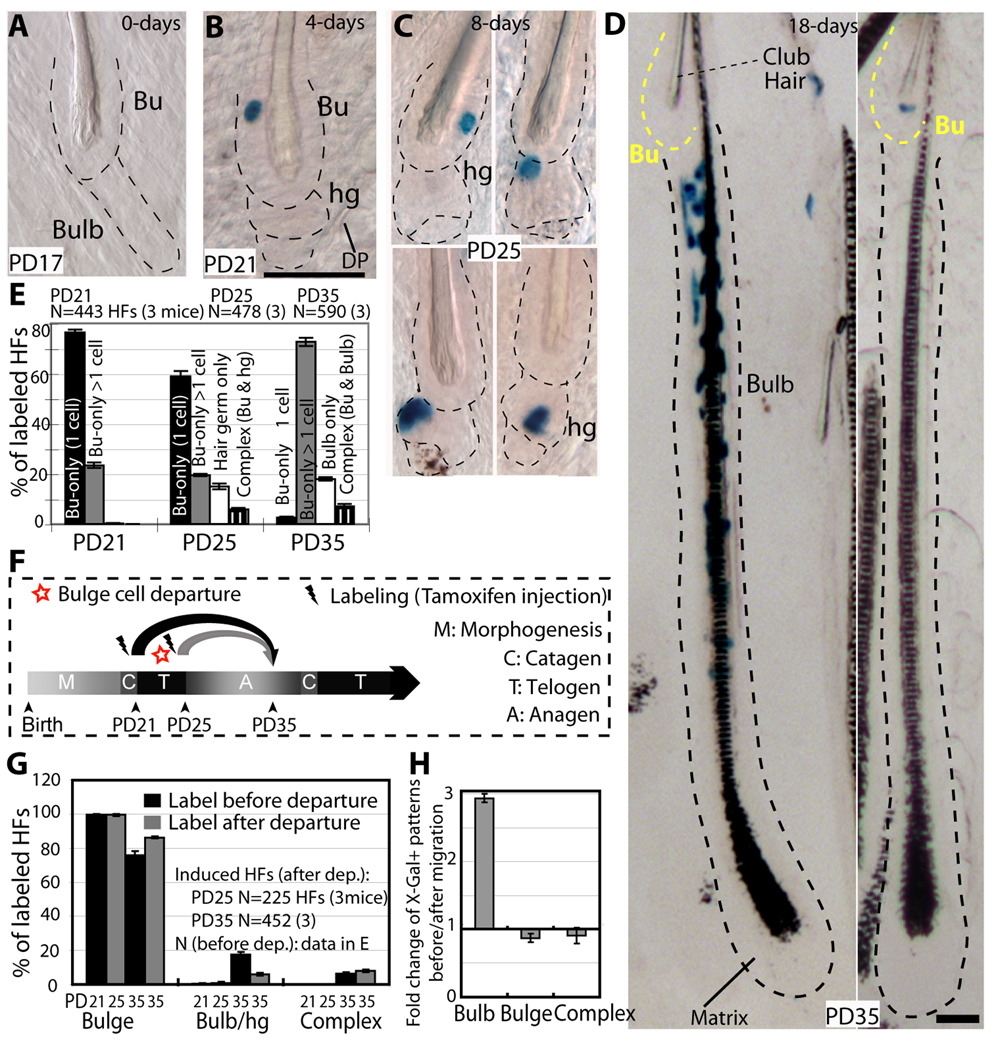
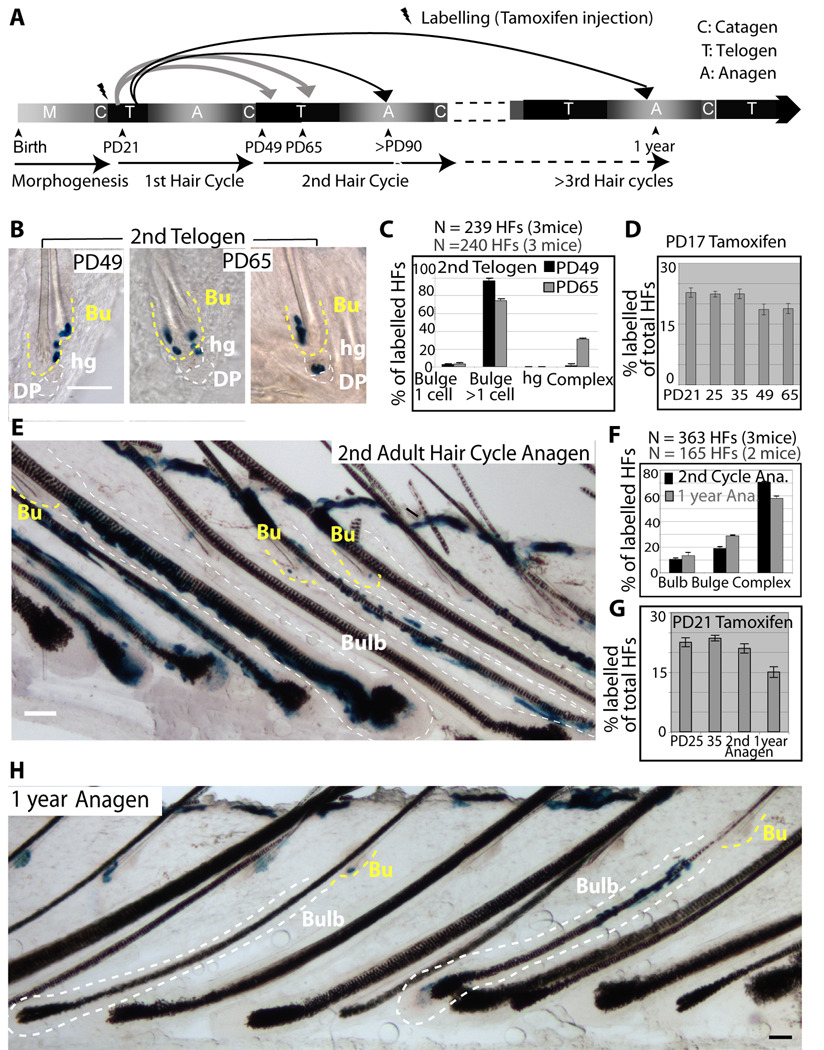
To map bulge-cell fate in vivo we genetically labelled single cells at quiescence and examined progeny distribution in HF compartments during hair growth. We chose the first telogen for labelling because it precedes the most synchronous hair cycle. We employ transgenic mice with keratin 14 (K14) epithelial expression of the tamoxifen inducible Cre-ER (Vasioukhin et al., 1999) crossed with Rosa26R reporter mice (Soriano, 1999). Tamoxifen injection into mice turns on β-Galactosidase in epithelial cells. The expression remains on in all the cell lineages descending from the originally marked cell, and is detected by X-Gal staining in skin sections. To increase our chance of labelling single bulge cells per HF we utilized the more inefficient mouse line of the two previously generated (Vasioukhin et al., 1999). First, we maximized X-Gal staining efficiency in a strong constitutively expressed epithelial K14-Cre line (Fig. S4C). Second, we established the appropriate dose of tamoxifen (Fig. S4A), the precise timing of hair cycle stages in this new mouse genetic background (Fig. S4B), and the minimum chase time (4 days) for reaching X-Gal labelling plateau during a prolonged quiescence phase to eliminate confounding proliferation effects (Fig. S4D). Finally, we injected mice with tamoxifen at the catagen/telogen transition (PD17), sacrificed them at different stages and analyzed skin tissue at telogen (PD17-PD21 chase), early anagen (PD17-PD25 chase) and late anagen (PD17-PD35 chase; Fig. S4E). Since the K14 promoter was active in several skin epithelial compartments in their basal cells, X-Gal+ cells were found in HFs, inter-follicular epidermis, and sebaceous glands. The latter, located above the bulge, do not contribute cells to normal HFs homeostasis (Horsley et al., 2006). X-Gal staining was undetectable in skin sections analyzed from 10 K14-CreER x Rosa26R mice injected with oil only (no tamoxifen control) at PD17 and sacrificed at PD25, 35, 90 and over 1 year (Fig. S4H). Moreover, the tamoxifen induced X-Gal labelling was highly inefficient (~20% of all HFs), and occurred as rare patches dispersed throughout the mouse back skin (Fig. S4H). To examine X-Gal distribution in full HFs, we used thick skin sections (60–90µm thick). We imaged them by phase contrast or bright field microscopy in skin sections with or without hematoxylin staining, which adds contrast to the tissue (Fig. 4A–D, Fig. S4F–H). To emphasize the X-Gal labelling, images are shown with little contrast. The K14 promoter is active in both bulge and HG cells, but given a cell ratio of bulge to HG cells of roughly 10:1, by random chance we expected ~90% X-Gal+ HFs with bulge-only labelling. In fact, we found ~99% of PD17-PD21 X-Gal+ HFs with bulge-only staining (76% as single and 23% as multiple cells; Fig. 4B,E). Some unexplained bias against HG marking by the CreER might have also contributed to specificity of labelling to the bulge.
Figure 4.
Single-bulge cells lineage tracing during 1st adult hair cycle. (A–D) X-Gal stained skin sections (60–90µm) from K14CreER x Rosa26R mice tamoxifen injected at PD17 and sacrificed at times indicated. Dotted lines delineate the bulge (Bu) and hair germ (hg) or bulb compartments. Scale bar, 50 µm. (E) Frequencies of X-Gal patterns shown as average with SEM (N.3 mice/stage). DP, dermal papillae. (F) Scheme of chases PD17-PD21 or PD21-PD25, and X-Gal patterns in HFs at PD35 in anagen (G), re-shown as fold changes (H).
If bulge-cell daughters followed a simple model of asymmetric fate decisions with respect to their niche, they would replenish the bulge (SC) pool (self-renew) and simultaneously generate the progenitor cell (differentiate), which would be expelled from the bulge and differentiate further into the bulb. Thus, after a growth period the X-Gal+ progeny of the original single-labeled bulge cell should be always detectable in both the bulge (stem cell) and the bulb (differentiated) compartments (complex labeling pattern). Strikingly, skin from PD17-PD25 chases showed only 6% HFs with a complex pattern, likely attributable to noise in the system due to 23% of the original labeled HFs with multiple X-Gal+ cells (PD17-PD21). The remaining 94% of labeled HFs had X-Gal+ cells exclusively in either one of the two HF compartments: 80% HFs with bulge-only X-Gal labeling pattern and 14% HFs with HG-only pattern (Fig. 4C,E). The bulge-resident cells remained largely undivided in the PD17-PD25 chase, as shown by ~60% of labeled HFs with a single X-Gal+ cell/bulge. In contrast X-Gal+ HG cells were found single or in pairs (indicative of divisions), sometimes in direct contact with the DP (Fig. 4C and S4G). These patterns detected in thick skin sections could be reconstituted from a random pool of 36-labelled HFs in serial thin (20µm) sections, ruling out potential issues of X-Gal penetration (Fig. S5A–C). Moreover, the fraction of labeled HFs in PD17-PD21, PD17-PD25, PD17-PD35 chases remained constant (Fig. 6D). This rules out the possibility that a significant fraction of single labeled bulge cells were missed in our analysis, or that the labeling may take longer to reveal in the HG or hair bulb cells. These data indicated that bulge cells exited their bulge residency without simultaneous division inside the niche, consistent with our previous H2B–GFP tracking data (Fig. 3). We also examined the distribution of single X-Gal+ cells within the bulge in PD17-PD21 chase and PD17-PD25 chase in > 100 HFs. The X-Gal+ cells marked preferentially the middle bulge at PD21 and the lower bulge at PD25, suggesting a generalized downwards bulge cellular displacement (Fig. S5D). Simple upward motion of the hair shaft can potentially explain these data, but might not account for the presence of bulge X-Gal+ cells away from the bulge base onto the DP (Fig. 4D&Fig. S4G). This latter observation suggests a possible active migration for the X-Gal+ bulge-cell through the pre-existing HG cells (which were X-Gal-) to reach the DP.
Figure 6.
Long-term single-bulge cell lineage tracing. (A) Scheme of 1st telogen labelling and long-term chases. (B,E&H) Illustrative images from thick (60–90µm) skin sections show X-Gal staining in bulge (Bu) (yellow line) and bulb/germ (white line) compartments. Image in (E) shows a rare patch of frequent HF induction illustrating the coexistence of all 3 staining patterns in one skin region. Image in H shows bulb-only and bulge-only patterns after one year and at least 3 hair cycles post-labelling. The predominant patterns were complex (see counts in F). Scale bars, 50µm. (C&F) Quantification with SEM of X-Gal patterns at stages indicated. (D&G) Frequency of all labelled follicles counted at each stage (from data in Fig 4&Fig 6) show significant decline from one hair cycle to another.
X-Gal+ cells at full anagen (PD35) remained segregated in either bulb or bulge compartments in 93% of the labeled HFs (Fig. 4D). HFs with bulb-only labelling pattern, represented 17% of all HFs labelled at PD35, and contained numerous X-Gal+ cells in all the differentiated hair lineages in thick sections and cross-sections, and in thin serial sections (Fig. 4D and Fig. 5). These data were consistent with previous transplantation and skin dermabrasion experiments (Cotsarelis, 2006; Fuchs, 2009), and indicated frequent division of the bulge-progeny in the bulb. The data also demonstrated multipotency in vivo of the originally marked single bulge-cells (Fig.5). Bulbs showed sectors of labelling as expected from their known polyclonal origin (Cotsarelis, 2006). Only a fraction of the bulb-labelled follicles displayed X-Gal+ Mx cells, most likely due to the cessation of self-renewal of this progenitor/transit amplifying cell population by the end of anagen at PD35, as expected (Legue and Nicolas, 2005). The bulge zone around the old hair shaft (club hair) completely lacked X-Gal staining in the bulb-only patterns (Fig. 4D and Fig. S4H), while the zone adjacent to it rarely displayed few X-Gal+ cells (~2% of the total labelled HFs) mostly in inner (differentiated) layers around the new hair shaft.
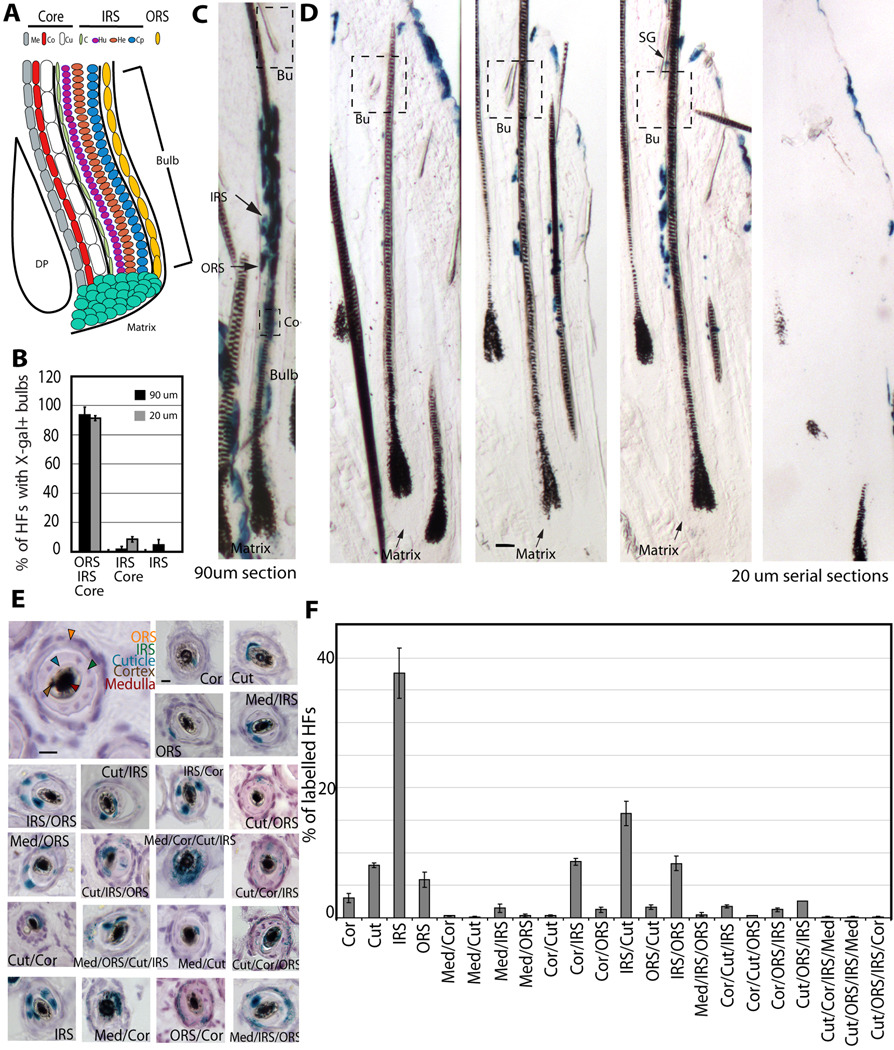
Figure 5.
Multipotency of bulge cells in vivo.
(A) Hair bulb with indicated differentiated lineages. Outer root sheath (ORS); IRS (inner root sheath made of cuticle (C), Huxley (Hu), and Henle (He) cell layers); hair shaft “core” made of 3 cell layers: (Cu) cuticle, (Co) cortex, (Me) medulla. Cp is ORS companion layer, included with the IRS category for simplification. (B) Quantification of PD35 labelling patterns in mice injected with tamoxifen at PD17. Lineages containing X-Gal+ cells are indicated at bottom, shown with SEM in 3 mice (>60 labelled HFs/mouse). (C&D) X-Gal bulb-only patterns in thick (90µm) or thin (20µm) serial sections. Scale bar, 50µm. (E) Cross-sectional images of X-gal stained skin from PD17-PD35 chase 20µm shows multiple differentiated layers in a majority of HFs. Scale bar, 10 µm. (F) Patterns of labelling, with SEM shown as % of a defined pattern from total labelled HFs (N= 3 mice, > 200 labelled HFs/mouse).
HFs with bulge-only labelling patterns at PD35, represented 76% of total labelled HFs, and contained 2–5 X-Gal+ cells clustered around the club hair (Fig. 4D and Fig. S4H). These data were consistent with rare cell divisions (mostly 1–3) within the niche, in line with previous results from quantitative proliferation history (Fig. 1 and Waghmare et al., 2008). The presence of multiple X-Gal+ cells confined to the bulge indicated that bulge cells divided within the niche and generated more bulge-cell progeny, but none of them left the niche within the first hair cycle. These data collectively provides evidence for divergent fate of individual bulge cells within one hair cycle to either: a) leave the niche, proliferate vigorously to generate many differentiated bulb cells at an earlier stage; b) remain in the SC niche (bulge), divide infrequently, generate more bulge cells and replenish the bulge pool (self-renew) at a latter stage.
To further define these differentiation and self-renewal stages, we marked cells in the bulge before or after some of them departed the bulge (Fig. 4F). To do this we injected tamoxifen at PD21, after the bulge cell migrated into the HG, and examined HF labelling in PD21-PD25 and PD21-PD35 chases. We found comparable labelling efficiency with that obtained from previous chases (PD17-PD21 and PD17-PD35; Fig. S4E). Moreover, 99% of the PD21-PD25 labelled HFs displayed bulge-only patterns, 76% as single cells (Fig. 4G). Importantly, the bulge-only (self-renewal) pattern was more prominent in PD21-PD35 chase than in previous PD17-PD35 chase, while the bulb-only (differentiation) pattern was diminished by 3-fold (Fig. 4H). These results indicated reduction in bulge-cell differentiation fate choice by the time of early anagen onset and supported a temporal segregation of differentiation and self-renewal phases of bulge cells in telogen/anagen transition versus anagen.
Differentiation or self-renewal of long-term bulge residents throughout life
Our data thus far suggested a spatial and temporal divergent fate of bulge resident cells to either differentiate or self-renew. However, the bulb-only pattern, indicative of unidirectional differentiation fate, might arise from direct X-Gal labelling of a putative short-lived progenitor cell temporarily residing in the bulge rather than from the SC itself. These progenitors could be generated in anagen by the asymmetric divisions of the true long-lived SCs, and could remain confined to the bulge until the next telogen-anagen transition. In this scenario, the true SCs might only exist among the X-Gal+ cells that self-renewed in anagen by divisions and survived in HFs through at least one hair cycle. If these SCs behaved by an asymmetric cell fate decision model, then we should always detect complex patterns in the subsequent hair cycles. However, if the SCs undergo unidirectional or symmetric fate decisions, we may still detect bulb-only patterns in which the originally labelled cell, and all of its bulge-progeny altogether decide to leave the bulge pool, without maintaining any replenishing descendants behind in the niche.
To follow the fate of bulge-derived cells in long-term we analyzed X-Gal patterns in mice sacrificed at different time points after the first hair cycle, up to 1 year of life (Fig. 6A). We first checked skin sections at PD49 in early telogen, which follows the massive bulb apoptosis that occurred in catagen. When we compared the overall frequency of marked HFs before catagen (PD35) and after catagen (PD49), we found a 4% overall drop, accounting for a loss of ~17% of the labelled HFs (Fig. 6D). Since at PD35 17% of HFs showed bulb-only labelling patterns, these data suggested that all X-Gal+ bulb cells died in the massive apoptosis detected in the bulb in catagen. If some X-Gal+ bulb cells in each HF survived catagen to make a new HG, as previously proposed (Jaks et al, 2008), we would expect a substantial fraction of HFs (17% of the PD35 bulb-only labelled follicles) to show exclusive X-Gal labelling in HGs at PD49. However, we found no HFs with exclusive HG labelling pattern (Fig. 6C). Furthermore, at PD49 the surviving X-Gal+ cells were found exclusively in the bulge in ~99% of labelled HFs and in bulge and HGs in the remaining ~1% (Fig. 6B,C). The X-Gal+ bulge clone size was on average ~ 4 cells (N=61 HFs) roughly comparable with the bulge clone size at PD35. By PD65, in late second telogen when bulge cells are still in divisional quiescence, we detected X-Gal+ cells in both bulge and HGs in ~24% of labelled HFs (complex pattern; Fig. 6B,C). The bulge-only pattern decreased proportionally at PD65 relative to PD49 to 74% of the labelled HFs. These results indicated that bulge-cells left the niche again in quiescence during the second telogen, re-iterating their earlier behaviour.
In second anagen (PD90) or after 1-year, ~70% of labelled HFs showed mainly complex patterns (Fig. 6E–H). These results demonstrated the contribution of the originally single labelled bulge cells to all hair compartments including their own, after repeated cycles of regeneration, a hallmark of SCs. The remaining HFs showed bulge-only (19–29%) or bulb-only (11–13%) labelling patterns, in thick (90µm) or thin serial sections (Fig. 6E,F,H; Fig. S6). The bulb-only pattern is indicative of unidirectional fate decision for the long-lived self-renewing bulge cells to leave their niche and differentiated into the bulb. The bulge-departed cells were likely lost in the bulb by differentiation and apoptosis as supported by the incremental decline in frequency of labelled HFs from one hair cycle to another (Fig. 6G).
Discussion
Single-cell fate tracing in intact vertebrate tissue confirms the “slow-cycling” SC model
The “slow-cycling” SC model states that cells with infrequent divisions contribute to normal homeostasis of adult tissues. Most evidence at the single cell supporting this model comes from blood and hair follicle in vitro transplantation assays (Fuchs, 2009). Recent evidence from unperturbed tissue employing lineage tracing in epidermis, hair follicle, and intestine seemed to challenge this model (Barker et al., 2007; Clayton et al., 2007; Jaks et al., 2008). Here we provide evidence from an intact vertebrate tissue, at the single cell level and in long-term assays, for the infrequent division model in hair follicle bulge stem cells.
We employed mice with repressible H2B–GFP expression (Tumbar et al., 2004) to determine proliferation kinetics, gene expression, and cell identity data. Furthermore, we used epithelial driven inducible K14-CreER transgenic mice (Vasioukhin et al., 1999) to genetically mark single bulge cells and follow their fate during adult hair regeneration. We found that in the bulge, single X-Gal+ cells generated ~2–5 progeny during anagen, which remained confined to the bulge in ~70% of labeled HFs, and were not exported in the bulb during that same anagen. This suggested infrequent (mostly 1–3) anagen divisions, in line with our H2B–GFP division counts (Fig. 1 and Waghmare et al., 2008). The newly generated bulge cells at anagen preserved the known bulge characteristics, as shown by location, kinetics of divisions, and gene expression profiles, therefore self-renewing and expanding their own pool. In the next two anagen phases analyzed here, ~70% of these cells generated progeny which contributed to all differentiated bulb lineages, while maintaining long-term progeny as bulge residents (complex X-Gal patterns; Fig. 6E,F). Thus, we conclude that a majority of these long-lived infrequently dividing (bulge) cells differentiate and self-renew (although not necessarily by asymmetric division; see below), a hallmark of multipotent SCs. In the bulb, bulge-derived progeny behaved as bona fide multipotent progenitor (transit amplifying) cells, generating large clones of hundreds of X-Gal+ cells in only 2 weeks (PD21-PD35). Finally, the bulb cells derived from the bulge did not survive in the bulb past the apoptotic (catagen) phase to make the hair germ (HG). This was suggested by lack of exclusive HG labelling patterns and little (<1%) HFs with complex bulge and HG labelling in the second telogen, and by the incremental decrease in overall labelled HFs from one hair cycle to another by a fraction numerically consistent with the frequency of bulb-only patterns detected in the previous cycle.
LGR5 was reported as a germ-specific gene in catagen, presumably marking preferentially frequently cycling cells (Jaks et al 2008). However, we detected LGR5 mRNA expression in CD34+/α6+ bulge cells at all hair cycle stages, although the levels were lower in anagen. In catagen, expression was variable in cells with 0–1 divisions, but strong in cells with only 2–3 divisions after one complete anagen (Fig. 2E). These observations confound the interpretation of recently published lineage-tracing data. That said, the bulge cells fate-mapped in our present study via Cre recombination contributed to Mx and inner hair lineages but not to the early telogen HG, suggesting that unlike the Mx, the HG might be a bulge-independent lineage. These data might suggest a possible direct lineage relationship of HG to make most of the ORS, and of bulge to make Mx and some ORS. ORS and Mx cells were maintained as independent progenitors as shown by lineage tracing in anagen (Legue et al., 2005). This possibility requires further experimental testing via a germ specific Cre-driven recombinase. Importantly, the majority of lower ORS cells dilute rapidly BrdU or H2B–GFP label, while surviving bulge and some germ cells retain significant label after one hair cycle (Cotsarelis, 2006; Greco et al., 2009; Tumbar et al., 2004). We quantified that here (Fig. S1A; Fig. 3B–G) and found that all HG cells divided in fact infrequently, only 2–5x during one hair cycle (Fig. 3G). Thus, if a putative lower bulb traveling HG “cycling” population existed (Jaks et al., 2008), would differ from a “dormant” bulge population by a mere 1–2 divisions, since bulge cells divide on average ~ 3x (Waghmare, 2008). Our previous experiments utilizing BrdU and H2B–GFP pulse-chase demonstrated that a few extra divisions are sufficient to dilute BrdU beyond detection (Waghmare, et al 2008), which can lead to miss-interpretation of the results from BrdU label retention experiments. We conclude that all the surviving cells in the permanent HG segment are in fact infrequently dividing. Given an estimate of ~ 10 hair cycles in a lifetime they divide altogether ~30–50 times, while some of them divided much fewer times.
Intriguingly, our H2B–GFP chases documented that most bulge divisions occur over a short period of only several days and bright H2B–GFP LRCs group on one side of the bulge (examples in Fig. 3). These data suggested a potentially short-lived inductive polarized signal, which may awaken bulge cells to rapidly proliferate for a few days and than return to quiescence. The activation signal has been proposed to travel as a wave across the body (Plikus et al., 2008) and may be relayed from follicle to follicle. It is possible that a few bulge cells, potentially similar to the BMI-1 expressing cells at the +4 position in the intestine, may remain relatively unused in normal tissue growth ( Sangiorgi, et al, 2008; Fuchs, 2009), and may account for a fraction of the bulge-only patterns detected in our long-term assays. These cells need not represent a distinct population and do not remain quiescent, but may simply divide fewer times due to their farther position relative to some diffusible activation and migration signals.
The HFSC niche displays distinct differentiation and self-renewal stages
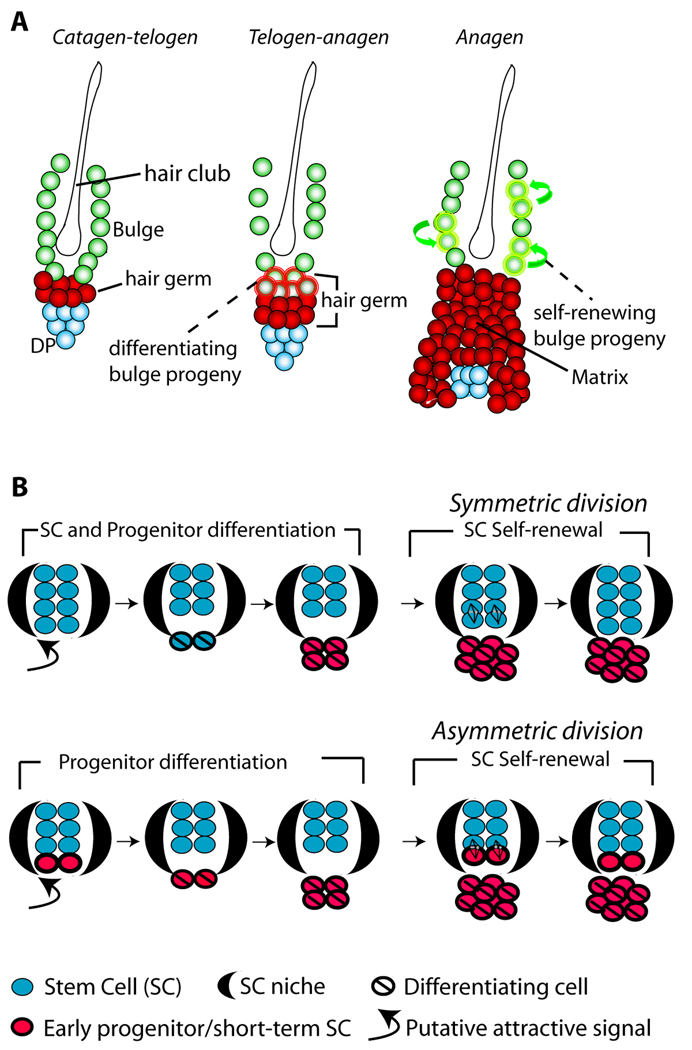
During telogen-anagen transition HG cells divide first while bulge cells follow later in anagen (Cotsarelis, 2006; Greco V, 2009; Fuchs, 2009). However, data have been lacking as to whether bulge cells would constantly export the newly generated putative progenitor cells into the bulb upon each division. Such export would be expected from a simple relationship of the stem and progenitor cells with the resident niche (bulge), as predicted from an asymmetric fate decision documented by early work in Drosophila (Morrison and Kimble, 2006). Our data clearly showed that upon division in anagen bulge-cell progeny were not immediately exported into the bulb, but rather remained confined to the niche (70% bulge-only patterns in first anagen) where they retained the bulge-specific characteristics, and finally returned to quiescence. Therefore in anagen the bulge cells divided and produced more of their kind, self-renewing their pool. To replenish the tissue, the bulge cells departed the niche at a different stage (telogen), in quiescence, and did not simultaneous self-renew the bulge pool by divisions. Once outside the niche the bulge-derived cells subsided to a fate of proliferation, differentiation, and death in the bulb as suggested collectively by all of our data including lineage-tracing experiments. From early anagen (PD25) to late anagen (PD35) the frequency of HFs with any germ/bulb labelling remained nearly constant, another indication that in anagen bulge cell divisions did not significantly contribute differentiated progeny cells to the bulb (Fig. 4E). Moreover, telogen resident bulge cells showed 3x higher propensity to depart the niche and differentiate than early anagen bulge cells (Fig. 4F–H). These data collectively suggest that individual cells within the HFSC niche, the bulge, are assigned distinct fates to either leave the niche and differentiate, or stay in the bulge and self-renew the pool at different time points in homeostasis (Fig. 7A). The HFSC niche displays distinct stages of differentiation and self-renewal, which occur in telogen and telogen/anagen transition and in anagen, respectively. Progenitor cell differentiation takes place at distinct times and tissue location from the self-renewal of SCs. This model is consistent with the previous results from long-term in vitro assays on whisker bulge cells (Oshima et al., 2001). We speculate that the spatial-temporal segregation of self-renewal and differentiation may ensure protection of the SC niche from penetration of cell differentiation tissue signals.
Figure 7.
Dynamics of hair follicle bulge stem cells during tissue homeostasis
(A) The bulge cells undergo distinct phases of differentiation and self-renewal. First, at the telogen-anagen transition bulge cells depart the niche, proliferate and begin to differentiate to matrix progenitors of inner hair lineages. Second, bulge cells replenish their pool by divisions in anagen (self-renew), when newly generated bulge cells remain in the niche during the same hair cycle. (B) Models for adult SC renewal by illustrating symmetric (top) and asymmetric (bottom) divisions, and the predicted organization and dynamic of stem/progenitor cells in the niche in each scenario. Loss of SC identity may occur outside the niche (bulge) before or after the first differentiating division.
Symmetric fate decisions for stem cell daughters: Future lessons for SCs dynamics throughout life
Our data provide information on the long-term dynamics and interplay of fate decision that SCs face in a complex tissue in adult life. Let us assume a rigid model of asymmetric bulge SC fate decisions during adult homeostasis (Morrison and Kimble, 2006), in which the SC is permanently retained inside the niche (bulge) and the progenitor is eventually (after a lag period in our system) exported outside the niche. Then, marked bulge cells, which previously self-renewed in the niche in the first anagen and survived catagen death, qualifying as SCs, should always replenish the bulge with cell progeny in the later hair cycles. Therefore we should not detect patterns in which all bulge SC descendants left the bulge niche to differentiate into the bulb. However, in our experiments we detected a significant fraction of bulb-only labelling patterns (11–13%) in the second anagen and even one year post-labelling. Since the original labelling was nearly 100% bulge specific, this indicated that a substantial fraction of self-renewing, long-lived bulge cells exited the niche and differentiated into the bulb without leaving any surviving descendants behind in the niche (Fig. 6). Bulb descendants of these cells most likely did not survive in the follicles to make the new HGs, as explained above. Bulge cell death in catagen (Cotsarelis, 2006) may also contribute indirectly in the long-term to the overall loss of labelled HFs, by reducing the frequency of X-Gal+ cells per bulge. Why would infrequently dividing bulge cells divide a few more times to generate more progeny than needed to repopulate the new bulge just to die later on? The explanation may be in a safety mechanism that insures enough bulge cells were generated prior to their return to quiescence. These cells would re-organize around the additional hair club in catagen, while the surplus cells would die. The collective loss of bulge cells by migration and differentiation documented here potentially combined with some apoptosis accounted for approximately one third of the original labelling by one year of chase. This suggested a massive loss of SCs from the pool via a unidirectional, or symmetric fate decision of SC daughters towards a non self-renewing fate. Given the magnitude of this loss, if the SC pool size were to be grossly maintained over lifetime, then a compensation mechanism for this incremental SC decline would be required.
First, the simplest compensation mechanism that best fits our data would implicate in part or entirely symmetric self-renewing SC daughter fates in adult homeostasis (Fig. 7B, top). Such a mechanism was also suggested for at least a fraction of adult inter-follicular epidermis SCs (Clayton et al., 2007), and for other SCs in embryogenesis or in injury (Morrison and Kimble, 2006). Our data are consistent with a population model for tissue homeostasis, similar with that proposed for the C. elegans germ line (Morrison and Kimble, 2006), another system besides HF with a cluster of relatively many SCs. In a population model individual HFSCs have equal potential to replenish the pool through symmetric division in the niche (in anagen) or to be exported (in telogen), proliferate, differentiate, and eventually die. Assignment to one of these two fates would likely depend upon the distance of individual SCs from the signalling centre (in this case the DP). In contrast, in systems where only few SCs are found in a niche, such as the Drosophila germ line, the tissue must insure that self-renewal occurs before a SC may leave the niche compartment to be lost in the differentiating region. The applicability of this model to other adult vertebrate tissues with clustering SCs will have to be addressed experimentally in the future.
Second, if asymmetric divisions were strictly imposed onto SCs in adult homeostasis (Fig. 7B, bottom), in light of our data, progenitors would be constantly required to de-differentiate at each regenerative cycle to compensate for the substantial incremental SC loss documented here. In this scenario, mutual interchange between progenitor and SC populations could be more extensive than previously recognized, potentially obscuring the distinction between these predicted cell types.
Conclusions
Here we provide evidence from an intact adult vertebrate tissue, at the single cell level and in long-term for the infrequently dividing “slow-cycling” stem cell model. In addition, we showed that differentiation and self-renewal in the hair follicle stem cell niche are spatially and temporally segregated during hair cycle. Finally, our data are consistent with a stem cell population fate deterministic model involving, at least in part, unidirectional fate decisions of stem cell daughters upon divisions.
Experimental Procedures
Mice
All experiments were approved by Cornell IACUC and carried out using standard procedure, as described (Waghmare et al, 2008). K5tTA (FvB) x pTREH2B-GFP (CD1) mice were fed doxy at 1g/KG of food (Bio-serv) to repress H2BGFP expression (Tumbar et al., 2004). K14-CreER (CD1) x Rosa26R (C57BL6) mice were intraperitoneally injected tamoxifen at 100 ug/g body weight.
Confocal, FACS, and Tissue Staining
were described (Waghmare et al., 2008), except skin sections were further incubated 3 hrs in 1M Na2CO3 to maximize X-gal staining sensitivity (Tanahashi and Tabira, 2000). Detailed protocols and antibodies are described in Supplementary Materials.
Microarray and (Q)RT–PCR
RNA isolation, cDNA generation and RT-PCR were described (Osorio et al., 2008); see Table S1 for primers. Quantitative (Q) RT-PCR performed on MyiQ thermocycler (Bio-Rad) were normalized to GAPDH. For microarrays 5 ng of high quality RNA (Bioanalyzer, Agilent) were amplified (Ovation Amplification; Nugene) in the Cornell Microarray Core Facility. GeneChip IVT labelling was followed by Gene Chip MOE 430 2.0 hybridization, GeneArray 3000 scanning, GCOS generation of present calls and signal values (Affymetrix). Heat map and correlation tree were generated by un-supervised hierarchical average linkage cluster analysis in Cluster 3.0 software (http://bonsai.ims.u-tokyo.ac.jp/).
For additional information on Experimental Procedures see Supplementary Materials.
Supplementary Material
Acknowledgements
We thank Dr. Jim Smith and Jie Zhao for help with FACS and microarray data generation. Dr. Wei Wang and Dr. Jeff Pleiss for help with microarray data analysis. We thank our colleagues for critical reading of manuscript. Funding: NIH/NIAMS AR053201 and NYSTEM (2008 Award) grants to T.T; Cornell Vertebrate Genomics Institute fellowship to Y.V.Z. The authors declare no financial competing interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Clayton E, Doupe DP, Klein AM, Winton DJ, Simons BD, Jones PH. A single type of progenitor cell maintains normal epidermis. Nature. 2007;446:185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- Cotsarelis G. Epithelial stem cells: a folliculocentric view. J Invest Dermatol. 2006;126:1459–1468. doi: 10.1038/sj.jid.5700376. [DOI] [PubMed] [Google Scholar]
- Fuchs E. The tortoise and the hair: Slow-cycling cells in the stem cell race. Cell 137 811–819. 2009 doi: 10.1016/j.cell.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco V, Chen T, Rendl M, Schober M, Pasolli HA, Stokes N, Dela Cruz-Racelis J, Fuchs E. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 2009;4:155–169. doi: 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V, O’Carroll D, Tooze R, Ohinata Y, Saitou M, Obukhanych T, Nussenzweig M, Tarakhovsky A, Fuchs E. Blimp1 defines a progenitor population that governs cellular input to the sebaceous gland. Cell. 2006;126:597–609. doi: 10.1016/j.cell.2006.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H, Toftgard R. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008 doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- Jones CL, Reiners JJ., Jr Differentiation status of cultured murine keratinocytes modulates induction of genes responsive to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Arch Biochem Biophys. 1997;347:163–173. doi: 10.1006/abbi.1997.0350. [DOI] [PubMed] [Google Scholar]
- Lansdorp PM. Immortal strands? Give me a break. Cell. 2007;129:1244–1247. doi: 10.1016/j.cell.2007.06.017. [DOI] [PubMed] [Google Scholar]
- Legue E, Nicolas JF. Hair follicle renewal: organization of stem cells in the matrix and the role of stereotyped lineages and behaviors. Development. 2005;132:4143–4154. doi: 10.1242/dev.01975. [DOI] [PubMed] [Google Scholar]
- Luong MX, van der Meijden CM, Xing D, Hesselton R, Monuki ES, Jones SN, Lian JB, Stein JL, Stein GS, Neufeld EJ, van Wijnen AJ. Genetic ablation of the CDP/Cux protein C terminus results in hair cycle defects and reduced male fertility. Mol Cell Biol. 2002;22:1424–1437. doi: 10.1128/mcb.22.5.1424-1437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- Oshima H, Rochat A, Kedzia C, Kobayashi K, Barrandon Y. Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell. 2001;104:233–245. doi: 10.1016/s0092-8674(01)00208-2. [DOI] [PubMed] [Google Scholar]
- Osorio KM, Lee SE, McDermitt DJ, Waghmare SK, Zhang YV, Woo HN, Tumbar T. Runx1 modulates developmental, but not injurydriven, hair follicle stem cell activation. Development. 2008;135:1059–1068. doi: 10.1242/dev.012799. [DOI] [PubMed] [Google Scholar]
- Plikus MV, Mayer JA, de la Cruz D, Baker RE, Maini PK, Maxson R, Chuong CM. Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature. 2008;451:340–344. doi: 10.1038/nature06457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendl M, Lewis L, Fuchs E. Molecular dissection of mesenchymal-epithelial interactions in the hair follicle. PLoS Biol. 2005;3:e331. doi: 10.1371/journal.pbio.0030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger E, Bijl JJ, van Oostveen JW, Soyer HP, Oudejans CB, Jiwa NM, Walboomers JM, Meijer CJ. Expression of the homeobox gene HOXC4 in keratinocytes of normal skin and epithelial skin tumors is correlated with differentiation. J Invest Dermatol. 1994;103:341–346. doi: 10.1111/1523-1747.ep12394888. [DOI] [PubMed] [Google Scholar]
- Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Sur I, Rozell B, Jaks V, Bergstrom A, Toftgard R. Epidermal and craniofacial defects in mice overexpressing Klf5 in the basal layer of the epidermis. J Cell Sci. 2006;119:3593–3601. doi: 10.1242/jcs.03070. [DOI] [PubMed] [Google Scholar]
- Tanahashi H, Tabira T. Alkaline treatment after X-Gal staining reaction for Escherichia coli beta-galactosidase enhances sensitivity. Anal Biochem. 2000;279:122–123. doi: 10.1006/abio.1999.4444. [DOI] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T. Epithelial skin stem cells. Methods Enzymol. 2006;419:73–99. doi: 10.1016/S0076-6879(06)19004-7. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci U S A. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waghmare SK, Bansal R, Lee J, Zhang YV, McDermitt DJ, Tumbar T. Quantitative proliferation dynamics and random chromosome segregation of hair follicle stem cells. Embo J. 2008;27:1309–1320. doi: 10.1038/emboj.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.