Abstract
RNA silencing in the zygomycete Mucor circinelloides exhibits uncommon features, such as induction by self-replicative sense transgenes and the accumulation of two size classes of antisense small interfering RNAs (siRNAs). To investigate whether this silencing phenomenon follows the rules of a canonical RNA-silencing mechanism, we used hairpin RNA (hpRNA)-producing constructs as silencing triggers and analyzed the efficiency and stability of silencing in different genetic backgrounds. We show here that the dsRNA-induced silencing mechanism is also associated with the accumulation of two sizes of antisense siRNAs and that this mechanism is not mediated by the previously known dcl-1 (dicer-like) gene, which implies the existence of an additional dicer gene. An M. circinelloides dcl-2 gene was cloned and characterized, and the corresponding null mutant was generated by gene replacement. This mutant is severely impaired in the silencing mechanism induced by self-replicative sense or inverted-repeat transgenes, providing the first genetic evidence of a canonical silencing mechanism in this class of fungus and pointing to a role for dcl-2 in the mechanism. Moreover, a functional dcl-2 gene is required for the normal accumulation of the two sizes of antisense RNAs, as deduced from the analysis of dcl-2− transformants containing hpRNA-expressing plasmids. In addition to its critical role in transgene-induced silencing, the dcl-2 gene seems to play a role in the control of vegetative development, since the dcl-2 null mutants showed a significant decrease in their production of asexual spores.
RNA-mediated gene silencing (RNA silencing) acts through transcriptional and posttranscriptional mechanisms to suppress gene expression in a sequence-specific manner. Widespread among plants, animals, and fungi, these mechanisms are involved in a wide variety of biological processes, including defense against viruses and transposons, the regulation of endogenous gene expression, and heterochromatin formation (reviewed in references 2, 5, 31, and 35). Although a number of specialized RNA-silencing pathways have been reported, a common mechanism has emerged, with homologous genes and proteins acting in different organisms. A central feature in all of these silencing phenomena is the processing of double-stranded RNA (dsRNA) (long dsRNA and pre-microRNAs [miRNAs]) into small duplexes of 21 to 25 nucleotides (nt) (small interfering RNAs [siRNAs] and miRNAs) by the RNase III enzyme Dicer. The small RNAs generated by Dicer are subsequently incorporated into the RNA-induced silencing complex (RISC), where they are used as a guide for the sequence-specific degradation of the target mRNA, the repression of its translation, or the inhibition of its transcription.
The Dicer endonucleases are multidomain proteins evolutionarily conserved in eukaryotes. In metazoans and plants, in addition to two copies of the RNase III domain and a dsRNA-binding domain at the carboxyl terminus, the Dicer proteins include an amino-terminal ATP-dependent RNA helicase domain, a small domain of unknown function found specifically in Dicer proteins and proposed to fold as a dsRNA-binding domain (the DUF283 domain), and a Piwi-Argonaut-Zwille (PAZ) domain, which binds specifically to the 3′ end of single-stranded RNA. In lower eukaryotes, Dicer proteins frequently have a less complex domain organization (9).
The number of Dicer proteins varies among organisms. Thus, mammalian genomes, Caenorhabditis elegans, and the fission yeast Schizosaccharomyces pombe contain only one dicer gene, which is involved in the siRNA and miRNA pathways. Two dicer genes have been identified in organisms such as Drosophila melanogaster, whereas plants, such as Arabidopsis thaliana and rice, contain four dicer-like (dcl) genes. These paralogous genes may have different roles in the siRNA/miRNA-silencing pathways. Thus, in D. melanogaster, distinct roles in RNA silencing have been established for Dicer-1 and Dicer-2, which generate miRNA and siRNA, respectively, although the functional separation is not absolute, since both enzymes are required for siRNA-directed target mRNA cleavage and gene silencing (12). The four Arabidopsis Dicer-like proteins also have specialized functions in small-RNA biogenesis: DCL1 processes miRNA precursors and is also involved in the production of small RNAs from endogenous inverted repeats, while DCL2 produces siRNAs associated with antiviral defense and from natural cis-acting antisense transcripts; DCL3, on the other hand, produces 24-nt siRNAs that are involved in chromatin modification and transcriptional silencing, and DCL4 generates 21-nt trans-acting siRNAs (8, 9). In fungi, sequence analyses have revealed that the number of paralogous Dicer proteins in the genome differs considerably among species, although the best-studied filamentous fungi contain two dicer genes (17). In the ascomycete Neurospora crassa, the two genes are functionally redundant for the silencing function, since defects in silencing can be observed only in the dcl-1 dcl-2 double mutants (3). However, DCL2 contributes most of the dicer activity in the siRNA pathway, whereas DCL1 is one of the components of the MSUD (meiotic silencing by unpaired DNA) machinery (1). In Magnaporthe oryzae, a fungus taxonomically close to N. crassa, only one of the two dicer genes, mdl-2, is required for siRNA biogenesis (10), and no role has been assigned to the mdl-1 gene. Also, two dicer genes have been functionally analyzed in the chestnut blight fungus, Cryphonectria parasitica, an ascomycota closely related to N. crassa, and it has been demonstrated that dcl-2 is essential for the RNA-silencing pathway involved in antiviral defense (29, 39).
The zygomycete Mucor circinelloides is one of the few fungi of this clade which is accessible to a molecular approach for the study of a number of processes, including RNA silencing. Moreover, the fact that it is evolutionarily distant from other fungal model organisms makes it an evolutionary indicator in the fungal kingdom. We have previously demonstrated that sense transgenes trigger an RNA-silencing response in M. circinelloides (18). One of the interesting features of the gene-silencing phenomenon in this organism is its induction by nonintegrative transgenes. As a result, transgene expression is not affected by position effects or host regulatory sequences at the insertion sites, which allows the effects of transgene expression on silencing frequency and stability to be analyzed (21). Notably, the gene silencing triggered by sense transgenes in M. circinelloides is associated with two different classes of antisense siRNA, 21 nt and 25 nt long, which are differentially accumulated during the vegetative growth of silenced strains (18). This makes the silencing mechanism in this fungus unique. To date, only one dicer-like gene (dcl-1) has been identified in M. circinelloides. However, phenotypic analysis of dcl-1 null mutants indicated that this gene is not involved in the silencing response and is not essential for the biogenesis of self-replicative transgene-derived siRNAs (19). Moreover, the dcl-1 mutants showed a reduced growth rate and alterations in hyphal growth, which suggested that the gene plays a role in the control of endogenous functions (19). Since there is no genetic evidence to date concerning the participation of silencing proteins in the self-replicative transgene-induced silencing pathway in M. circinelloides, one of the questions we pose in this paper is whether a canonical RNA-silencing mechanism operates in M. circinelloides. We show here that silencing triggered by hairpin RNA (hpRNA)-expressing constructs is also associated with the two size classes of antisense siRNAs and that dcl-1 is not involved in this mechanism, which suggests the existence of an additional dicer gene in the M. circinelloides genome. By coupling two different strategies, we cloned a dcl-2 gene. The disruption of this gene resulted in severe impairment of the gene silencing triggered by hpRNA or aberrant transgenic RNA, as well as in the production of the two classes of siRNAs, which provides direct evidence of the crucial role of Dcl-2 in the RNA-silencing pathway. In addition, we found that the dcl-2 null mutants showed a significant decrease in the production of asexual spores, which suggests a role for the gene, and therefore for RNA silencing, in the regulation of vegetative development.
MATERIALS AND METHODS
Strains, growth, and transformation conditions.
The leucine auxotroph R7B (26), derived from M. circinelloides f. sp. lusitanicus CBS 277.49 (syn. Mucor racemosus ATCC 1216b), was used as the wild-type strain. Strain MU402 (19), a uracil and leucine auxotroph derived from R7B, was used as the recipient strain to knock out the dcl-2 gene. Strain MU406 is a dcl-1 null mutant derived from MU402 (19). Strain MU241 (kindly provided by M. D. Quiles-Rosillo, University of Murcia) is a prototrophic strain derived from R7B after replacement of the leuA1 mutant allele by the wild-type allele. Cultures were grown at 26°C in minimal YNB medium, complete YPG medium, or MMC medium as described previously (19). The media were supplemented with l-leucine (20 μg/ml) or uridine (200 μg/ml) when required. The pH was adjusted to 4.5 and 3.2 for mycelial and colonial growth, respectively. Transformation was carried out as described previously (18). Transformants were grown in selective medium for several vegetative cycles to increase the proportion of transformed nuclei, since primary transformants are heterokaryons due to the presence of several nuclei in the protoplasts. Illumination conditions were as previously described (25). Escherichia coli strain DH5α was used for all cloning experiments and strain LE392 (Promega, Madison, WI) for the propagation of M. circinelloides genomic lambda clones.
Plasmids.
Plasmid pMAT1253, which expresses a carB hpRNA under the control of the M. circinelloides gpd1 promoter, was constructed by inserting an inverted-repeat sequence corresponding to the carB gene (accession number AJ238028) into the M. circinelloides expression vector pEUKA4 (37), which uses the M. circinelloides leuA gene as a selectable marker. To obtain the inverted-repeat transgene, a 1,189-bp PCR-generated fragment (primers dsrna5 forward and dsrna3a reverse; see Table S1 in the supplemental material) was digested with SalI and ligated, in the opposite orientation, to an XhoI-digested 840-bp PCR fragment generated with the same primer at the 5′ end and the reverse primer dsrna5b (see Table S1 in the supplemental material). The resulting inverted repeat was digested with BamHI and cloned into the pBluescript vector to obtain plasmid pMAT1251. The 2-kb inverted-repeat fragment was excised from plasmid pMAT1251 with XhoI and NotI and cloned into pEUKA4 digested with the same enzymes.
Plasmids pMAT1254 and pMAT1255 express the carB hpRNA under the control of the light-inducible promoter of the carB gene. To construct these plasmids, the 2-kb inverted-repeat fragment isolated from pMAT1251 by BamHI digestion was cloned, in opposite orientations with respect to the carB promoter, into plasmid pMAT678. This plasmid consists of a 4.4-kb PstI fragment that includes the leuA gene of M. circinelloides and a 492-bp PCR-generated fragment amplified with primers carRP and carBSpe (see Table S1 in the supplemental material), which contains the light-inducible carB promoter. Plasmid pMAT1254 expresses the hpRNA in the required orientation for processing the intron included in the spacer region between the inverted-repeat sequences, whereas orientation of the inverted-repeat fragment with respect to the promoter in pMAT1255 produces an unspliceable transcript.
Plasmid pMAT1203 contains an 880-bp PCR-amplified fragment of the dcl-2 gene. This fragment was amplified using the degenerate oligonucleotides dicertrun2 and dicer5 (see Table S1 in the supplemental material), corresponding to sequences of the PAZ and RNase III domains of Dicer proteins (PI[SN]ASVY and EFLGDAIF, respectively), and DNA of the null dcl-1 mutant as a template. The primers were designed taking into account the DNA sequences of the M. circinelloides dcl-1 gene and the dicer-like genes of the zygomycete Rhizopus oryzae, whose genomic sequence is available at http://www.broad.mit.edu/annotation/genome/rhizopus_oryzae/Home.html. PCR amplifications were carried out at 52°C using 50 ng of the dcl-1 null mutant MU406 as a template and 2.5 U of EcoTaq Plus (Ecogen, Spain).
Plasmid pMAT1208 contains the 5.5-kb EcoRI genomic fragment that includes a 3′-truncated version of the dcl-2 gene, isolated from a M. circinelloides-positive lambda clone and cloned into the pBluescript SK vector.
Plasmid pMAT1214 contains a 480-bp fragment that was obtained in a 3′ rapid amplification of cDNA ends (RACE) experiment designed to identify the 3′ end of the dcl-2 gene. The fragment was cloned into the pGEM-T Easy vector (Promega).
Plasmid pMAT1220 includes a genomic fragment of 7,941 bp that comprises the complete dcl-2 sequence. It was constructed by cloning the 4.9-kb XhoI fragment isolated from a positive lambda clone, which expands the 3′ region of dcl-2, between the two XhoI sites of pMAT1208. This plasmid, together with pMAT1208, was used to obtain the complete genomic sequence of the dcl-2 gene.
Plasmid pMAT1218 was constructed to disrupt the dcl-2 gene. In brief, the 5.5-kb EcoRI fragment of the dcl-2 gene, cloned into the pGEMT Easy vector, was digested with BamHI and ligated to a 3.2-kb BamHI fragment isolated from plasmid pEPM1 (kindly provided by A. P. Eslava, University of Salamanca, Salamanca, Spain), which includes a wild-type allele of the M. circinelloides pyrG gene. The resulting plasmid has a deletion of 1.7 kb in the dcl-2 coding region and contains the selective marker pyrG+ flanked by sufficient sequences of the dcl-2 gene (1.35 kb upstream and 2.45 kb downstream) to allow homologous recombination. The 7-kb disruption fragment was released from pMAT1218 by EcoRI digestion and introduced into MU402 protoplasts by transformation. Only transformants showing a stable PyrG+ phenotype, as an indicator of exogenous DNA integration, were considered for further analysis.
Plasmid pMAT1223 was used to complement the dcl-2− mutation. This plasmid contains a complete version of the wild-type dcl-2 gene and regulatory sequences and was constructed by cloning a 7.13-kb SpeI fragment of the dcl-2 locus, isolated from pMAT1220, into the pyrG+-containing plasmid pEPM1. This fragment expands 1,300 bp upstream of the putative ATG initiation site and 758 bp downstream of the TAG stop codon.
Plasmid pMAT1228 was designed to obtain a dcl-1 null mutation by gene replacement, using the leuA gene of M. circinelloides as a selective marker. This plasmid contains a 4.4-kb PstI fragment isolated from pLEU4, which contains the leuA gene and regulatory sequences, flanked by 1.3-kb and 1.5-kb PCR-amplified fragments of the dcl-1 locus. SacI digestion of this plasmid produced a 7.2-kb disruption fragment that allowed homologous recombination at the dcl-1 locus, leading to replacement of 4.7 kb of the dcl-1 coding region by the leuA gene.
Plasmid pMAT689 contains the complete dcl-1 coding region and adjacent sequences. It expands 7.4 kb of the dcl-1 locus and was constructed by cloning the 2.2-kb and 5.2-kb SacI fragments of the dcl-1 locus into the pBluescript SK vector.
Plasmid pMAT1330 contains the same inverted-repeat carB transgene as pMAT1253 and the pyrG gene as a selective marker. It was obtained by isolating the complete inverted-repeat construct from pMAT1253, including the promoter and terminator regions, and cloning it in the pyrG+ plasmid pEPM1.
Selection for dcl-2− pyrG− mutants.
Spores of the MU410 (dcl-2− pyrG+ leuA−) and MU411 (dcl-1− dcl-2− pyrG+ leuA+) strains were treated with N-methyl-N ′-nitro-N-nitrosoguanidine (0.1 mg/ml) as described previously (19). After completion of a cycle of vegetative growth on complete-medium plates containing 200 μg/ml uridine, the resulting spores were inoculated on minimal medium containing 3 mg/ml 5-fluoroorotic acid and 200 μg/ml uridine. Leucine (20 μg/ml) was added when required. 5-Fluoroorotic acid-resistant colonies were tested for uracil auxotrophy, and several independent Ura− mutants were assayed for orotidine 5′-phosphate decarboxylase activity. Seven pyrG− mutants obtained in independent mutagenesis experiments were isolated from MU410, and one pyrG− mutant was derived from MU411. PyrG+ transformants of the dcl-2− pyrG− leuA− mutant were selected in MMC medium.
Nucleic acid manipulation and analysis.
Standard recombinant DNA manipulations were performed as described previously (28). Genomic DNA from M. circinelloides was prepared as described previously for Phycomyces blakesleeanus (27). A LambdaGEM-11 genomic library of the M. circinelloides wild-type strain CBS 277.49 (25) was screened with three distinct probes under two different conditions to isolate recombinant plaques containing the complete dcl-2 gene. Screening of 36,000 plaques with a 2.5-kb fragment of the dcl-1 gene cloned in plasmid pMAT660 (19) was performed using low-stringency conditions. Filters (Hybond N+; GE Healthcare) were hybridized to the radioactively labeled probe at 60°C in 0.9 M NaCl, 1% SDS, 0.1 g/ml dextran sulfate and washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS) at 60°C for 20 min; 1× SSC, 0.1% SDS at 60°C for 20 min; and 0.5× SSC, 0.1% SDS at 60°C for 10 min. Twenty-seven positive recombinant plaques were further screened with an 880-bp fragment of the dcl-2 gene isolated from plasmid pMAT1203 under high-stringency conditions (19). A positive clone containing a 5.5-kb EcoRI fragment with a 3′-truncated version of the dcl-2 gene was isolated. A second screening of 9,000 plaques of the genomic library was carried out under stringent conditions to isolate the complete dcl-2 gene, using as a probe a 480-bp DNA fragment cloned in plasmid pMAT1214, which corresponds to the 3′ end of the dcl-2 gene. Probes were labeled with [α-32P]dCTP using Ready-to-Go DNA-labeling beads (GE Healthcare), following the instructions of the supplier.
For Southern blot analysis, genomic DNA (1 μg) isolated from the wild-type strain and the dcl-2− or dcl-1− dcl-2− mutant was digested with the appropriate restriction enzymes, fractionated on a Tris-acetate-EDTA-agarose gel, blotted onto positively charged nylon filters (Hybond-N+; GE Healthcare), and hybridized at 65°C to radioactively labeled dcl-2 or dcl-1 probes. The hybridization and washing conditions were as described previously (18).
For Northern blot hybridization, total RNA was isolated using the guanidinium thiocyanate-cesium chloride method. About 20 μg of total RNA was hybridized to radioactively labeled probes as previously described (19). The dcl-2 probe corresponded to a 2.5-kb XhoI-EcoRI fragment isolated from plasmid pMAT1208. The pyrG probe was a 1.5-kb BamHI-SalI fragment isolated from plasmid pEPM1.
Reverse transcription (RT)-PCR amplifications were performed to identify introns in the dcl-2 genomic sequence. Total RNA was isolated from cultures grown for 24 h on minimal liquid medium (pH 4.5) and reverse transcribed with dcl-2-specific primers. Primers DBlong5 and DB3 (see Table S1 in the supplemental material) were used to identify exon-intron boundaries in introns 1 and 2. To identify intron 3, primers DBs5 and DBex3 (see Table S1 in the supplemental material) were used. Intron 4 and the 3′ end of the dcl-2 mRNA were identified by 3′-RACE experiments, using the dcl-2 sense-specific primers DB10 and DB9 (see Table S1 in the supplemental material) to amplify the cDNA reverse transcribed with the RACE-T primer (Roche), using as a template total RNA isolated from cultures of a silenced transformant grown for 24 h on minimal liquid medium. To characterize dcl-2 transcripts in the dcl-2− mutant, RT-PCR amplifications were performed using total RNA isolated from 24-h cultures of the dcl-2− mutant and primers DBex3 and DBs5 to amplify the RNase III catalytic domains and pyrG3 and pyrG9 to amplify the pyrG sequence (see Table S1 in the supplemental material). Expand reverse transcriptase (Roche) and EcoTaq Plus (Ecogen, Spain) were used in all the experiments. The amplified fragments were cloned in pGEM-T Easy vectors (Promega) and sequenced.
To identify transformants that correctly integrated the knockout vector designed to disrupt the dcl-2 gene, a rapid protocol for isolating the DNA to be used in PCR amplifications was utilized (19). Primers pyrG10 and DB16 (see Table S1 in the supplemental material) were used in the PCRs to identify homologous integrations at the dcl-2 locus. Primers leua-P2 and dicer19 were used to identify transformants of the dcl-2− mutant that correctly integrated the knockout vector designed to disrupt the dcl-1 gene.
Computer analysis of the sequence was carried out using European Bioinformatic Institute server software (EMBL Outstation, Hinxton, United Kingdom); the National Center for Biotechnology Information server (NCBI, Bethesda, MD); the ExPASy Molecular Biology server (Swiss Institute of Bioinformatics); and the Baylor College of Medicine search launcher (Houston, TX).
Small-RNA analysis.
For siRNA analysis, total RNA was extracted from frozen mycelia of wild-type and silenced strains grown under illumination with white light (blue light component at 4 W/m2) using Trizol reagent, following the recommendations of the supplier (Invitrogen). Low-molecular-weight RNA was prepared from total RNA, separated by electrophoresis, and hybridized to radioactive riboprobes as previously described (18). Filters were washed as described previously (19), except that treatment with the RNase A-RNase T1 cocktail was carried out at 37°C for 60 min.
The carB antisense-specific riboprobe was prepared by in vitro transcription of the linearized pMAT652 plasmid (18), using the Maxiscript transcription kit (Ambion) following supplier-recommended protocols. Riboprobes were treated as described previously (18) to be broken to an average size of 50 nt. Oligonucleotides corresponding to sense and antisense sequences of the carB gene were used as size and polarity controls.
Sporulation measurements.
To quantify the sporulation efficiencies of dcl-2− mutants, spores were harvested from dcl-2−, dcl-1− dcl-2−, and wild-type mycelia as described previously (20). Briefly, small colonies (2-mm diameter; n = 10) of the different strains were transplanted to MMC solid medium, pH 3.2, and grown for 72 h under dark or light conditions. After incubation, the diameter of each colony was determined, and the complete colony was harvested in 1.0 ml distilled water and vortexed to allow suspension of the spores. The spores were counted with a hemocytometer.
Nucleotide sequence accession number.
The nucleotide sequence of the dcl-2 gene of M. circinelloides has been deposited in the EMBL/GenBank/DDBJ databases under accession number FN421463.
RESULTS
HpRNA efficiently induces stable gene silencing.
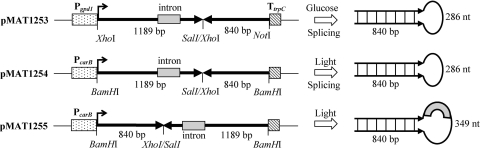
Previous studies demonstrated that the introduction of self-replicative sense transgenes in M. circinelloides triggers a gene-silencing response in a low percentage of the transformants (18, 19). To investigate whether a canonical RNA-silencing mechanism, which is induced by dsRNA, operates in M. circinelloides, we constructed an inverted-repeat transgene that produces, upon transcription, an hpRNA corresponding to the carB gene. The silencing of carB, a gene required for carotenoid biosynthesis, results in albino colonies that strongly contrast with the bright-yellow phenotype of the wild-type strain in the light. Two different endogenous promoters were used to drive transcription of the carB transgene, the strong gpd1 promoter, which is active during vegetative growth and is regulated by the carbon source (37), and the light-induced carB promoter (36). Plasmid pMAT1253 expresses a carB hpRNA comprising an 840-bp stem and a 349-bp loop under the control of the gpd1 promoter and the Aspergillus nidulans trpC terminator (Fig. 1). Plasmid pMAT1254 expresses the same construct under the control of the carB promoter. In both cases, a spliceable intron was included in the loop region, since the presence of introns has been reported to increase silencing efficiency in plants and fungi (6, 34). We expected that splicing of the primary RNA would produce a 286-bp loop in the mature transcripts derived from both constructs. Plasmid pMAT1255 expresses the carB inverted-repeat sequence in the reverse orientation with respect to the carB promoter. Thus, this construct would produce an hpRNA with an unspliceable intron within the loop region (Fig. 1).
FIG. 1.
Schematic representation of inverted-repeat transgenes used in this study. Promoters (P), terminators (T), and introns are represented as boxes and coding regions as thick arrows. The sizes of the stem-and-loop regions of the hpRNAs obtained upon transcription are shown. Only relevant restriction sites are indicated.
To evaluate the silencing efficiencies of the three constructs in M. circinelloides, the wild-type strain for carotenogenesis, R7B, was transformed with these plasmids, and the color phenotypes of the transformants grown in glucose medium (YNB) and under light conditions were analyzed (Table 1). The highest frequency of albino transformants was obtained using plasmid pMAT1253, which was able to trigger silencing of the carB gene in more than 85% of transformants. Most of the silenced primary transformants showed albino and yellow patches, but after a cycle of vegetative growth in selective medium to increase the proportion of transformed nuclei, more than 95% of the mycelia were uniformly albino. Transgenes expressing hpRNA under the control of the light-inducible carB promoter were also able to induce silencing, although the frequency of albino transformants was lower than that obtained with the gpd1 promoter (Table 1). The lowest efficacy as a silencing trigger was displayed by the hpRNA-expressing transgene in which the intron included in the spacer region could not be spliced out (pMAT1255). In fact, only 32% of transformants showed an albino phenotype after a cycle of vegetative growth, compared with the 54% obtained in transformations with the construct containing a correctly oriented intron. These results suggest that, similarly to what has been reported for plants and ascomycetes, the presence of a spliceable intron enhances the silencing triggered by hpRNA in M. circinelloides.
TABLE 1.
Efficiencies of hpRNA-expressing constructs as silencing triggersa
| Plasmid | No. of transformants
|
Silencing frequency (%) | ||
|---|---|---|---|---|
| Albino | Wild type | Total | ||
| pMAT1253 | 143 | 25 | 168 | 85.1 |
| pMAT1254 | 85 | 71 | 156 | 54.5 |
| pMAT1255 | 69 | 146 | 215 | 32.1 |
| pLEU4 | 0 | 1,758 | 1,758 | 0 |
Plasmid pLEU4 was used as a control. The colors of the transformants were observed after 48 h under illumination with blue light.
In addition to the silencing efficiency, the stability of the silenced phenotype was also dependent on the construct used as a silencing trigger. To measure the phenotypic reversion rate, spores of albino transformants harboring different constructs were grown in glucose medium under light conditions for 48 h, and the proportion of nonsilenced (bright-yellow) colonies was calculated. Under these conditions, the silenced phenotype of transformants containing sense transgenes was unstable, since they segregated to the wild-type phenotype after a cycle of vegetative growth (18). Results with the hpRNA constructs demonstrated that 100% of the descendants of pMAT1253 albino transformants showed a silenced phenotype after a cycle of vegetative growth, whereas silenced transformants expressing hpRNA driven by the light-induced carB promoter maintained the albino phenotype in only 77% of descendants, on average. Moreover, the stability of the silenced phenotype in the latter transformants was reduced to 12 to 30% when incubation was carried out for 48 h in the dark followed by 24 h under illumination. These results suggest that the frequency and stability of gene silencing in M. circinelloides are highly dependent on the level of transcription of the hpRNA, and they agree with previous results obtained using sense transgenes as silencing triggers (21). Due to its high silencing efficiency and stability, plasmid pMAT1253, expressing hpRNA driven by the gpd1 promoter, was used as a silencing trigger in the subsequent research.
Gene silencing induced by hpRNA is associated with two classes of small antisense RNAs and does not require the dcl-1 gene.
Gene silencing in M. circinelloides is correlated with the accumulation of two classes of antisense siRNAs, 25-nt and 21-nt long, which are differentially accumulated during the vegetative cycle (18). These two classes of siRNAs may be specific to sense transgene-induced silencing and may be generated via a different pathway from canonical RNA silencing, since, to date, no genetic evidence has been provided to show that RNA-silencing proteins (such as Dicer, Argonaute, or RdRP), actually participate in the sense transgene-induced gene-silencing pathway in M. circinelloides. To verify that gene silencing induced by hpRNA-expressing plasmids is also associated with two different size classes of siRNA, samples of low-molecular-weight RNAs were isolated from albino transformants of the wild-type strain harboring sense or inverted-repeat transgenes. RNA blot hybridization with a carB antisense-specific riboprobe indicated that hpRNA-triggered silenced strains were able to accumulate the two size classes of antisense RNAs, as did the wild-type strain transformed with sense transgenes (Fig. 2A, lanes 4 and 6). These results indicate that accumulation of two size classes of siRNAs does not depend on the silencing trigger but is a specific feature of the M. circinelloides silencing mechanism.
FIG. 2.
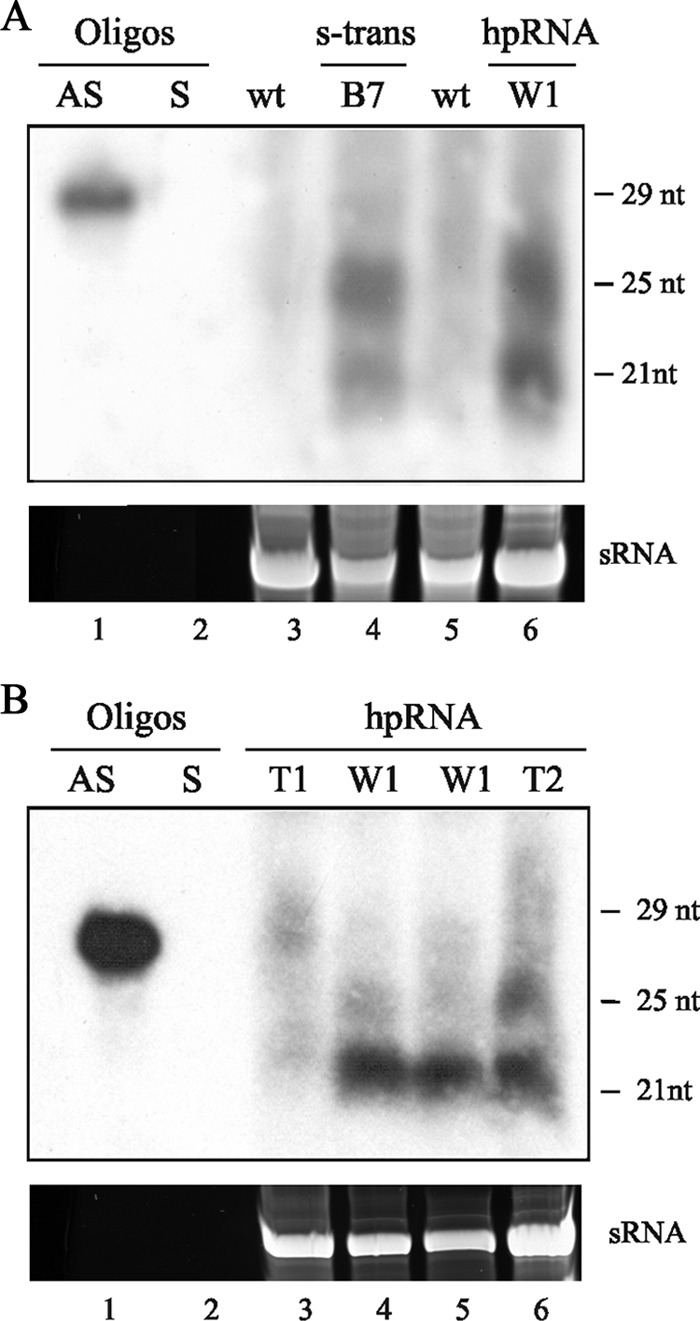
siRNA production in hpRNA-induced gene silencing. (A) Northern blot analysis of low-molecular-weight RNAs (50 μg) isolated from the wild-type strain R7B (lanes 3 and 5), a silenced albino transformant obtained by introduction of the sense transgene (s-trans) expressing plasmid pMAT647 into the wild-type strain (transformant B7; lane 4), and a silenced albino transformant obtained by introduction of the hpRNA-expressing plasmid pMAT1253 into the wild-type strain (transformant W1; lane 6). The cultures were grown for 24 h in liquid medium under continuous-illumination conditions. Oligos, oligonucleotides; AS, antisense; S, sense; wt, wild type. (B) Production of siRNAs in the dcl-2− genetic background. Northern blot analysis of low-molecular-weight RNAs (50 μg) isolated from cultures grown for 24 h in liquid medium from transformants of the dcl-2− mutant (transformant T1; lane 3) or the wild-type strain (transformant W1; lane 4) carrying the hpRNA-expressing transgene pMAT1253. Analysis of other dcl-2− transformants gave the same results. To detect siRNAs in the dcl-2− genetic background, cultures of one of the 15 dcl-2− transformants with a partially silenced phenotype (T2) and a wild-type silenced strain (W1) were grown for 48 h in solid minimal medium pH 3.2 (lanes 5 and 6). Ten picomoles per lane of 29-mer DNA oligonucleotide in antisense orientation (AS; lane 1) and 25-mer DNA oligonucleotide in sense orientation (S; lane 2) were used as size markers and to control the hybridization specificity. The RNA blot was hybridized with a hydrolyzed carB antisense-specific riboprobe, which corresponded to a 1,662-bp DNA fragment of the carB sequence that extends from position +863 to the end of the carB gene (18). The predominant RNA species in the small-RNA samples (sRNA) were stained with ethidium bromide after size separation by agarose gel electrophoresis.
Previous results had indicated that gene silencing triggered by self-replicative sense transgenes was not impaired in the dcl-1− strain MU406, which has a null mutation in a dicer-like 1 gene (19). To investigate whether this mutant was able to silence gene expression using hpRNA as a trigger, plasmid pMAT1253 was introduced into MU406 and the color phenotypes of transformants were scored. As a control, plasmid pMAT647 (18), which expresses a sense carB transgene, including the complete genomic sequence and promoter region, was also used (Table 2). The results indicated that silencing efficiency in the dcl-1− mutant was identical to that of the wild- type strain, regardless of the construct used as a silencing trigger. The stability of the silenced phenotype in the dcl-1− genetic background after a cycle of vegetative growth was also similar to that in the wild-type strain, as was the accumulation of the two size classes of antisense siRNAs (data not shown). These results confirm that dcl-1− mutants are silencing competent and suggest that the dcl-1 gene is not essential to the silencing mechanism triggered by sense transgenes or inverted-repeat transgenes.
TABLE 2.
Gene silencing by hpRNA-expressing constructs in dcl-1− mutantsa
| Plasmidb | Strain | No. of transformants
|
Silencing frequency (%) | ||
|---|---|---|---|---|---|
| Albino | Wild type | Total | |||
| pMAT1253 (hpRNA) | wtc | 543 | 96 | 639 | 85.1 |
| dcl-1− | 544 | 82 | 626 | 86.9 | |
| pMAT647 (s-transgene) | wt | 26 | 489 | 515 | 5.0 |
| dcl-1− | 93 | 1,513 | 1,606 | 5.7 | |
The colors of the transformants were observed after 48 h under illumination with white light.
hpRNA, construct expressing hpRNA; s-transgene, construct expressing the sense transgene.
wt, wild type.
Cloning an M. circinelloides dcl-2 gene.
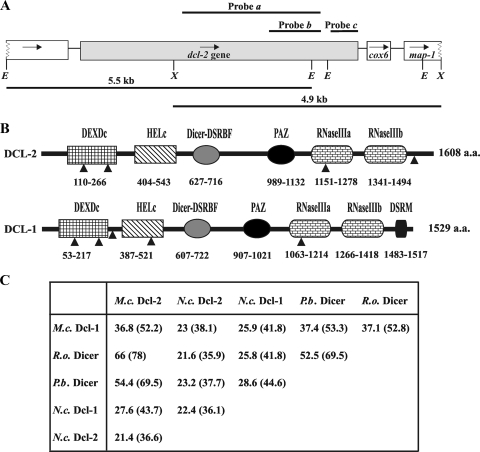
The dcl-1 gene is not required for the biogenesis of siRNAs, since dcl-1− mutants are perfectly able to produce the 25-nt and 21-nt siRNAs associated with gene silencing in M. circinelloides (19). These results imply that at least one additional dicer gene would have to exist in M. circinelloides to produce the siRNA molecules. A dcl-2 gene was cloned by screening a genomic LambdaGEM-11 library of the wild-type strain of M. circinelloides with three different probes, a 2.5-kb fragment of the dcl-1 gene corresponding to the HELc-RNase III domains (Fig. 3A, probe a), an 880-bp fragment of dcl-2 amplified with degenerate oligonucleotides (Fig. 3A, probe b), and a 480-bp fragment of dcl-2 obtained in a 3′-RACE experiment (Fig. 3A, probe c) (see Materials and Methods). A nucleotide sequence of 7,941 bp was obtained by sequencing the two DNA strands of the 5.5-kb EcoRI and 4.9-kb XhoI fragments isolated from positive lambda clones (Fig. 3A). This sequence included the complete 5,073-bp structural region of the dcl-2 gene and located the polyadenylation site 23 bp downstream of the AAUAAA polyadenylation sequence found 71 bp downstream of the stop codon. This gene was flanked downstream by a highly conserved sequence corresponding to the cytochrome c oxidase chain VI precursor (COX6 gene), followed by a sequence similar to that of the map-1 gene of N. crassa (accession number AF323912), which encodes an ATP-dependent Zn protease that belongs to the AAA superfamily of ATPases. The same genomic structure is found downstream of the single dicer-like sequence of P. blakesleeanus, which has recently been made available (http://genome.jgi-psf.org/Phybl1/Phybl1.home.html), and the complete dicer-like sequence of R. oryzae, which denotes a high degree of synteny among zygomycete fungi in this region. The sequence upstream of the predicted ATG initiation codon of the M. circinelloides dcl-2 gene contains pyrimidine-rich tracks that are common to many fungal promoters (7). This region has been involved in the correct initiation of transcription in some A. nidulans promoters (23).
FIG. 3.
Isolation of the dcl-2 gene. (A) Schematic representation of the dcl-2 genomic region. The probes used in the screening of the genomic library are shown above the scheme. Probe a corresponds to a dcl-1 sequence. Probes b and c correspond to dcl-2. The arrows indicate the directions of transcription. Only relevant restriction sites are indicated. E, EcoRI; X, XhoI. (B) Domain organization of the Dcl-2 protein. Dicer domains are shown by boxes with the starting and stopping amino acid (a.a.) of each domain indicated. For comparison, the domain organization of the Dcl-1 protein is included. The black bars correspond to the full protein sequences. DEXDc, DEAD-like helicase superfamily domain; HELc, helicase C domain; Dicer-DSRBF, a dsRNA-binding domain found in members of the Dicer family; PAZ, Piwi/Argonaut/Zwille domain; RNase III a and b, dsRNA-specific endonuclease domains; DSRM, dsRNA-binding motif. The arrowheads indicate the positions of introns. (C) Identity and similarity (in parentheses) between the deduced amino acid sequences of M. circinelloides Dcl-2 and other Dicer proteins. M.c., M. circinelloides (Dcl-1, accession no. AM269947); R.o., R. oryzae (http://www.broad.mit.edu/annotation/genome/rhizopus_oryzae/Home.html; protein ID, RO3G_15434.3); P.b., P. blakesleeanus (http://genome.jgi-psf.org/Phybl1/Phybl1.home.html; protein ID, 24670); N.c., N. crassa (Dcl-1, accession no. Q7S8J7; Dcl-2, accession no. Q7SCC1).
Comparison of the dcl-2 cDNA and the genomic sequence identified four introns in the gene (Fig. 3B). The protein sequence deduced from the dcl-2 gene contains 1,608 amino acids, and its analysis revealed the presence of all the structural domains normally found in Dicer proteins, including two helicase domains (DEXDc-DEAD-DEAH and HELc) at the N terminus of the protein, a Dicer-specific dsRNA-binding domain (Dicer-DSRBF), a PAZ domain, and two RNase III domains. The C-terminal end of the protein contains a sequence similar to that found in the same region of the M. circinelloides Dcl-1 protein, where a canonical dsRNA-binding domain (DSRM) was detected. However, all the computer programs tested failed to identify such a domain at the C-terminal end of the Dcl-2 protein (Fig. 3B). The predicted protein sequence of M. circinelloides Dcl-2 showed a high degree of similarity to the Dicer-like proteins of zygomycete and, to a lesser extent, ascomycete fungi (Fig. 3C). Among these proteins, the similarity to the complete Dicer-like protein of R. oryzae (78% similarity) was much higher than that observed to the M. circinelloides paralogous protein, Dcl-1 (52.2% similarity). The high similarity among M. circinelloides Dcl-2 and the P. blakesleeanus and R. oryzae putative Dicer proteins, together with the analogous sequence organization found among the corresponding genomic regions, suggests that the dicer-like sequences of Phycomyces and Rhizopus correspond to the M. circinelloides dcl-2 gene.
Expression analysis of the M. circinelloides dcl-2 gene.
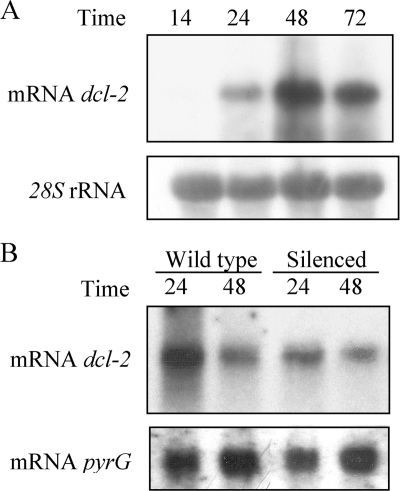
In order to analyze the expression pattern of the dcl-2 gene during vegetative growth and the response of this expression to the induction of the silencing mechanism, the transcript abundance of the corresponding mRNA was determined by Northern blot experiments. Total RNA was isolated from the wild-type strain MU241 grown for different periods of time in minimal liquid medium. Hybridization with a dcl-2-specific probe corresponding to a 2.5-kb XhoI-EcoRI fragment identified a 5-kb transcript that matched the expected size of the dcl-2 mRNA (Fig. 4A). The amount of transcripts increased during the vegetative cycle, with maximum accumulation after 48 h of growth, when the liquid cultures reached the end of the exponential phase. Accumulation of the dcl-2 mRNA at early times in vegetative growth (14 h) was observed when cultures were grown in complete medium or after overexposure of the membranes (data not shown).
FIG. 4.
Expression of the dcl-2 gene. (A) Northern blot analysis of total RNA (20 μg) isolated from the wild-type strain MU241 grown for different times (hours) in liquid minimal medium under continuous illumination. The filter was hybridized with a 2.5-kb dcl-2 fragment corresponding to the central region of the gene. The membrane was reprobed with a 28S rRNA probe to check loading. (B) Northern blot analysis of total RNA (20 μg) isolated from the wild-type strain R7B transformed with the control plasmid pLEU4 (leuA+) and a carB silenced transformant harboring plasmid pMAT1253 (carB leuA+) grown for different times (hours) in solid minimal medium under continuous illumination. The filter was hybridized with the same probe as for panel A. The membrane was reprobed with a 1.5-kb BamHI-SalI fragment of the pyrG gene to check loading.
To investigate whether expression of the dcl-2 gene was regulated by activation of the silencing mechanism, Northern blot experiments were carried out using total-RNA samples isolated from solid cultures of the wild-type strain and a silenced transformant obtained with plasmid pMAT1253, which expresses a carB hpRNA (Fig. 4B). The expression of the dcl-2 gene was unaffected by the presence of an active silencing mechanism, since the levels of transcripts detected with the dcl-2-specific probe were similar in wild-type and silenced strains. This result suggests that induction of the silencing mechanism is not required for expression of the dcl-2 gene. Differences in the mRNA accumulation patterns of the wild-type and silenced strains with respect to the data shown in Fig. 4A were due to the different growth profiles of M. circinelloides in liquid and solid media.
Generation of knockout mutants for the dcl-2 gene.
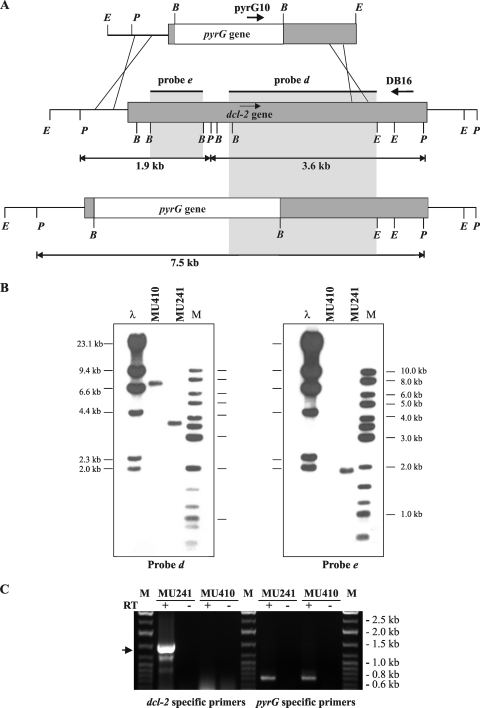
To investigate the role of the dcl-2 gene in the silencing mechanism, a knockout vector, named pMAT1218, was designed to disrupt the gene. A 7-kb EcoRI fragment from this plasmid, containing the pyrG gene and sufficient sequences of the dcl-2 gene to allow homologous recombination (see Materials and Methods for details), was used to transform the MU402 strain. Integration of this fragment by homologous recombination into the dcl-2 locus was expected to result in the replacement of 1.71 kb of the endogenous dcl-2 gene by the pyrG gene (Fig. 5). One out of 14 homokaryotic transformants PCR analyzed to distinguish between homologous and ectopic integration amplified the expected 4-kb fragment. Disruption of the dcl-2 gene in this strain, named MU410, was confirmed by Southern analysis. Hybridization of digested DNA from MU410 with a dcl-2 probe that recognized the wild-type and the disrupted dcl-2 alleles but could discriminate between them (Fig. 5A, probe d) and with a probe corresponding to the DNA region deleted in the knockout vector (Fig. 5A, probe e) showed the expected fragments, indicating that the dcl-2 wild-type allele had been successfully replaced (Fig. 5B). Since the knockout mutant maintained the 3′ region of the dcl-2 gene downstream of the pyrG sequence, an RT-PCR experiment was performed to confirm that the disrupted gene could not generate a fusion protein containing a catalytic RNase III domain. Unsurprisingly, primers flanking the RNase III domains were able to amplify dcl-2-specific transcripts in the wild-type strain, but not in the MU410 dcl-2− mutant (Fig. 5C), while pyrG-specific primers, used as controls, amplified pyrG transcripts in both the dcl-2− and wild-type strains. These results demonstrate that the disrupted mutant was unable to synthesize a Dicer protein fragment with RNase III activity, confirming the loss of function of the dcl-2− mutant.
FIG. 5.
Disruption of the dcl-2 gene. (A) Schematic representation of the wild-type dcl-2 locus (middle) and after homologous recombination with the disruption fragment (below). Dark-gray boxes, genomic dcl-2 loci; white boxes, pyrG selectable markers. The positions of the probes used and the expected sizes of the PstI restriction fragments are indicated. P, PstI; E, EcoRI; B, BamHI. The primers used to identify homologous integration events are shown (pyrG10 and DB16). (B) Southern blot analysis of the dcl-2+ strain MU241 and the dcl-2− mutant MU410. Genomic DNA (1 μg) was digested with PstI and hybridized with probe d, which corresponds to a 2.5-kb XhoI-EcoRI fragment of the dcl-2 coding region (left), and with probe e, a 1.2-kb BamHI fragment isolated from plasmid pMAT1208 (right). The positions and sizes of the λ-HindIII (λ) and the GeneRuler DNA ladder mixture (M) (Fermentas) size markers are indicated. (C) RT-PCR analysis of dcl-2 expression in the dcl-2− mutant. Total RNA isolated from the wild-type strain MU241 and the dcl-2− mutant MU410 was retrotranscribed using an oligo(dT) primer and amplified with primers corresponding to dcl-2 sequences flanking the RNase III catalytic domains (left side). The arrow marks the size of the expected fragment. As a positive control, pyrG-specific primers were used to amplify pyrG transcripts (right side). Total RNA without RT was used as a negative control. M, GeneRuler DNA ladder mixture (Fermentas).
The dcl-2 gene is essential for efficient gene silencing.
To investigate the involvement of dcl-2 in transgene-induced gene silencing, mutant MU410 (dcl-2− pyrG+ leuA−) was transformed with plasmids pMAT1253, which produces a carB hpRNA upon transcription, and pMAT647 (19), which carries a sense carB transgene, and the frequency of albino transformants was compared with that in the wild-type strain. Transformation of the strain R7B (dcl-2+ pyrG+ leuA−) with these self-replicative plasmids caused silencing of the carB function in a high proportion of the transformants (Table 3). However, the frequency of albino transformants was severely impaired in the dcl-2− mutant, in which only a few colonies with patches of partially silenced phenotype were obtained when the hpRNA-expressing transgene was used as a silencing trigger. The stability of the silenced phenotype in the dcl-2− genetic background was also affected, as seen from a comparison with the wild-type strain. Whereas 100% of the descendants of wild-type silenced colonies obtained with the hairpin transgene maintained the albino phenotype after a cycle of vegetative growth, only 10% of the colonies derived from the silenced dcl-2− mutant showed an albino phenotype. This indicates that the residual silencing activity shown by the dcl-2− mutant was not sufficient to maintain a silenced phenotype and highlights the need for the dcl-2 gene in transgene-induced gene silencing.
TABLE 3.
Gene silencing by self-replicative sense and inverted-repeat transgenes in the dcl-2− mutant MU410 and a MU410 transformant carrying a wild-type allele of dcl-2a
| Plasmidb | Strain | No. of transformants
|
Silencing frequency (%) | ||
|---|---|---|---|---|---|
| Albino | Wild type | Total | |||
| pMAT1253 (hpRNA) | wtc | 275 | 49 | 324 | 84.8 |
| dcl-2− | 15 | 500 | 515 | 3.47 | |
| dcl-2−dcl-2+ | 270 | 90 | 380 | 71.8 | |
| pMAT647 (s-transgene) | wt | 25 | 82 | 107 | 23.4 |
| dcl-2− | 0 | 129 | 129 | 0 | |
| pLEU (control) | wt | 0 | 340 | 340 | 0 |
| dcl-2− | 0 | 130 | 130 | 0 | |
| dcl-2−dcl-2+ | 0 | 50 | 50 | 0 | |
The colors of the transformants were observed after 48 h under illumination with white light.
hpRNA, construct expressing hpRNA; s-transgene, construct expressing the sense transgene.
wt, wild type.
The phenotype shown by the MU410 mutant was exclusively due to the absence of the dcl-2 gene, as was confirmed by transformation of the dcl-2 null mutant with a plasmid containing a dcl-2 wild-type allele and analysis of the silencing efficiency of the resultant strain. This strategy required induction of a pyrG− mutation in the MU410 genetic background, since two distinct auxotrophic markers were needed, one for introduction of the dcl-2 wild-type allele and the other one for introduction of silencing vectors. Seven independent uracil auxotroph strains (pyrG−) were derived from the MU410 mutant. As a control, one of them was transformed with plasmid pMAT1330, which contained the inverted-repeat carB transgene and the pyrG+ selective marker, to confirm its silencing phenotype. In fact, none of the 50 transformants analyzed showed an albino phenotype. This pyrG− derivative of MU410 was then transformed with plasmid pMAT1223, which contains a complete version of the wild-type dcl-2 gene and the pyrG gene as the selectable marker. To investigate the silencing efficiency of the dcl-2− dcl-2+ transformants, one of them was further transformed with the silencing plasmid pMAT1253, which contains the leuA gene as a selectable marker and expresses a carB hpRNA. The silencing frequency and the stability of the albino phenotype of the dcl-2− dcl-2+ strain were similar to those obtained in the wild-type strain (Table 3), indicating that the wild-type dcl-2 allele fully complemented the mutant phenotype of the MU410 strain. Similar results were obtained when different pyrG− derivatives of MU410 were used, the small differences observed in the silencing frequency being due to differences in the plasmid copy numbers of the analyzed transformants (data not shown).
Disruption of dcl-2 impaired dsRNA dicing.
To determine the biochemical role of Dcl-2 in the RNA-silencing pathway, the ability of the dcl-2− mutant to accumulate the two classes of siRNAs described in M. circinelloides (18) was tested. Samples of low-molecular-weight RNAs were isolated from transformants expressing the hpRNA, in a wild-type or dcl-2− genetic background, after being grown for 24 h in liquid culture. RNA blot hybridization with a carB antisense-specific RNA probe detected the accumulation of siRNAs in the wild-type transformant (W1), but neither of the two size classes of siRNA was identified in a dcl-2− transformant (T1) (Fig. 2B, lanes 3 and 4). This is consistent with the idea that dcl-2 plays a pivotal role in the transgene-induced silencing mechanism and that it is required for efficient siRNA production. However, the partially silenced phenotype shown by a reduced number of dcl-2− transformants must require some amount of siRNAs, the effector molecules of the silencing mechanism. Therefore, we carefully examined siRNA accumulation in the 15 partially silenced dcl-2− transformants (Table 3), which presented patches of albino and wild-type phenotypes. Due to the instability of the silenced phenotype in the dcl-2− genetic background, it was impossible to obtain albino mycelia from those transformants when they were grown for 24 h in liquid medium. To circumvent this problem, small albino patches of each transformant were transferred to solid YNB medium, pH 3.2, and grown for 48 h. Under these conditions, the expansion of the colonies was reduced and the stability of the silenced phenotype increased (our unpublished results). After growth, only one of the 15 partially silenced transformants gave rise to enough albino mycelium for Northern analysis. Overloading low-molecular-weight RNA samples isolated from that mycelium allowed us to detect the two size classes of siRNAs in the dcl-2− transformant (T2) (Fig. 2B, lane 6). As a control, small-RNA samples of an albino transformant of the wild-type strain grown under the same conditions were used (Fig. 2B, lane 5). Differences in the accumulation of the two size classes of siRNAs between the wild-type and dcl-2− transformants were probably due to growth differences of the cultures, since the two classes of siRNA are differentially accumulated during the vegetative growth of the silenced transformants, with the 21-nt class being predominant at the end of vegetative growth (18).
The results described above strongly suggest that the Dcl-2 protein is required for efficient dsRNA dicing in the RNA-silencing pathway. They also indicate that a residual dicing activity is present in a dcl-2− genetic background. Since we have demonstrated that the dcl-2− strain MU410 carries a null mutation (Fig. 5), the residual dicing activity must be provided by a different Dicer enzyme. To test whether the Dcl-1 protein is responsible for this activity, we sought a double dcl-1− dcl-2− mutant, using as the recipient strain the pyrG− derivatives of MU410 (dcl-2− leuA− pyrG−), since we needed two auxotrophic markers, one for the disruption of dcl-1 and the other for the introduction of silencing vectors. Thus, three independent pyrG− derivatives of MU410 were transformed with a 5.8-kb PuvII-SpeI fragment isolated from the knockout vector pMAT661, which was designed to disrupt the dcl-1 gene (19). The fragment contained the pyrG gene of M. circinelloides flanked by 1.8 kb and 0.76 kb of the dcl-1 gene, and its integration by homologous recombination provoked replacement of the dcl-1 sequence by the pyrG gene. However, all the pyrG− derivatives of MU410 spontaneously reverted to the PyrG+ phenotype with a frequency higher than that of homologous recombination. These strains contain two mutant copies of the pyrG gene, one at its endogenous locus and the other at the dcl-2 locus, which can generate a wild-type pyrG sequence by gene conversion or homologous recombination. Whatever the case, PCR analysis of 138 stable PyrG+ transformants obtained in different transformation experiments indicated no integration of the pyrG gene in the dcl-1 locus.
To check whether the difficulty of obtaining a double dcl-1− dcl-2− mutant was due to the lethality of this condition, we tried a different approach and searched for a double mutant using the dcl-2− mutant MU410 (dcl-2− leuA−) as the recipient strain and generating a knockout mutation at the dcl-1 locus by disruption with the leuA+ gene. This strategy provided a strain that did not retain any auxotrophic marker, which meant that it could not be used as a recipient of silencing vectors to investigate silencing efficiency. To generate such a strain, a 7.2-kb SacI fragment isolated from plasmid pMAT1228 was used to transform MU410. The disruption fragment contained the leuA gene of M. circinelloides flanked by 1.3 kb and 1.5 kb of the dcl-1 genomic region. Five LeuA+ transformants showing more than 35% leuA+ spores were PCR analyzed to detect homologous recombination at the dcl-1 locus. One of the homokaryotic transformants amplified the expected 1.9-kb fragment and was subjected to Southern analysis to confirm the correct disruption of the dcl-1 gene (see Fig. S1 in the supplemental material). The results indicated that the dcl-1 gene in this strain, named MU411, had been successfully disrupted and demonstrated that the double dcl-1− dcl-2− mutation is not lethal.
One uracil auxotroph derivative (pyrG−) obtained from the double mutant MU411 was used as the recipient in transformation experiments with the silencing vector pMAT1330, which contains the inverted-repeat carB transgene and the pyrG+ selective marker. The pyrG− strain showed a high rate of spontaneous reversion to the PyrG+ phenotype, as described above for the pyrG− derivatives of the dcl-2− mutant MU410. However, we were able to analyze the phenotypes of 137 pMAT1330-containing transformants, none of which showed a silencing phenotype, suggesting that the silencing mechanism was completely blocked in the double dcl-1− dcl-2− mutant.
Impaired asexual sporulation in dcl-2 mutants.
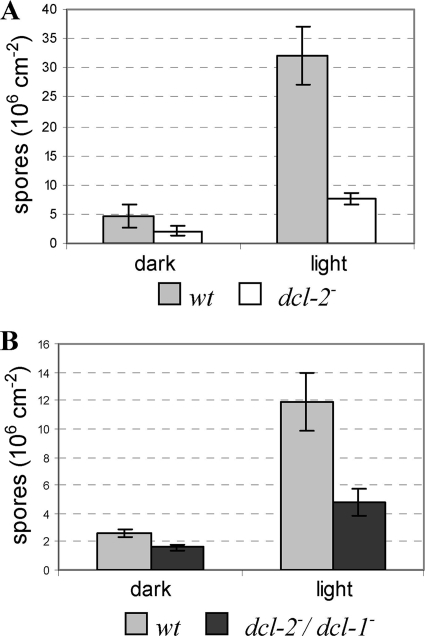
We investigated whether, apart from its resistance to gene silencing, the dcl-2− mutant showed any major phenotypic changes during vegetative growth with respect to the wild-type strain. The growth rate was similar to that of the wild type, as was its hyphal morphology. However, a reduction in spore production was consistently detected in the dcl-2− genetic background. To quantify the defective phenotype for sporulation of the dcl-2− mutant, spores from wild-type and MU410 mycelia (n = 10) grown on MMC plates under dark or light conditions were harvested and counted (Fig. 6A). Sporulation in the dcl-2− mutant was stimulated by light, as occurs in the wild-type strain (20). However, the results pointed to a significant difference in spore production between the two strains, since there was less than 0.001 probability (Student t test) that the average spore production of the dcl-2− mutant and that of the wild-type strain would be identical. Similar results were obtained when the double dcl-1− dcl-2− mutant was analyzed (Fig. 6B) (t < 0.001), suggesting a role for the dcl-2 gene in the control, direct or indirect, of asexual development in M. circinelloides.
FIG. 6.
Effect of the dcl-2− mutation on asexual-spore production. (A) Production of asexual spores in mycelia of the wild-type strain R7B (leuA−) and the dcl-2− mutant MU410 (leuA−) grown under white light and dark conditions for 3 days. (B) Production of asexual spores in mycelia of the prototrophic wild-type strain MU241 and the dcl-1− dcl-2− double mutant MU411 (leuA+) grown under the same conditions as for panel A. The values are means and standard errors of 10 independent measurements.
DISCUSSION
The canonical RNA-silencing mechanism is triggered by dsRNA molecules and results in the production, mediated by the Dicer proteins, of siRNAs of defined sizes, which act as effectors for the suppression of gene expression. We demonstrated here that the gene-silencing phenomenon operating in M. circinelloides follows a canonical mechanism with some peculiarities, since it is efficiently activated by hpRNA, as occurs in other organisms, but is associated with the production of two size classes of antisense siRNAs, most probably generated by a single Dicer activity.
We had previously seen that the dcl-1 gene is not required for the biogenesis of the siRNAs associated with sense transgene-induced gene silencing in M. circinelloides (19). Here, we showed that it is also not necessary for the silencing triggered by hpRNA-expressing plasmids, which implies the existence of at least one additional dicer gene. The demonstration that the silencing triggered by hpRNA or aberrant transgenic RNA is severely impaired in dcl-2− mutants provides direct evidence of the pivotal role of Dcl-2 in the RNA-silencing pathway. Our findings contrast with results in N. crassa, where single dcl-1 or dcl-2 mutants are perfectly able to silence gene expression (3), pointing to the redundant functions of these proteins in vivo, even though the N. crassa Dicer enzymes preferentially act in different silencing pathways (1, 30). The phenotype of the M. circinelloides dcl-2− strain resembles that of the D. melanogaster Dicer-2 mutants, which exhibit a large reduction in hpRNA-derived siRNA accumulation in vivo, but not a complete loss (12). Similarly, residual Dicer activity could be detected in the M. circinelloides dcl-2− mutant, although this activity is not sufficient to maintain the silenced phenotype. The residual activity is most probably due to the Dicer-1 protein, since RNA silencing is completely abolished in the double dcl-1− dcl-2− mutant, and the recent release of the M. circinelloides genome sequence by JGI (http://genome.jgi-psf.org/Mucci1/Mucci1.home.html) reveals the absence of additional putative dicer genes.
Most of the transformants harboring hpRNA-expressing plasmids in a dcl-2− genetic background do not show a silenced phenotype and do not accumulate any of the two different sizes of antisense siRNAs, suggesting that both siRNAs are generated by the Dcl-2 activity. This is also the case with the ascomycete M. oryzae, in which only one of the two dicer genes is involved in the production of different sizes of siRNAs (10). Only a small number of M. circinelloides dcl-2− transformants exhibited a partially silenced phenotype, probably due to residual Dicer activity of the Dicer-1 protein in the dcl-2− mutant, and this activity is also able to produce small amounts of the two size classes of antisense siRNAs. Therefore, the situation in a number of fungi seems to be divergent from that described in plants, where distinct Dicer enzymes are responsible for the generation of different-size siRNAs that act in different silencing pathways (38).
The unique spatial arrangement of the binding PAZ domain and the RNase III domains within Dicer suggests that it acts as a molecular ruler, generating siRNA products of defined lengths by anchoring the 3′ end of the substrate dsRNA in the PAZ domain and cleaving it at a fixed distance from that end (13). Thus, differences in siRNA sizes may arise from intrinsic structural characteristics of the Dicer enzymes, although interaction of these enzymes with specific dsRNA-binding proteins may provoke slight differences in the sizes of the siRNA molecules (2). The results discussed above suggest that a single Dicer enzyme is able to produce, directly or indirectly, two different sizes of antisense siRNAs. We have tried to characterize the dicing activity of recombinant Dcl-2 in vitro. However, the Dcl-2 protein expressed in insect cells failed to exhibit significant dicing activity toward dsRNA in vitro (our unpublished results), suggesting that the functional enzyme requires interaction with other proteins. Dcl-2 could form a complex with a dsRNA-binding protein that it is required for the RISC loading of small RNA in plants and animals (24), and this interaction may result in the production of different sizes of siRNAs. However, taking into account that only 25-nt sense siRNAs are accumulated in silenced strains (18), we cannot rule out the possibility that the intrinsic dicing activity of Dcl-2 generates only the 25-nt duplex siRNA, with the antisense 21-nt class being produced by cutting 4 bp off the 25-nt antisense siRNA within the RISC. This unusual siRNA processing may require a very specific Dicer protein and, possibly, additional components, perhaps an exonuclease, such as the N. crassa QIP, which converts the duplex siRNA into single strands within the RISC by removing the passenger strand (14). No specific roles have been assigned to the M. circinelloides 21-nt and 25-nt antisense siRNAs, although overaccumulation of the 21-nt class has been associated with an increase in silencing efficiency and stability (21). This suggests a role for this siRNA class as effectors of silencing, taking part in the degradation of the target mRNA and thus contributing to the efficient induction and maintenance of silencing in M. circinelloides.
Our attempts to analyze the silencing efficiency of a double dcl-1− dcl-2− mutant were hampered by the high frequency of spontaneous PyrG+ reversion in strains carrying two mutated copies of the pyrG gene (pyrG− derivatives of the strains MU410 and MU411). Seven different pyrG− mutants obtained from MU410 in independent mutagenesis experiments showed the same behavior, with the frequency of spontaneous reversion ranging between 1.28 × 10−7 and 4.68 × 10−7 spores. This indicates that the observed reversion is not related to a particular pyrG− mutation. The high frequency of reversion is probably a consequence of gene conversion events occurring between the duplicated copies of the pyrG sequence. A high frequency of gene conversion has been described in the zygomycete R. oryzae, a fungus closely related to M. circinelloides, in which the integration of linearized plasmids is typically associated with gene conversion or the replacement of the selectable marker (15, 33). Gene conversion among homologous unlinked sequences is more than 10 times more frequent than homologous recombination in R. oryzae (32), which supports the idea that repairing one of the nonfunctional pyrG sequences in the pyrG− derivatives of strains MU410 and MU411 could account for the high frequency of spontaneous reversion in those strains.
Dicer and other enzymes with critical roles in the silencing mechanism have been described as being upregulated by long dsRNA in N. crassa as part of a dsRNA response (4). We investigated the expression pattern of the dcl-2 gene and found that its expression is very low at the beginning of the vegetative cycle and increases considerably during the exponential phase. The level of expression did not change in the silenced strains with respect to the nonsilenced strains, which indicates that induction of the silencing mechanism by hpRNA-producing transgenes is not necessary to activate dcl-2 expression. This could be explained if the dcl-2 gene has a constitutive expression pattern as a consequence of its role in the control of endogenous functions. In fact, we demonstrated here that dcl-2− mutants show a significant reduction in the production of asexual spores, a complex developmental process in which many different genes are involved (20, 22). Other fungal dicer mutants, such as mdl-1 and mdl-2 of M. oryzae and dcl-1 of M. circinelloides, have been reported to be affected in their spore morphology or colony growth (16). In addition, the dcl-2 gene is strictly required for the production of a high number of endogenous small RNAs in M. circinelloides (J. P. de Haro, F. E. Nicolás, S. Moxon, S. Torres-Martínez, T. Dalmay, and R. M. Ruiz-Vázquez, unpublished data). Several types of endogenously produced small RNAs have been identified in plants and animals (5), where they have important roles in gene regulation, transposon silencing, and germ cell maintenance. However, relatively little is known about the physiological roles of the fungal RNA-silencing pathways (16). A new type of endogenous siRNA induced by DNA damage has recently been identified in N. crassa (11). An in-depth analysis of the dcl-2-dependent endogenous small RNAs normally accumulated in M. circinelloides will open the way to clearly establishing the roles of the dcl-2 gene and the RNA-silencing mechanism in the control of endogenous functions in this fungus.
Supplementary Material
Acknowledgments
We thank V. Garre for his critical reading of the manuscript and helpful discussion and J. A. Madrid for technical assistance.
This work was supported by the Spanish Dirección General de Investigación (BFU2006-02408). J.P.D.H. and S.C. were funded by graduate fellowships from the Fundación Séneca (Murcia, Spain) and the Spanish Ministerio de Educación, respectively.
Footnotes
Published ahead of print on 7 August 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alexander, W. G., N. B. Raju, H. Xiao, T. M. Hammond, T. D. Perdue, R. L. Metzenberg, P. J. Pukkila, and P. K. T. Shiu. 2008. DCL-1 colocalizes with other components of the MSUD machinery and is required for silencing. Fungal Genet. Biol. 45:719-727. [DOI] [PubMed] [Google Scholar]
- 2.Brodersen, P., and O. Voinnet. 2006. The diversity of RNA silencing pathways in plants. Trends Genet. 22:268-280. [DOI] [PubMed] [Google Scholar]
- 3.Catalanotto, C., M. Pallota, P. ReFalo, M. S. Sach, L. Vaysie, G. Macino, and C. Cogoni. 2004. Redundancy of the two dicer genes in transgene induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell. Biol. 24:2536-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choudhary, S., H. C. Lee, M. Maiti, Q. He, P. Cheng, Q. Liu, and Y. Liu. 2007. A double-stranded-RNA response program important for RNA interference efficiency. Mol. Cell. Biol. 27:3995-4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghildiyal, M., and P. D. Zamore. 2009. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10:94-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldoni, M., G. Azzalin, G. Macino, and C. Cogoni. 2004. Efficient gene silencing by expression of double stranded RNA in Neurospora crassa. Fungal Genet. Biol. 41:1016-1024. [DOI] [PubMed] [Google Scholar]
- 7.Gurr, S. J., S. E. Unkles, and J. R. Kinghorn. 1987. The structure and organization of nuclear genes of filamentous fungi, p. 93-139. In J. R. Kinghorn (ed.), Gene structure in eukaryotic microbes. IRL Press, Oxford, United Kingdom.
- 8.Henderson, I. R., X. Zhang, C. Lu, L. Johnson, B. C. Meyers, P. J. Green, and S. E. Jacobsen. 2006. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38:721-725. [DOI] [PubMed] [Google Scholar]
- 9.Jaskiewicz, L., and W. Filipowicz. 2008. Role of Dicer in posttranscriptional RNA silencing. Curr. Top. Microbiol. Immunol. 320:77-97. [DOI] [PubMed] [Google Scholar]
- 10.Kadotani, N., H. Nakayashiki, Y. Tosa, and S. Mayama. 2004. One of the two Dicer-like proteins in the filamentous fungi Magnaporthe oryzae genome is responsible for hairpin RNA-triggered RNA silencing and related small interfering RNA accumulation. J. Biol. Chem. 279:44467-44474. [DOI] [PubMed] [Google Scholar]
- 11.Lee, H. C., S. S. Chang, S. Choudhary, A. P. Aalto, M. Maiti, D. H. Bamford, and Y. Liu. 2009. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature 459:274-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, Y. S., K. Nakahara, J. W. Pham, K. Kim, Z. He, E. J. Ontheimer, and R. W. Carthew. 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117:69-81. [DOI] [PubMed] [Google Scholar]
- 13.MacRae, I. J., K. Zhou, F. Li, A. Repic, A. N. Brooks, W. Z. Cande, P. D. Adams, and J. A. Doudna. 2006. Structural basis for double-stranded RNA processing by Dicer. Science 311:195-198. [DOI] [PubMed] [Google Scholar]
- 14.Maiti, M., H. C. Lee, and Y. Liu. 2007. QIP, a putative exonuclease, interacts with the Neurospora Argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev. 21:590-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michielse, C. B., K. Salim, P. Ragas, A. F. Ram, B. Kudla, B. Jarry, P. J. Punt, and C. A. van den Hondel. 2004. Development of a system for integrative and stable transformation of the zygomycete Rhizopus oryzae by Agrobacterium-mediated DNA transfer. Mol. Genet. Genomics 271:499-510. [DOI] [PubMed] [Google Scholar]
- 16.Nakayashiki, H., and Q. B. Nguyen. 2008. RNA interference: roles in fungal biology. Curr. Opin. Microbiol. 11:494-502. [DOI] [PubMed] [Google Scholar]
- 17.Nakayashiki, H., N. Kadotani, and S. Mayama. 2006. Evolution and diversification of RNA silencing proteins in fungi. J. Mol. Evol. 63:127-135. [DOI] [PubMed] [Google Scholar]
- 18.Nicolás, F. E., S. Torres-Martínez, and R. M. Ruiz-Vázquez. 2003. Two classes of small antisense RNAs in fungal RNA silencing triggered by non-integrative transgenes. EMBO J. 22:3983-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicolás, F. E., J. P. de Haro, S. Torres-Martínez, and R. M. Ruiz-Vázquez. 2007. Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet. Biol. 44:504-516. [DOI] [PubMed] [Google Scholar]
- 20.Nicolás, F. E., S. Calo, L. Murcia-Flores, V. Garre, R. M. Ruiz-Vázquez, and S. Torres Martínez. 2008. A RING-finger photocarotenogenic repressor involved in asexual sporulation in Mucor circinelloides. FEMS Microbiol. Lett. 280:81-88. [DOI] [PubMed] [Google Scholar]
- 21.Nicolás, F. E., S. Torres-Martínez, and R. M. Ruiz-Vázquez. 2009. Transcriptional activation increases RNA silencing efficiency and stability in the fungus Mucor circinelloides. J. Biotechnol. 142:123-126. [DOI] [PubMed] [Google Scholar]
- 22.Ocampo, J., L. Fernandez Nuñez, F. Silva, E. Pereyra, S. Moreno, V. Garre, and S. Rossi. 1 May 2009. A subunit of protein kinase A regulates growth and differentiation in the fungus Mucor circinelloides. Eukaryot. Cell. doi: 10.1128/EC.00026-09. [DOI] [PMC free article] [PubMed]
- 23.Punt, P. J., M. A. Dingemanse, A. Kuyvenhoven, R. D. Soede, P. H. Pouwels, and C. A. van der Hondel. 1990. Functional elements in the promoter region of the Aspergillus nidulans gpdA gene encoding glyceraldehyde-3-phosphate dehydrogenase. Gene 93:101-109. [DOI] [PubMed] [Google Scholar]
- 24.Qi, Y., and G. J. Hannon. 2005. Uncovering RNAi mechanisms in plants: biochemistry enters the foray. FEBS Lett. 579:5899-5903. [DOI] [PubMed] [Google Scholar]
- 25.Quiles-Rosillo, M. D., S. Torres-Martínez, and V. Garre. 2003. cigA, a light-inducible gene involved in vegetative growth in Mucor circinelloides is regulated by the carotenogenic repressor crgA. Fungal Genet. Biol. 38:122-132. [DOI] [PubMed] [Google Scholar]
- 26.Roncero, M. I. G. 1984. Enrichment method for the isolation of auxotrophic mutants of Mucor using the polyene antibiotic N-glycosyl-polyfungin. Carlsberg Res. Commun. 49:685-690. [Google Scholar]
- 27.Ruiz-Pérez, V. L., F. J. Murillo, and S. Torres-Martínez. 1995. PkpA, a novel Phycomyces blakesleeanus serine/threonine protein kinase. Curr. Genet. 28:309-316. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Segers, G. C., X. Zhang, F. Deng, Q. Sun, and D. L. Nuss. 2007. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. USA 104:12902-12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shiu, P. K. T., N. B. Raju, D. Zickler, and R. L. Metzenberg. 2001. Meiotic silencing by unpaired DNA. Cell 107:905-916. [DOI] [PubMed] [Google Scholar]
- 31.Siomi, H., and M. C. Siomi. 2009. On the road to reading the RNA-interference code. Nature 457:396-404. [DOI] [PubMed] [Google Scholar]
- 32.Skory, C. D. 2004. Repair of plasmid DNA used for transformation of Rhizopus oryzae by gene conversion. Curr. Genet. 45:302-310. [DOI] [PubMed] [Google Scholar]
- 33.Skory, C. D., and A. S. Ibrahim. 2007. Native and modified lactate dehydrogenase expression in a fumaric acid producing isolate Rhizopus oryzae 99-880. Curr. Genet. 52:23-33. [DOI] [PubMed] [Google Scholar]
- 34.Smith, N. A., S. P. Singh, M. B. Wang, P. A. Stoutjesdijk, A. G. Green, and P. M. Waterhouse. 2000. Total silencing by intron-spliced hairpin RNAs. Nature 407:319-320. [DOI] [PubMed] [Google Scholar]
- 35.Tomari, Y., and P. D. Zamore. 2005. Perspective: machines for RNAi. Genes Dev. 19:517-529. [DOI] [PubMed] [Google Scholar]
- 36.Velayos, A., J. L. Blasco, M. I. Alvarez, E. A. Iturriaga, and A. P. Eslava. 2000. Blue-light regulation of phytoene dehydrogenase (carB) gene expression in Mucor circinelloides. Planta 210:938-946. [DOI] [PubMed] [Google Scholar]
- 37.Wolff, A. M., and J. Arnau. 2002. Cloning of glyceraldehyde-3-phosphate dehydrogenase-encoding genes in Mucor circinelloides (syn. racemosus) and use of the gpd1 promoter for recombinant protein production. Fungal Genet. Biol. 35:21-29. [DOI] [PubMed] [Google Scholar]
- 38.Xie, Z., L. K. Johansen, A. M. Gustafson, K. D. Kasschau, A. D. Lellis, D. Zilberman, S. E. Jacobsen, and J. C. Carrington. 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2:E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, X., G. C. Segers, Q. Sun, F. Deng, and D. L. Nuss. 2008. Characterization of hypovirus-derived small RNAs generated in the chestnut blight fungus by an inducible DCL-2-dependent pathway. Virology 82:2613-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.