Abstract
A major pathway for HIV-1 resistance to nucleoside reverse transcriptase inhibitors (NRTIs) involves reverse transcriptase (RT) mutations that enhance ATP-dependent pyrophosphorolysis, which excises NRTIs from the end of viral DNA. We analyzed novel NRTIs for their ability to inhibit DNA synthesis of excision-proficient HIV-1 RT mutants. D-carba T is a carbocyclic nucleoside that has a 3′ hydroxyl on the pseudosugar. The 3′ hydroxyl group allows RT to incorporate additional dNTPs, which should protect D-carba TMP from excision. D-carba T can be converted to the triphosphate form by host cell kinases with moderate efficiency. D-carba T-TP is efficiently incorporated by HIV-1 RT; however, the next dNTP is added slowly to a D-carba TMP at the primer terminus. D-carba T effectively inhibits viral vectors that replicate using NRTI-resistant HIV-1 RTs, and there is no obvious toxicity in cultured cells. NRTIs based on the carbocyclic pseudosugar may offer an effective approach for the treatment of HIV-1 infections.
Keywords: HIV-1, Reverse Transcriptase, NRTI, Resistance, Kinase
Introduction
A major class of drugs used to treat HIV-1 infection is nucleoside reverse transcriptase inhibitors (NRTIs). The NRTIs currently approved for the treatment of HIV-1 infections differ from normal nucleosides in that the NRTIs lack a 3′ OH group and, as a consequence, incorporation of an NRTI by HIV-1 reverse transcriptase (RT) leads to the termination of viral DNA synthesis. NRTIs have proven to be highly effective against wild-type (wt) HIV-1 RT, but treatment with NRTIs can lead to the development of resistance (see 1 and references therein). Two types of NRTI resistance mechanisms are known; both involve an increase in the ability of HIV-1 RT to discriminate between normal nucleosides and the NRTI. In one resistance mechanism, exclusion, RT develops an increased ability to discriminate between normal dNTPs and the NRTI-TP at the time of incorporation. The other resistance mechanism, excision, depends on HIV-1 RT developing the ability to selectively excise the analog after it has been incorporated using ATP-dependent pyrophosphorolysis.
We have previously described nucleoside analogs that have modifications to the sugar ring that were specifically designed to limit their excision by excision-proficient HIV-1 RT variants 2, 3. These compounds inhibited DNA synthesis by different mechanisms than immediate chain termination seen with NRTIs such as 3′-azido 3′-deoxythymidine (AZT) 2, 3. Unfortunately there were drawbacks to the compounds we have tested thus far. As we had hoped, the compounds were not efficiently excised by the RT variants proficient at ATP-dependent pyrophosphorolysis 2, 3; however, the M184V mutation, which causes resistance to 2′,3′-dideoxy-3′ thiacytidine (3TC) by exclusion, did cause significant resistance to the compounds we previously tested. These compounds were not efficiently converted to the triphosphate form by cellular kinases due to an unfavorable conformation of the sugar ring 4, 5; the analogs were efficiently phosphorylated in cells that express the Herpes Simplex Virus thymidine kinase (HSV-TK). In addition, when the compounds were phosphorylated in the HSV-TK expressing cells, the compounds appeared to be toxic to the cells.
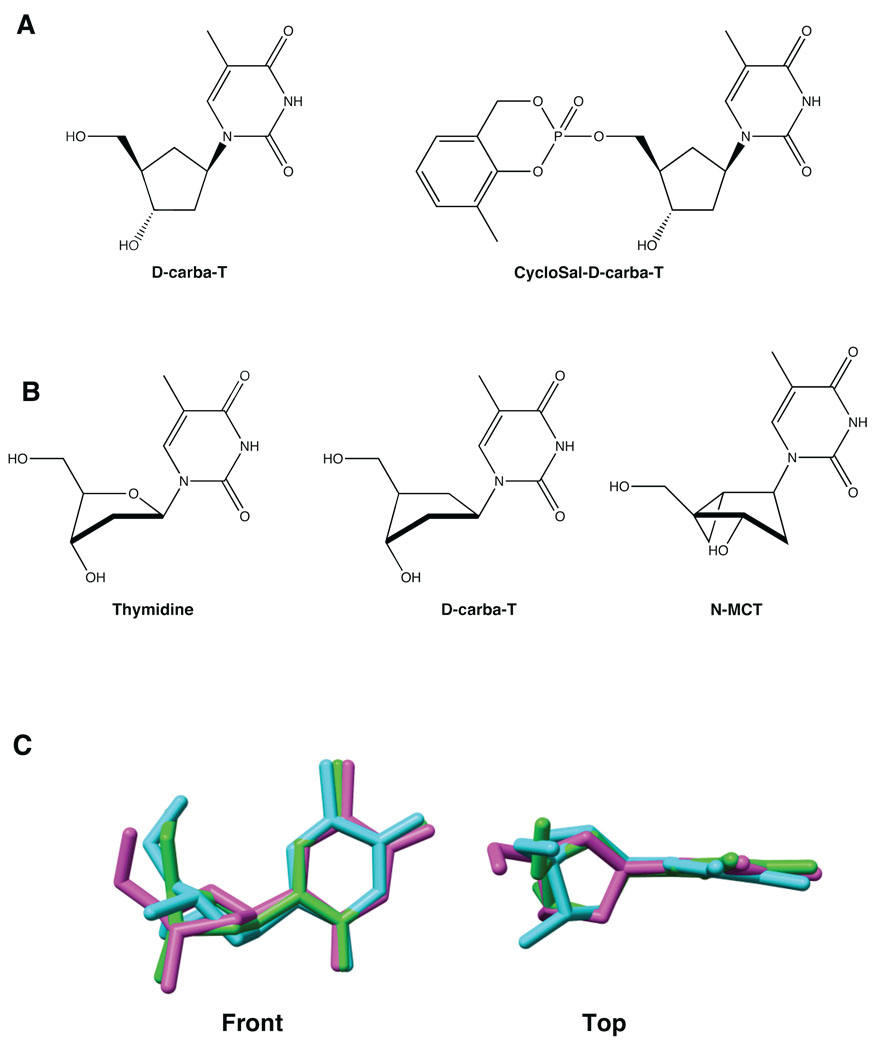
To address these problems, we examined another compound, D-carba T, a carbocyclic nucleoside in which the furanose oxygen has been replaced with a methylene group, creating a cyclopentane ring (Fig. 1A). This causes a change in conformation of the pseudosugar ring compared to the conformation of the deoxyribose ring of normal nucleosides, which are primarily in either a 3′-endo (North) or 2′-endo (South) conformation 4. Carbocyclic nucleosides tend to adopt an atypical 1′-exo conformation that is principally governed by the steric bulk of the nucleobase, which prefers an equatorial orientation 6. Kinases, in general, prefer nucleosides that are in the South conformation 2, 3, 5. As can be seen in Figure 1B and 1C, D-carba T has a structure that is closer to the South conformation of thymidine, suggesting that this compound would be phosphorylated by cellular kinases. D-carba T has a hydroxyl group on the 3′ carbon, which could allow the next incoming dNTP to be added to the primer strand if the geometry at the active site is approximately correct (Fig. 1B, C). The orientation of the 3′-OH is critical for the recognition of the nucleosides by both kinases and polymerases. The crystal structure of D-carba T shows that it has a P value of 118.6°, suggesting that the position of the 3′-OH is, in some sense, a compromise between the orientations of the locked North and South nucleosides. Because the structure of the pseudosugar of D-carba T is intermediate between the North and South conformations, it might be an acceptable substrate for both kinases and polymerases 6. In addition, the cyclopentane ring is flexible, which might allow for minor adjustments in the conformation of D-carba T that could allow it to satisfy the structural requirements of both kinases and polymerases.
Figure 1.
Structures of D-carba T and related compounds. A) Structures of the carbocyclic nucleoside D-carba T and the carbocyclic produg CycloSal-D-carba T. Note the presence of a 3′ OH group. B) Comparison of the D-carba T structure with the thymidine and North –Methanocarbathymidine (N-MCT) structures. C) Superposition of three thymidine compounds (Cambridge Database Structures). For clarity, no hydrogen atoms are shown. Both the view from the front and from the top are shown. Green: Thymidine. Cyan: N-MCT. Magenta: D-carba T. The root mean square deviation (RMSD) of thymidine with N-MCT and D-carba T are 0.3902 and 0.2550 respectively; the maximum difference between superimposed atom positions of thymidine relative to N-MCT and D-carba T are 1.0097 and 0.6372, indicating that the similarity between the conformations of thymidine and D-carba T is much higher than that between thymidine and N-MCT. The 3′-OH groups of thymidine and D-carba T are in similar positions, indicating that D-carba T has the proper conformation for recognition by cellular kinases. The 3′-OH of N-MCT is not aligned with the 3′-OH groups of the other compounds and N-MCT is not readily phosphorylated by cellular kinases.
We examined D-carba T and CycloSal-D-carba T in cell-based assays, and D-carba T- triphosphate (TP) in biochemical assays. The results are promising; both the compound and the prodrug version are active against HIV-1 in cultured cells, and the triphosphate form of D-carba T blocks DNA synthesis by HIV-1 RT in vitro. D-carba T is moderately well phosphorylated by cellular kinases and inhibits the replication of both wild-type HIV vectors and vectors containing drug-resistant RT variants, including the M184V variant. D-carba T-TP is efficiently incorporated by HIV-1 RT during DNA synthesis; however, incorporation of the compound does not cause the complete termination of DNA synthesis as do conventional NRTI-TPs such as AZT-TP. After the incorporation of D-carba T-TP, the next dNTP is added slowly to a D-carba T-monophosphate (MP) terminated primer. We have called this mechanism kinetic chain termination; it is the mechanism by which the analog 4′ C-methyl thymidine-triphosphate (4′ C -Me T-TP) inhibits DNA synthesis by HIV-1 RT 2. D-carba T appears to be a promising lead for the development of anti-HIV therapeutic agents that will be effective against the known NRTI-resistant viruses.
Results
Polymerase Assays
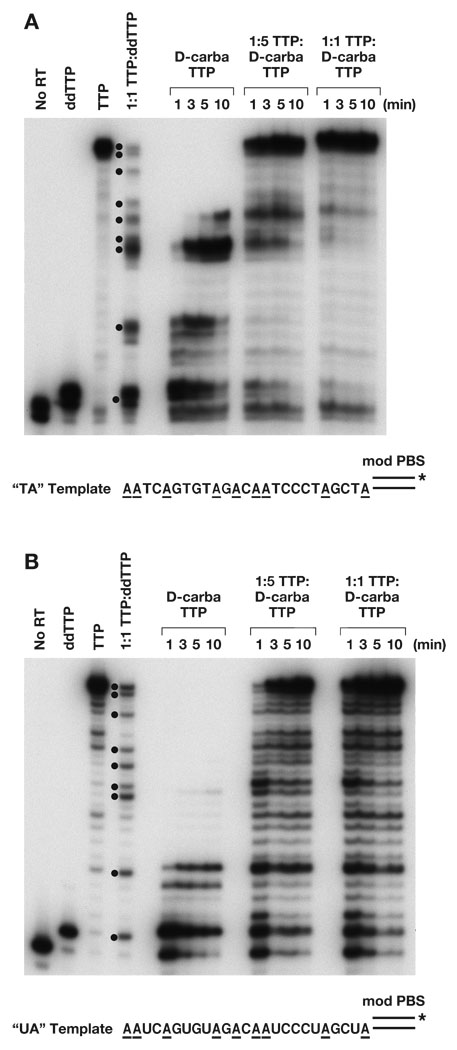
D-carba T has good anti-HIV-1 activity and only modest toxicity in CEM/0 cells, which implies that D-carba T can be converted to the triphosphate form, at least to some extent, in these cells, and that D-carba T-TP inhibits viral DNA synthesis by RT7, 8. We examined the effects of D-carba T-TP on DNA synthesis by wild-type (wt) HIV-1 RT. A DNA primer was 5′ end-labeled, hybridized to a DNA or RNA template, both having the same sequence, and the labeled template-primer was extended by HIV-1 RT in the presence of 10.0 µM each dCTP, dGTP, and dATP and 10.0 µM total of TTP or a TTP analog (ddTTP or D-carba T-TP) (Figure 2). The “No RT” control lane shows the position of migration of the unextended primer, while the TTP lane shows the full-length product. The “ddTTP” lane shows that DNA synthesis terminated immediately after the addition of the ddTTP and indicates the position of migration of a “primer + 1” product. The “1:1 TTP:ddTTP” lane has a mixture of the normal TTP and the ddTTP analog. If TTP was incorporated, DNA synthesis continued, while if ddTTP was incorporated, DNA synthesis was terminated. This lane indicates all the positions at which TTP, or a TTP analog, can be incorporated; these sites correspond to the highlighted nucleotides in the template sequence in Figure 2. We used a template whose sequence specified that TTP, or a thymidine analog such as D-carba T-TP, was the first nucleotide added to the primer strand (Figure 2). If D-carba T-TP cannot be readily incorporated by HIV-1 RT, the product bands will be one nucleotide smaller than the corresponding ddTTP product. If D-carba T-TP acts as a conventional chain terminator and causes a complete termination of DNA synthesis immediately upon incorporation, the product would be the same length as the primer + 1 product in the ddTTP lane, and no larger products will be synthesized. If, like the 4′ C -Me T-TP analog previously described 2, D-carba T-TP causes DNA synthesis to pause after the analog is incorporated by HIV-1 RT, the products would be the same length as the products in the 1:1 lane.
Figure 2.
Inhibition of DNA synthesis by D-carba T-TP. As described in Experimental, the primer was 5′ end-labeled, then annealed to either a DNA template (2A) or an RNA template (2B). The “No RT” lane shows the size of the starting primer. All reactions had 10.0 µM each of dCTP, dGTP, dATP, and the indicated TTP or T-TP analog. The “ddTTP” lane shows the primer with the addition of a single ddTTP residue (N + 1), while the “TTP” lane shows the primer extended to the end the template (full-length product). The 1:1 TTP:ddTTP had a mixture of 5.0 µM TTP and 5.0 µM ddTTP in the reaction. Because ddT-TP is an immediate chain terminator, DNA synthesis carried out with this mixture generates products corresponding to all of the sites at which a TTP or TTP analog can be incorporated. The positions at which the addition of ddTTP terminated DNA synthesis are indicated by dots. The reactions were carried out either with 10.0 µM D-carba T-TP (no TTP present) or with different ratios of TTP:D-carba T-TP (1:5 or 1:1) where the final concentration totaled 10.0 µM. The reactions were incubated at 37° for the indicated length of time and the products fractionated by gel electrophoresis. The unusual pause site discussed in the text for an RNA template is marked with a star. The sequence of the template extension is shown below the reactions, and the positions of all “A” residues are highlighted
When the primer was extended in the presence of D-carba T-TP and dATP, dCTP, and dGTP, products larger than primer + 1 were detected, showing that D-carba T-TP is not an absolute chain terminator like ddTTP (Fig. 2A, B). This effect was seen with both the DNA and the RNA templates. It appears that the lengths of the products generated by D-carba T-TP are similar in size to the products in the “1:1 TTP:ddTTP” marker lane. This suggests that D-carba T-TP is readily incorporated by HIV-1 RT, and that its effect on DNA synthesis by HIV-1 RT is similar to 4′ C -Me T-TP, in that D-carba T causes HIV-1 RT to pause immediately after the analog has been incorporated. With the RNA template, there was one position where there was both a pause site one nucleotide before the position where the TTP analog was incorporated and another pause site where the analog was incorporated (Fig. 2B). This suggests, at this position, that the RT either has some problem binding D-carba T-TP or a problem incorporating it into the growing primer strand. The position where RT has a problem incorporating D-carba T-TP occurs before a run of three cytidine residues, a location which appears to alter the localized structure of the template/primer 3. At longer incubation times, wild-type HIV-1 RT was able to extend primers that have D-carba TMP at the 3′ end, generating longer products, but no full-length product was detected (Fig. 2A, B). This analog appears to be another example of a temporal (or kinetic) NRTI, which are readily incorporated by HIV-1 RT, but which, after incorporation, slow the addition of the next dNTP.
We also tested the ability of D-carba T-TP to compete with the normal substrate, TTP. Two ratios of TTP:D-carba T-TP (1:5 and 1:1) were tested. With a DNA template, some pausing was detectable, suggesting that D-carba T-TP is incorporated to some extent. However, with the DNA template, D-carba T-TP did not appear to be able to compete effectively with the normal TTP, since most of the products are full length (Fig. 2A). With the RNA template, there was more pausing than with the DNA template, suggesting that D-carba T-TP is better able to compete with TTP with an RNA template (Fig. 2B). Because D-carba T is able to inhibit viral replication in cell culture (discussed below), these data suggest that most of the inhibitory effect of this compound occurs during first strand DNA synthesis.
Cell-Based Assays
We previously described an HIV-1 based vector that lacks a functional Env coding region; infectious virions are produced by complementation with vesicular stomatitis virus-G (VSV-G) 9, 10. The VSV-G complementation strategy limits the vector to a single replication cycle. The original vector contained a wt HIV-1 RT, but the vector has been modified to contain mutant forms of HIV-1 RT, and the resulting mutant vectors were used to study the effects of the mutations on viral replication and resistance to NRTIs. We also described HOS cells that were modified to contain, and express, the thymidine kinase (TK) from herpes simplex virus (HSV); these cells are designated HOS 313 2. TK catalyzes the conversion of T (nucleobase plus sugar) to TMP (the monophosphate form). HSV-TK also has some thymidylate kinase activity 11, and can convert some of the monophosphate to the diphosphate, but the generation of the triphosphate requires cellular kinases. Because HSV-TK is able to phosphorylate a wider range of thymidine analogs than the corresponding cellular kinases, and because the first step in phosphorylation is often the rate limiting step, cells that express HSV-TK can be used to determine whether nucleoside analogs that are not readily phosphorylated by cellular kinases are able to inhibit HIV-1 replication 11, 12. These cell lines were previously tested against AZT 2. In an assay with the wt vector, infections carried out in 313 cells were more sensitive to treatment with AZT than in HOS cells lacking HSV-TK.
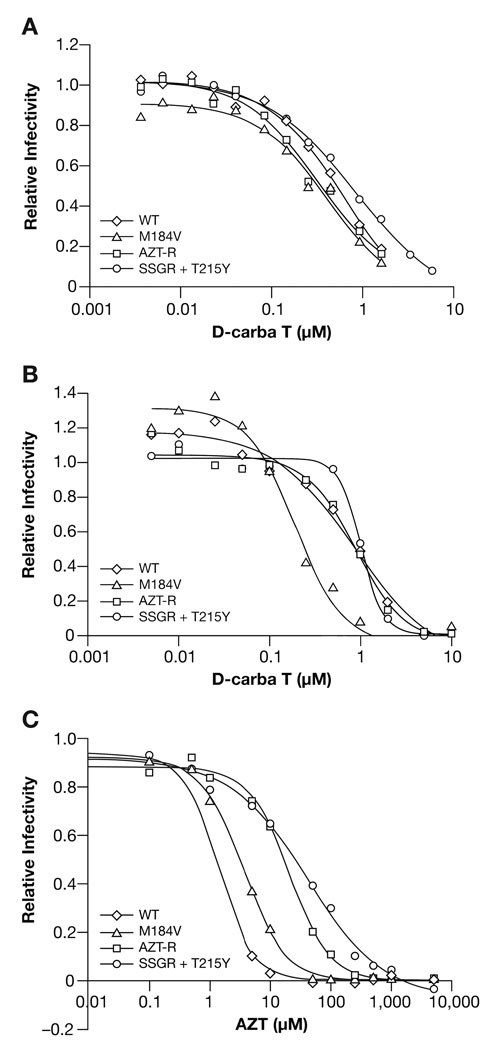
HIV vectors containing wt RT or different drug-resistant RT variants were used to infect HOS cells in the presence of various concentrations of D-carba T. As shown in Figure 3A, D-carba T effectively blocked the replication of the vector containing wt RT, and has an IC50 value of approximately 3.2 µM (Table 1). This indicates that D-carba T can be phosphorylated, at least to some extent, by cellular kinases, and that once it is phosphorylated, it is efficiently incorporated into viral DNA by HIV-1 RT, which agrees with the reports that D-carba T has anti-HIV activity in CEM/0 cells 7, 8. For comparison, AZT was also tested against the HOS cells (Fig. 3C). The IC50 value for AZT against the replication of the vector containing wt RT was 1.4 nM (Table 1) When D-carba T was tested in the HSV-TK expressing HOS 313 cell line against the HIV-1 vector, the IC50 of D-carba T was 0.83 µM (Fig. 3B; Table 1). The fact that there is approximately a 4-fold reduction in the concentration of D-carba T needed to block the replication of the vector in the presence of HSV-TK (Fig. 3A,B; Table 1) indicates that the normal cellular kinases do not efficiently carry out the early steps in the conversion of D-carba T to the triphosphate form. We have also done single-cycle inhibition assays in TZM cells (a HeLa-derived indicator cell line described below) using D-carba T and the replication competent wild-type HIV-1 strain LAI. In this assay, the IC50 value for D-carba T was 0.05–0.07 µM (data not shown). In general, assays in which there are multiple rounds of viral replication give lower IC50s than single round assays; there may also be differences in the ability of the two different cell lines to take up and convert D-carba T to the triphosphate.
Figure 3.
Cell-based assay comparing the effects of D-carba T on the replication of an HIV-1 vector containing wt HIV-1 RT and vectors that replicate using NRTI-resistant HIV-1 RTs. AZT-R and SSGR+T215Y are efficient at ATP-dependent pyrophosphorolysis (excision), while M184V interferes with the incorporation of NRTI-TPs. The vectors also express luciferase from the nef reading frame. A) HOS cells were infected with the various HIV-1 vectors in the presence of increasing levels of D-carba T. Luciferase activity in the absence of D-carba T was considered to be 100 and the luciferase activity in the presence of the different concentrations of D-carba T was normalized to this value to give relative infectivity. B) Assay done in HOS 313 cells (which express HSV-TK). C) HOS cells were infected with the various HIV-1 vectors in the presence of increasing levels of AZT.
Table 1.
Anti-viral efficacy of AZT and D-Carba T versus wild-type and select NRTI resistant HIV-1 viral vectors.
| D-Carba T (µM) |
AZT (nM) |
||
|---|---|---|---|
| HOS | 313 | HOS | |
| Wild-type | 3.2 ± 1.7 | 0.83 ± 0.21 | 1.4 ± 0.1 |
| M184V | 1.8 ± 1.0 | 0.19 ± 0.03 | 3.6 ± 0.3 |
| AZT-R | 1.3 ± 0.4 | 0.87 ± 0.13 | 20.1 ± 1.8 |
| SSGR + T215Y | 4.6 ± 1.2 | 1.0 ± 0.1 | 39.6 ± 7.7 |
D-carba T effectively inhibited the replication of vectors containing a number of different NRTI-resistant RT variants. The M184V mutation confers resistance by causing enhanced discrimination between normal dNTPs and the analogs 3TC-TP and FTC-TP by steric hindrance at the polymerase active site, reducing the incorporation of these NRTI-TPs (see 1 and references therein). AZT-R (M41L/D67N/K70R/T215Y/K219Q) and SSGR/T215Y, which contains several amino acid substitutions in addition to the insertion of two amino acids in the thumb subdomain of RT (T69K70/T215 → S69SGR70/T215Y), are RT variants that confer resistance to NRTIs due to their enhanced ability to excise an NRTI monophosphate (NRTI-MP) from the 3′ end of a primer using ATP-dependent pyrophosphorolysis (excision), thus freeing the blocked end of the primer for continued DNA synthesis 1. As expected, vectors containing AZT-resistance mutations AZT-R and SSGR/T215Y showed a reduce susceptibility to AZT. While vectors containing the wt RT had an IC50 value for AZT of 1.4 nM, the AZT-R containing vector had an IC50 value of 20.1 nM, and the SSGR/T215Y containing vector had an IC50 value of 39.6 nM (Fig.3C; Table 1). The vector containing M184V, which does not confer significant resistance to AZT, had an IC50 value of 3.6 nM (Fig. 3C; Table 1). Whitcomb et al., who also used an HIV vector system, showed that the M184V mutation usually conferred a small increase in susceptibility to AZT 13. We do not have an explanation for this apparent discrepancy; however, in our experiments the difference in the IC50 for wt and the M814V mutant was small, and the data of Whitcomb et al. showed considerable scatter, so it is possible that the exact sequence of the RT in the vector could have influenced the outcome. In contrast, the vectors containing the AZT-R and SSGR/T215Y mutations appeared to have approximately the same sensitivity to D-carba T as vectors containing wt RT (Fig. 3A,B). The IC50 values for D-carba T are shown in Table 1. The IC50 for the SSGR/T215Y mutant is slightly higher than wt; however, the difference is within the error of the assay. We also showed that the Q151M complex (A62V/V75I/F77L/F116Y/Q151M), a set of mutations that cause NRTI-resistance by an exclusion mechanism, remains sensitive to D-carba T (data not shown).
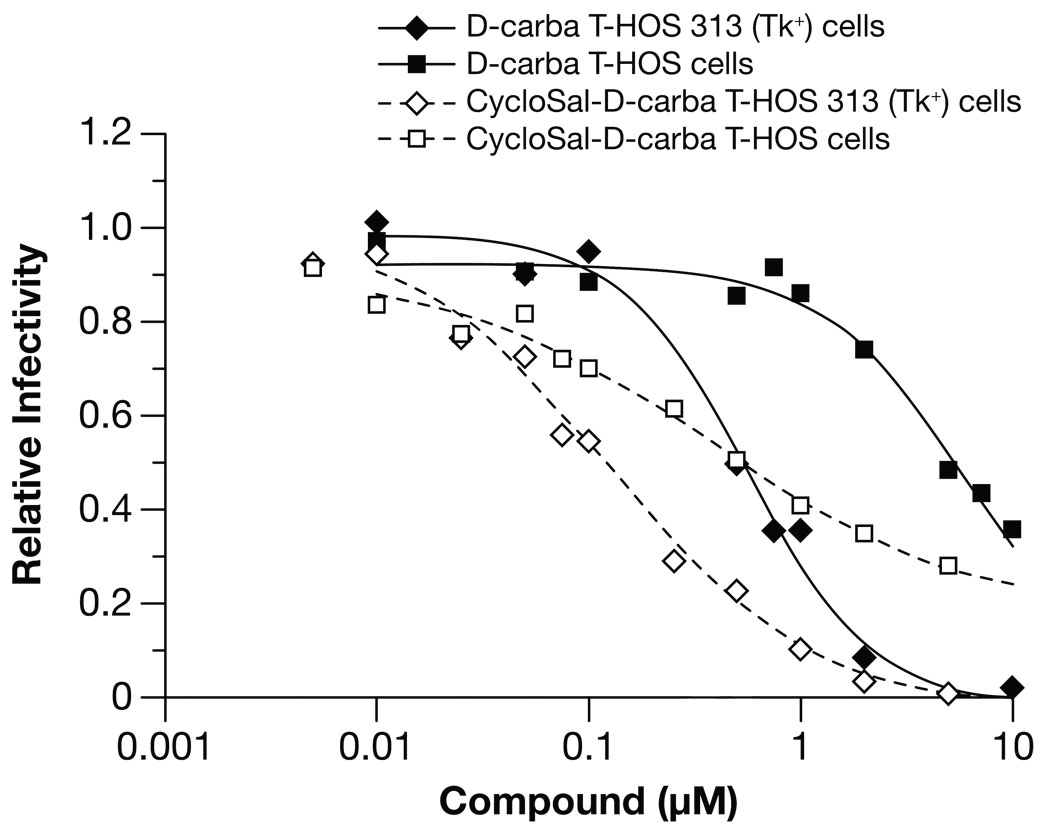
While HSV-TK can convert thymidine analogs into the monophosphate and the diphosphate forms 11, 12, which is useful for determining whether the compounds have anti-HIV activity, this approach cannot be used in patients. However, there is a method that can be used to introduce phosphorylated nucleosides into cells that can be used in patients. As has already been mentioned, for many nucleosides, the rate-limiting phosphorylation step is the step that generates the 5′-monophosphate. Phosphorylated analogs are not readily taken up by cells, presumably because the charged phosphate prevents uptake. If the charged phosphate is shielded by a hydrophobic residue, then the modified phosphorylated nucleoside can enter cells. If the modifying hydrophobic group is appropriately designed, once the compound enters a cell, the modifying group can be removed by spontaneous chemical degradation and/or cellular enzymes, yielding the free 5′ dNMP, which can then be converted to the triphosphate by cellular kinases 14, 15. If one phosphate is already present on the starting compound, only two cellular phosphorylation steps are needed to produce the active triphosphate (for review, see 15). A prodrug (pronucleotide) form of D-carba T, a CycloSal derivative developed by Meier16–18 (Fig. 1A), has been described. We asked whether the prodrug form of CycloSal-D-carba T effectively blocked the replication of an HIV vector in normal HOS cells and the HSV-TK expressing HOS 313 cells. The modified analog inhibited the replication of an HIV vector, showing that the prodrug is converted to D-carba TMP, which can then be converted to the triphosphate form (Fig. 4). CycloSal-D-carba T has a lower IC50 in HOS 313 cells than in the parental HOS cells (0.38 µM in HOS cells versus 0.12 µM in HOS 313 cells). This could be explained if the thymidylate kinase activity of HSV-TK helps in the conversion of the monophosphate to the diphosphate; however, there are other possible explanations (see Discussion).
Figure 4.
Cell-based assay comparing the effects of D-carba T versus the D-carba T prodrug, CycloSal-D-carba T, on the replication of an HIV-1 vector containing wild-type HIV-1 RT. HOS or HOS 313 cells were infected with the HIV-1 vector in the presence of increasing levels of D-carba T or CycloSal-D-carba T. Luciferase activity in the absence of analog was considered to be 100 and the luciferase activity in the presence of the different concentrations of D-carba T or CycloSal-D-carba T was normalized to this value to give relative infectivity.
Cellular Toxicity
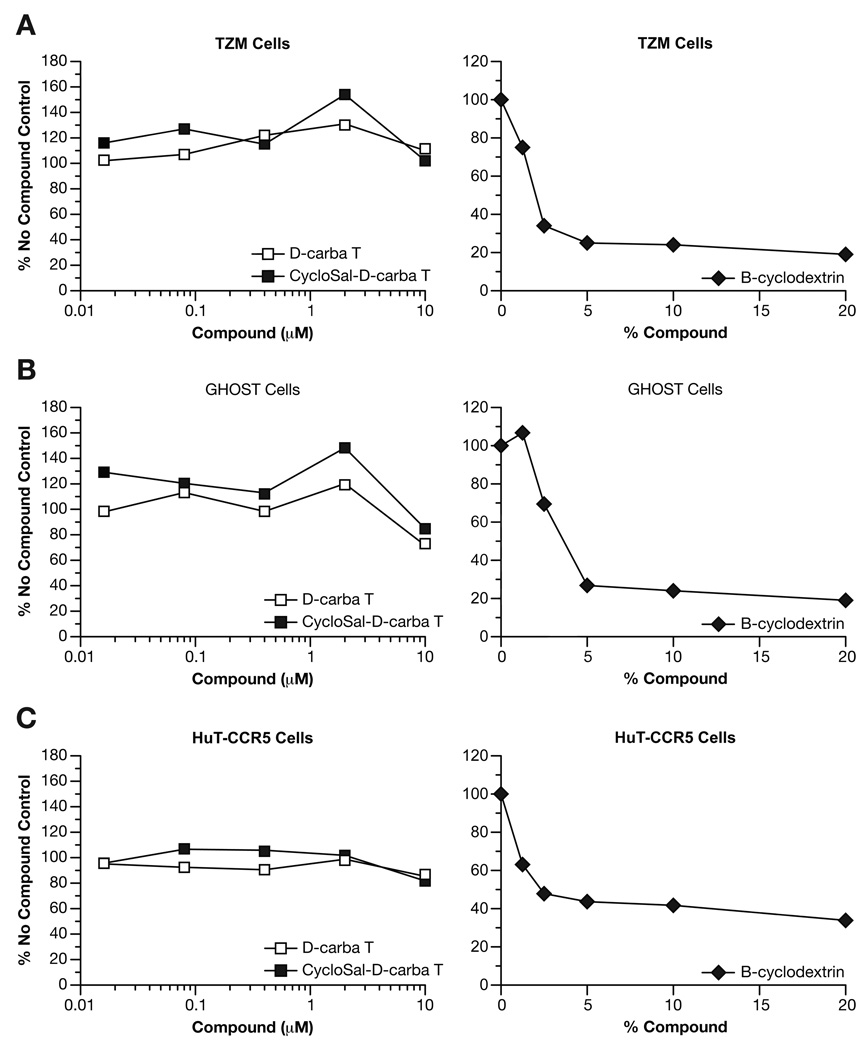
One potential problem with treating HIV-1 infections with NRTIs is toxicity to the host. D-carba T and CycloSal-D-carba T were tested in several cell lines to determine whether these compounds are toxic. Hydroxypropyl-β-cyclodextrin (BCD) was used as a positive control because previous studies showed that more than 50% of cells are killed when BCD is present at 2–5% (w/v) (Ambrose et al., unpublished observations). Three cell lines, TZM (a HeLa cell-derived indicator cell line), GHOST (a HOS cell-derived indicator cell line), and HuT-CCR5 (a human T-cell line), were incubated with different concentrations of D-carba T, CycloSal-D-carba T, and BCD and subsequently assayed for viability using the Roche Cell Proliferation kit (XTT). As described in Experimental, only viable cells can convert the precursor dye to a measurable metabolite. For all cell types tested, up to 10.0 µM of either D-carba T or CycloSal-D-carba T was not toxic (Fig. 5). In contrast, 5% BCD was sufficient to kill more than 50% of the cells (Fig. 5).
Figure 5.
Cytotoxicity of D-carba T and CycloSal-D-carba T in various cell lines. As described in Experimental, cell viability was determined using the XTT Cell Viability Assay kit from Roche. Hydroxypropyl-β-cyclodextrin (BCD) was used as a positive control for cell death since previous studies have shown that more than 50% of cells will be killed when BCD is present at 2–5% (w/v). A) Viability of TZM cells in the presence of increasing concentrations of D-carba T, CycloSal-D-carba-T, or BCD. B) Viability of GHOST cells in the presence of increasing concentrations of D-carba T, CycloSal-D-carba-T, or BCD. C) Viability of HuT-CCR5 cells in the presence of increasing concentrations of D-carba T, CycloSal-D-carba-T, or BCD.
Discussion
There are many NRTIs that effectively inhibit the replication of wt HIV-1. However, the appearance of RT variants resistant to these NRTIs in patients, and the subsequent transmission of the resistant viruses, is a major concern. All currently approved NRTIs used to treat HIV infections lack a 3′-OH group, which makes them vulnerable to the excision resistance mechanism. We have been examining nucleoside analogs that retain a 3′-OH group in an effort to identify NRTIs that can inhibit not only wt RT, but are also effective against known drug-resistant RT variants. We previously examined, North-methanocarbathymidine (N-MCT), North-methanocarbadeoxyadenosine (N-MCdA), 4′ C-Me T and 4′ C-ethyl thymidine (4′ C-Et T), all of which efficiently inhibit wt RT and RT variants that are proficient in ATP-dependent pyrophosphorolysis 2, 3. However, these compounds had several problems. None of the compounds we previously tested were efficiently converted to the triphosphate form by cellular kinases and testing the compounds against HIV-1 and HIV-1 based vectors in cell culture required the expression of HSV-TK in the target cells. The compounds we tested previously, although effective against excision-proficient RTs, appeared to be less effective against the exclusion variant M184V, and the compounds also appeared to be toxic in cell lines that express HSV-TK 2, 3.
We analyzed the carbocyclic nucleoside D-carba T to see if this compound circumvented these problems. D-carba T-TP is readily incorporated by HIV-1 RT during DNA synthesis, but the next dNTP is added relatively slowly; in this regard, the results we obtained with D-carba T-TP were similar to our previous results with 4′ C -Me T-TP 2. The mechanism of inhibition by 4′ C -Me T and D-carba T is unusual. These NRTIs do not prevent chain elongation, but rather delay it. It is likely that the conformation of the carbocyclic pseudosugar affects the position of the 3′-OH group at the polymerase active site, which reduces the efficiency with which HIV-1 RT is able to add the next dNTP to the primer strand. How does this delay block HIV replication? As we previously proposed, it is possible that there is a crucial period of time in which the single-strand RNA genome must be converted to double-strand DNA and, if the compounds delay this conversion, the ability of the virus to complete the synthesis of the viral DNA genome in the infected cells may be compromised 2, 3. For example, partially completed viral DNAs may be subject to degradation by the host cell. It is also possible that even if the viral DNA genome is completed, the presence of the incorporated analogs in the DNA provirus might provoke host DNA repair machinery to degrade the viral DNA once it enters the nucleus 2, 3.
In an in vitro polymerase assay, D-carba T-TP was better able to compete with the normal substrate, TTP, when the template was RNA rather than DNA. The two templates had the same sequence, so the difference in the ability of the analog to compete with the normal nucleoside triphosphate is not due to a difference in the sequence of the templates. The nucleotides in both an RNA and a DNA template assume a North (A-form) conformation near the polymerase active site of RT 1, 19 so, when these templates are bound to RT, there appear to be no major differences in their overall structure. However, even though the structural differences may be subtle, there are differences, and these differences are sufficient to allow D-carba T-TP to compete more effectively with TTP on an RNA vs. a DNA template. If we assume that D-carba T is also selectively incorporated on an RNA template during viral replication in infected cells, then D-carba T would be expected to preferentially inhibit the first strand of viral DNA synthesis. Whether or not this selectivity exists in infected cells, D-carba T does effectively block the replication of both wt and NRTI-resistant HIV in cultured cells.
The biochemical assays with D-carba T-TP show that this compound is an effective inhibitor of HIV-1 RT in vitro. However, as has been discussed, a major drawback of the nucleoside analogs we previously studied is that cellular kinases are not able to convert these compounds to the triphosphate form, which is necessary for their incorporation into DNA by RT. D-carba T appears to have a structure that allows kinases to bind and phosphorylate it with moderate efficiency. In comparing D-carba T to N-MCT, D-carba T is not constrained in the North conformation, a conformation that is unfavorable for cellular kinases. In contrast to the 4′ C-Me T and 4′ C-Et T compounds, D-carba T does not have extra substituents on the pseudosugar ring that could cause steric clashes with the kinases (Fig 1A). As described in Results, D-carba T was able to inhibit the replication of an HIV vector containing wt RT in HOS cells, indicating that this compound was converted, at least to some extent, to the triphosphate form by cellular kinases. However, it is clear that the presence of HSV-TK reduces the IC50 for D-carba T, suggesting that this analog is not an optimal substrate for the cellular kinases. D-carba T was effective against the excision competent HIV RT variants and, unlike the nucleoside analogs we have previously characterized, D-carba T also appears to be effective against the drug-resistant RT variant M184V, which causes resistance to 3TC-TP and FTC-TP by interfering with their incorporation into viral DNA. One of the particular advantages of the carbocyclic nucleoside analog D-carba T is that its pseudosugar ring has no extra substituents that could cause a steric clash with a β- branched amino acid at position 184.
While D-carba T is phosphorylated to some extent by cellular kinases, there appears to be a pool of unphosphorylated and/or partially phosphorylated compound in the cells. This does not necessarily render an NRTI unsuitable for anti-HIV therapy. For example, AZT is not efficiently phosphorylated within cells, yet remains one of the NRTIs most widely used for the treatment of HIV infection (see 1 and references therein). In general, improving the level of a NRTI-TP within a cell will increase the effectiveness of the parent compound by reducing the dose required to treat patients. To assist the conversion of D-carba T to the triphosphate form, we tested a prodrug which has the first phosphate group added to the pseudosugar, CycloSal-D-carba T (Fig. 1A). Ludek et al. tested both D-carba and CycloSal-D-carba T against HIV-1 using CEM/0 cells, which express the normal cellular thymidine kinase, and CEM/TK− cells, which do not express TK 20. In their experiments, both compounds had similar activities when TK was present, which suggests that the prodrug did not confer an advantage in this system. However, in their experiments, neither compound showed much antiviral activity in the absence of cellular TK 20. In contrast to these results, our experiments showed that, in the presence of a cellular TK (HOS cells), the prodrug was more active than the unphosphorylated compound, which suggests that phosphorylation is only moderately efficient. The results we obtained with the unphosphorylated compound in the presence of HSV-TK support this interpretation. (Fig. 4). The fact that the presence of HSV-TK lowered the IC50 for the unphosphorylated compound shows that, although the cellular TK in HOS cells can convert the compound to the monophosphate, this process is inefficient. Differences in activity of both compounds in CEM versus HOS based cell lines may reflect differences in drug uptake by the individual cell lines. Nevertheless, these results were encouraging, although the fact that the IC50 for the prodrug was lower in cells that expressed HSV-TK (0.38 µM in HOS vs. 0.12 µM in 313 cells) suggested the possibility that cellular kinases may still have some difficulty in converting the D-carba T from the monophosphate to the diphosphate. If this explanation is correct, then it would be the thymidylate synthetase activity of HSV-TK (the activity that converts the monophosphate to the diphosphate) that causes the presence of the HSV-TK to reduces the IC50 for the prodrug. However, there is a possible alternative explanation: If CycloSal-D-carba T loses some of its potency by extracellular cleavage of some of the prodrug, some of the monophosphate could be generated externally rather than within the cell. This external monophosphate would probably be converted back to the parent nucleoside by phosphatases outside the cell. The fact that CycloSal-D-carba T has a lower IC50 than the parent compound, D-carba T, in our assays indicates that much of the prodrug enters the cell, where it is cleaved to the monophosphate as expected. However, a modest amount of cleavage outside the cell might explain the observed increase in the potency of the prodrug in the presence of HSV-TK. If the prodrug is cleaved extracellularly and the monophosphate is then converted to D-carba T by phosphatases, some of the nucleoside could enter the cell. This pool of D-carba T could then be phosphorylated by the HSV-TK, which could explain why the prodrug form of D-carba T is more potent in the presence of HSV-TK.
All nucleoside analogs have the potential to be cytotoxic; all of the analogs we previously tested were cytotoxic in the presence of HSV-TK. However, D-carba T and CycloSal-D-carba T show little toxicity up to 10.0 µM in several cell lines. Because the prodrug was effective in inhibiting the one-round HIV vectors at much lower concentrations, and D-carba T itself was effective against replication competent HIV-1 in a multi-round assay at much lower concentrations, these compounds should continue to be evaluated for their potential to become anti-HIV drugs. We are attempting to select HIV-1 variants that are resistant to D-carba T, and there needs to be more extensive toxicity studies, both in cells and in animals, before D-carba T and CycloSal D-carba T can be put forward for clinical development.
Experimental
Nucleoside analogs
D-carba T and CycloSal D-carba T were synthesized as previously described8, 16–18. For the D-carba T triphosphate, D-carba T was converted into the 5′-CycloSal nucleotide, then converted into the corresponding triphosphate 21.
Molecular Modeling
The three superimposed molecules were obtained from the Cambridge Structural Database (CSD) 22 through the program ConQuest version 1.9 23. The superposition was done by the Superposition tool in Maestro 8.0. The corresponding heavy atoms (eleven atoms) in the thymine ring and the five-member sugar ring of N-MCT and D-carba T were the elements superimposed.
Polymerase assays
The expression and purification of HIV-1 RT and the polymerase assays were done as previously described 2, 3. For the extension assays, 7.0 µl of a 2.0 µM stock of synthetic DNA oligonucleotide (5′-GTCACTGTTCGAGCACCA-3′; Biosource, CA) was 5′ end-labeled, then annealed to an excess of either a DNA template (5′-AATCAGTGTAGACAATCCCTAGCTATGGTGCTCGAACAGTGAC-3′) or an RNA template (5′-AAUCAGUGUAGACAAUCCCTAGCUAUGGTGCUCGAACAGUGAC-3′; Dharmacon, CO). The A residue is the first nucleotide past the 3′ end of the primer. The annealed template-primer (T/P) was suspended in a final concentration of 25.0 mM Tris (pH 8.0), 75 mM KCl, 8.0 mM MgCl2, 100.0 µg/ml BSA, 10.0 mM CHAPS, and 10.0 µM each of dATP, dCTP, and dGTP. For the RNA template reactions, the mixture was also supplemented with 1 U/µl of SuperAsin (Ambion). The reaction buffer was aliquoted to individual tubes, then supplemented with a total of 10.0 µM TTP and/or thymidine triphosphate analog (as described in the figure legend). The reactions were initiated by the addition of 1.0 µg RT and the reaction was incubated at 37° for the various indicated time intervals. Reactions were stopped by the addition of EDTA and the reaction products were precipitated by the addition of isopropyl alcohol. The products were resuspended in formamide gel loading buffer II (Ambion), heated at 70° for 5 min to denature the products, and fractionated on a 15% polyacrylamide sequencing gel.
Cell-based Assays
The human embryonic kidney cell culture line 293 was obtained from the American Type Culture Collection (ATCC). The human osteosarcoma cell line, HOS, was obtained from Dr. Richard Schwartz (Michigan State University, East Lansing, MI). Cell lines were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA) supplemented with 5% (v/v) fetal bovine serum, 5% newborn calf serum, and penicillin (50 units/mL) plus streptomycin (50 µg/mL) (Quality Biological, Gaithersburg, MD). VSV-G –pseudotyped HIV vectors were produced by transfection of 293 cells. On the day prior to transfection, 293 cells were plated in 100 mm dishes at a density of 9×105 cells per plate. 293 cells were transfected with 10 µg of pNLNgoMIVR− ΔEnv.LUC (containing wild-type RT or NRTI resistant RT variants) 10 and 3 µg of pHCMV-g (obtained from Dr. Jane Burns, University of California San Diego) using calcium phosphate precipitation. After 48 hours, virus-containing supernatants were harvested, clarified by low-speed centrifugation, filtered, and diluted 1-to-5 for infection assays. HOS cells were plated in 96-well luminescence cell culture plates at a density of 4000 cells in 100 µL per well the day prior to infection. On the day of infection, cells were pretreated with the target compounds for 3 h. Infections were carried out by adding 100 µL of virus-containing supernatants to each well and incubating for 48 h. Infectivity was determined using a luciferase reporter assay. Luciferase activity was measured by adding 100 µL of steadylite plus reagent (PerkinElmer, Waltham, MA) directly to the cells and measuring luminescence using a microplate reader. Activity was normalized to infections in the absence of target compounds for the appropriate NRTI variant. Regression analysis on the data was performed using a four-parameter sigmoidal binding model, f(x) = a + b/(1+(x/c)d) and IC50 values were determined from the fit.
Cell Toxicity Assay
Cell viability was measured using the XTT Cell Viability Assay kit from Roche. Three cell types were used to analyze the toxicity of D-carba T and CycloSal D-carba T. TZM cells are a HeLa cell-derived indicator cell line, GHOST cells are a HOS cell-derived indicator cell line, and HuT-CCR5 cells are a human T cell line. Five dilutions were tested for D-carba T, CycloSal D-carba T, and the compound hydroxypropyl-β-cyclodextrin (BCD). The BCD was used as a positive control for cell killing; more than 50% of cells will be killed when BCD is present at concentrations of 2–5% (w/v) (Ambrose et al., unpublished observations). Cells were cultured with the various compounds for 48h, then the XTT viability substrate was added for an additional 4–5h.
Acknowledgements
We thank Pat Clark for preparing purified enzymes, Tom Martin for cell sorting, Al Kane with help with figures, and Teresa Burdette for help with the manuscript. This research has been funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. NO1-CO-12400, and by the Intramural program (Center for Cancer Research) of the National Cancer Institute
Glossary
- 3TC
4-amino-1[(C2R, 5S)-2-(hydroxymethyl)-1,3-oxathiolan-5-y1]-1,2-dihydropyrimidin-2-one
- 4′ C-Me T
1-[(4S,2R,5R)-4-hydroxy-5-(hydroxymethyl)-5-methyloxolan-2-yl]-5-methyl-1,3-dihydropyrimidine-2,4-dione
- 4′ C-Et T
1-[(4S,2R,5R)-5-ethyl-4-hydroxy-5-(hydroxymethyl)oxolan-2-yl]-5-methyl-1,3-dihydropyrimidine-2,4-dione
- AZT
1-((2R,4S,5S)-4-Azido-5-(hydroxymethyl)tetrahydrofuran-2-yl)-5-methylpyrimidine-2,4(1H,3H)-dione
- D4T
1-[(2R,5S)-5-(hydroxymethyl)-2,5-dihydrofuran-2-yl]-5- methylpyrimidine-2,4-dione
- D-carba-T
1-((1R,3S,4R)-3-Hydroxy-4-(hydroxymethyl)cyclopentyl)-5-methylpyrimidine-2,4(1H,3H)-dione
- HSV
Herpes simplex virus
- N-MCT
1-((1S,2S,4S,5R)-4-hydroxy-5-(hydroxymethyl)bicyclo[3.1.0]hexan-2-yl)-5-methylpyrimidine-2,4(1H,3H)-dione
- CycloSal-D-carba-T
1-{(3S,1R,4R)-3-hydroxy-4-[(8-methyl-2-oxo(4H-benzo[d]1,3,2-dioxaphosphan-2-yloxy))methylcyclopentyl}-5-methyl-1,3-dihydropyrimidine-2,4(1H,3H)-dione
- FTC
4-amino-5-fluoro-1-(C2R,5S)-2- (hydroxymethyl)-1,3-oxathiolan-5-y17-1,2-dihydropyrimidin-2-one.
- N-MCdA
(1S,2S,4S,5S)-4-(6-aminopurin-9-yl)-1-(hydroxymethyl)bicyclo[3.1.0]hexan-2-ol
- NRTI
Nucleoside reverse transcriptase inhibitor
- MP
Monophosphate
- RT
Reverse transcriptase
- TK
Thymidine kinase
- TP
Triphosphate
- VSV-G
Vesicular Stomatitis Virus G
References
- 1.Sarafianos SG, Das K, Hughes SH, Arnold E. Taking Aim at a Moving Target: Designing Drugs to Inhibit Drug-Resistant HIV-1 Reverse Transcriptases. Curr Opin Struct Biol. 2004;14:716–730. doi: 10.1016/j.sbi.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 2.Boyer PL, Julias JG, Ambrose Z, Siddiqui MA, Marquez VE, Hughes SH. The Nucleoside Analogs 4′ C-Methyl Thymidine and 4#x02032; C-Ethyl Thymidine Block DNA Synthesis by Wild-Type HIV-1 RT and Excision Proficient NRTI Resistant RT Variants. J Mol Biol. 2007;371:873–882. doi: 10.1016/j.jmb.2007.05.043. [DOI] [PubMed] [Google Scholar]
- 3.Boyer PL, Julias JG, Marquez VE, Hughes SH. Fixed Conformation Nucleoside Analogs Effectively Inhibit Excision-Proficient HIV-1 Reverse Transcriptases. J Mol Biol. 2005;345:441–450. doi: 10.1016/j.jmb.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 4.Saenger W. Principles of Nucleic Acid Structure. New York, Berlin, Heilderberg: Springer-Verlag; 1984. pp. 9–28. [Google Scholar]
- 5.Marquez VE, Ben-Kasus T, Barchi JJ, Jr., Green KM, Nicklaus MC, Agbaria R. Experimental and Structural Evidence That Herpes 1 Kinase and Cellular DNA Polymerase(s) Discriminate on the Basis of Sugar Pucker. J Am Chem Soc. 2004;126:543–549. doi: 10.1021/ja037929e. [DOI] [PubMed] [Google Scholar]
- 6.Kálman A, Koritsánszky T, Béres J, Sági G. Crystal and Molecular Structure of (+)-Carba-Thymidine, C11H16N2O6. Nucleosides Nucleotides. 1990;9:235–243. [Google Scholar]
- 7.Ludek OR, Kramer T, Balzarini J, Meier C. Divergent Synthesis and Biological Evaluation of Carbocyclic α, Iso- and 3′-Epi-Nucleosides and Their Lipophilic Nucleotide Prodrugs. Synthesis. 2006:1313–1324. [Google Scholar]
- 8.Ludek OR, Meier C. New Convergent Synthesis of Carbocyclic Nucleosides Analogues. Synthesis. 2003:2109. doi: 10.1081/NCN-120022700. [DOI] [PubMed] [Google Scholar]
- 9.Julias JG, Boyer PL, McWilliams MJ, Alvord WG, Hughes SH. Mutations at Position 184 of Human Immunodeficiency Virus Type-1 Reverse Transcriptase Affect Virus Titer and Viral DNA Synthesis. Virology. 2004;322:13–21. doi: 10.1016/j.virol.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Oh J, McWilliams MJ, Julias JG, Hughes SH. Mutations in the U5 Region Adjacent to the Primer Binding Site Affect tRNA Cleavage by Human Immunodeficiency Virus Type 1 Reverse Transcriptase in Vivo. J Virol. 2008;82:719–727. doi: 10.1128/JVI.02611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen MS, Summers WP, Walker J, Summers WC, Prusoff WH. Characterization of Pyrimidine Deoxyribonucleoside Kinase (Thymidine Kinase) and Thymidylate Kinase as a Multifunctional Enzyme in Cells Transformed by Herpes Simplex Virus Type 1 and in Cells Infected with Mutant Strains of Herpes Simplex Virus. J Virol. 1979;30:942–945. doi: 10.1128/jvi.30.3.942-945.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen MS, Walker J, Prusoff WH. Kinetic Studies of Herpes Simplex Virus Type 1-Encoded Thymidine and Thymidylate Kinase, a Multifunctional Enzyme. J Biol Chem. 1979;254:10747–10753. [PubMed] [Google Scholar]
- 13.Whitcomb JM, Parkin NT, Chappey C, Hellmann NS, Petropoulos CJ. Broad Nucleoside Reverse-Transcriptase Inhibitor Cross-Resistance in Human Immunodeficiency Virus Type 1 Clinical Isolates. J Infect Dis. 2003;188:992–1000. doi: 10.1086/378281. [DOI] [PubMed] [Google Scholar]
- 14.Meier C, Renze J, Ducho C, Balzarini J. Cyclosal-D4TMP Pronucleotides Structural Variations, Mechanistic Insights and Antiviral Activity. Curr Top Med Chem. 2002;2:1111–1121. doi: 10.2174/1568026023393183. [DOI] [PubMed] [Google Scholar]
- 15.Fung HB, Stone EA, Piacenti FJ. Tenofovir Disoproxil Fumarate: A Nucleotide Reverse Transcriptase Inhibitor for the Treatment of HIV Infection. Clin Ther. 2002;24:1515–1548. doi: 10.1016/s0149-2918(02)80058-3. [DOI] [PubMed] [Google Scholar]
- 16.Meier C. Advances in Antiviral Drug Design. Vol. 4. Elsevier; 2004. pp. 147–213. [Google Scholar]
- 17.Meier C, Balzarini J. Application of the Cyclosal-Prodrug Approach for Improving the Biological Potential of Phosphorylated Biomolecules. Antiviral Res. 2006;71:282–292. doi: 10.1016/j.antiviral.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 18.Meier C, Meerbach A, Balzarini J. Cyclosal-Pronucleotides--Development of First and Second Generation Chemical Trojan Horses for Antiviral Chemotherapy. Front Biosci. 2004;9:873–890. doi: 10.2741/1283. [DOI] [PubMed] [Google Scholar]
- 19.Jacobo-Molina A, Ding J, Nanni RG, Clark AD, Jr., Lu X, Tantillo C, Williams RL, Kamer G, Ferris AL, Clark P, Amnon H, Hughes SH, Arnold E. Crystal Structure of Human Immunodeficiency Virus Type 1 Reverse Transcriptase Complexed with Double-Stranded DNA at 3.0 Å Resolution Shows Bent DNA. Proc Natl Acad Sci U S A. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ludek OR, Balzarini J, Meier C. Synthesis and Antiviral Evaluation of Carbocyclic 3′-Azidothymidine (AZT) Analogues and Their Cyclosal-Phosphate Triesters. European Journal of Organic Chemistry. 2006:932–940. [Google Scholar]
- 21.Warnecke S, Meier C. Synthesis of Nucleoside Di- and Triphosphates and Dinucleoside Polyphosphates with Cyclosal-Nucleotides. J Org Chem. 2009;74:3024–3030. doi: 10.1021/jo802348h. [DOI] [PubMed] [Google Scholar]
- 22.Allen FH. The Cambridge Structural Database: A Quarter of a Million Crystal Structures and Rising. Acta Crystallogr B. 2002;58:380–388. doi: 10.1107/s0108768102003890. [DOI] [PubMed] [Google Scholar]
- 23.Bruno IJ, Cole JC, Edgington PR, Kessler M, Macrae CF, McCabe P, Pearson J, Taylor R. New Software for Searching the Cambridge Structural Database and Visualizing Crystal Structures. Acta Crystallogr B. 2002;58:389–397. doi: 10.1107/s0108768102003324. [DOI] [PubMed] [Google Scholar]