Abstract
Protein O-mannosyltransferases (Pmts) initiate O-mannosyl glycan biosynthesis from Ser and Thr residues of target proteins. Fungal Pmts are divided into three subfamilies, Pmt1, -2, and -4. Aspergillus nidulans possesses a single representative of each Pmt subfamily, pmtA (subfamily 2), pmtB (subfamily 1), and pmtC (subfamily 4). In this work, we show that single Δpmt mutants are viable and have unique phenotypes and that the ΔpmtA ΔpmtB double mutant is the only viable double mutant. This makes A. nidulans the first fungus in which all members of individual Pmt subfamilies can be deleted without loss of viability. At elevated temperatures, all A. nidulans Δpmt mutants show cell wall-associated defects and increased sensitivity to cell wall-perturbing agents. The Δpmt mutants also show defects in developmental patterning. Germ tube emergence is early in ΔpmtA and more frequent in ΔpmtC mutants than in the wild type. In ΔpmtB mutants, intrahyphal hyphae develop. All Δpmt mutants show distinct conidiophore defects. The ΔpmtA strain has swollen vesicles and conidiogenous cells, the ΔpmtB strain has swollen conidiophore stalks, and the ΔpmtC strain has dramatically elongated conidiophore stalks. We also show that AN5660, an ortholog of Saccharomyces cerevisiae Wsc1p, is modified by PmtA and PmtC. The Δpmt phenotypes at elevated temperatures, increased sensitivity to cell wall-perturbing agents and restoration to wild-type growth with osmoticum suggest that A. nidulans Pmts modify proteins in the cell wall integrity pathway. The altered developmental patterns in Δpmt mutants suggest that A. nidulans Pmts modify proteins that serve as spatial cues.
Filamentous fungi use highly polar growth to explore their environments. Except for a brief period of isotropic expansion just after spores break dormancy, filamentous fungi add new cell wall material exclusively at the tips of tubular hyphal cells. Such polar growth involves a high degree of coordination between signals from the environment and the secretory apparatus. In fungi, O mannosylation of specific target proteins has been shown to be important for sensing environmental stress, stabilizing the cell wall, and proper development (18, 28). The assembly of protein linked O-mannosyl glycans in the endoplasmic reticulum lumen is catalyzed by protein O-mannosyltransferases (Pmts), which transfer a single mannosyl residue to the hydroxyl group of serine or threonine residues to form an α-d-mannosyl linkage (30). The addition of further carbohydrate residues to the first O-linked mannose occurs in the Golgi apparatus and involves a range of enzymes (35). Modification by Pmts seems to be specific to proteins that are synthesized and sorted in the secretory pathway; however, the only motif so far identified is that Ser/Ter-rich membrane-bound proteins are O mannosylated by Pmt4 in Saccharomyces cerevisiae (15). This lack of a clear motif makes identification of Pmt targets by computational methods challenging. All of the fungal Pmt-modified proteins identified so far are localized to the cell membrane or cell wall or are secreted. At least 23 target proteins have been described in yeasts (15). Only three Pmt target proteins have been described in filamentous fungi (12, 23, 37).
Pmts have been found in both prokaryotes and eukaryotes (33), but not in plants (8). The lengths and compositions of O-mannosyl glycans are different among species. In fungi, O-glycosyl chains range from 2 to 7 residues. In S. cerevisiae, the mannosyl chain can be modified by mannosyl phosphate (6). In Schizosaccharomyces pombe, the O-linked glycan is capped with 1 or 2 galactose residues (6). In the filamentous fungi so far examined, O-glycans are linear and branched, with 3 to 5 monosaccharide residues (4).
In fungi, the Pmts are classified into the Pmt1, Pmt2, and Pmt4 subfamilies, with each species having three to seven members. S. cerevisiae and Candida albicans Pmts are the most redundant, with subfamilies 1 and 2 containing two or three members (7, 26). S. pombe and many filamentous fungi, including Aspergillus nidulans, have one representative from each subfamily. In S. cerevisiae, the enzymatic activity of Pmts requires interaction among members of the Pmt1 and Pmt2 subfamilies, while Pmt4 forms homomeric complexes (8). Heteromeric complexes between Pmt1 and Pmt2 subfamily members have also been reported in S. pombe (34).
O mannosylation appears to be required for the stability, localization, and function of target proteins (18, 28, 32), and in vivo consequences of Pmt loss range from limited to lethal. In S. cerevisiae, O mannosylation is essential for cell integrity and cell wall rigidity (7). In C. albicans and Cryptococcus neoformans, Pmt mutation affects morphogenesis and virulence (24, 26, 27). In S. cerevisiae, strains with single Pmt subfamily representatives deleted are viable; however, deletion of subfamily 2 representatives is lethal in S. pombe and C. albicans (7, 34). In filamentous fungi, deletion of individual Pmts has been reported. Deletions of Trichoderma reesei pmtI, Aspergillus fumigatus pmt1, A. nidulans pmtA, and Aspergillus awamori pmtA were not lethal but affected growth and development (10, 22, 23, 37).
In previous work, we identified the swoA mutant from a collection of temperature-sensitive polarity mutants and showed that the swoA allele encoded a Pmt2 subfamily member (PmtA) (21, 29). In this study, we use Δpmt strains to show that each of the three Pmts in A. nidulans (pmtA, pmtB, and pmtC) is nonessential but that all play distinct roles in cell wall integrity and developmental patterning. We also demonstrate that PmtA and PmtC modify an ortholog of S. cerevisiae Wsc1, a known Pmt target. Because of redundancy, all Pmt1 and Pmt2 subfamily members have not been deleted in S. cerevisiae. Because of lethality, the effects of loss of the Pmt2 subfamily cannot be addressed in S. pombe or C. albicans. This makes A. nidulans the first fungus in which the phenotypes of deleted strains for each Pmt subfamily have been reported.
MATERIALS AND METHODS
Aspergillus strains and media.
The strains used in this study are listed in Table 1. Complete medium (CM) consisted of 1% glucose, 2% peptone, 1% yeast extract, 1% Casamino Acids, and 0.01% vitamins and supplements. Minimal medium contained 1% glucose and supplements. Nitrate salts solution, trace elements, vitamins, and amino acid supplements were based on the appendices to Kafer (16) and Hill and Kafer (14). The pHs of media were adjusted to 6.5 with 1.0 N NaOH. CM was used for phenotypic studies and mycelium production. Minimal medium was used for marker selection and maintaining routine stocks. For solid media, 1.8% agar was added, and 0.6 M KCl or 1 M sorbitol was used as an osmostabilizer.
TABLE 1.
A. nidulans strains and plasmids
| Strain or plasmid | Genotype/phenotype | Source or reference |
|---|---|---|
| Strains | ||
| A850 | argB2::trpC_B methG | FGSC |
| A773 | pyrG89 wA3 pyroA4 | FGSC |
| TNO2A7/A1145 | pyrG89 ΔnkuA::argB pyroA4 riboB2 | 21a |
| ATK08 | pyrG89 argB2::trpC_B pyroA4 ΔpmtA::AfpyrG | This study |
| ATK16 | pyrG89 ΔpmtB::AfpyrG argB2 pyroA4 | This study |
| ATK38 | pyrG89 wA3 argB2 pyroA4 ΔpmtC::AfpyrG | This study |
| ATK40 | pyrG89 wA3 argB2::trpC_B pyroA4 | This study |
| ATK45 | pyrG89 argB2::trpC_B nkuA::Afpyro pyroA4 | This study |
| ATK57 | pryG89 ΔpmtB::Afpyro nkuA::argB pyroA4 riboB2 | This study |
| ATK88 | pyrG89 wA3 argB2::trpC_B pyroA4 ΔpmtC::AfargB | This study |
| ATK94 | pyrG89 argB2::trpC_B ΔpmtB::AfpyrG pyroA4 ΔpmtA::Afpyro | This study |
| ATK104 | pryG89 wA3 ΔpmtB::Afpyro argB2 pyroA4 | This study |
| ATK147 | A850::AfargB-gpd(P)-AN5660-S-tag | This study |
| ATK148 | ATK16::AfargB-gpd(P)-AN5660-S-tag | This study |
| ATK149 | ATK08::AfargB-gpd(P)-AN5660-S-tag | This study |
| ATK160 | ATK38::AfargB-gpd(P)-AN5660-S-tag | This study |
| Plasmids | ||
| pDV2 | AmprargB-gpd(p)-ccdB-sgfp | 32a |
| pFNO3 | Kanr GA5-GFP AfpyrG | 36 |
| pHL85 | Ampr Kanr GA5-mCherry Afpyro | B. R. Oakley |
| pAO81 | GA4-S-Tag AfpyrG | 36 |
| pAfargB2 | AmprargB2 | G. S. May |
| pTK59 | pENTR/D-TOPO::AN5660-S-tag | This study |
| pTK64 | pDV2::AN5660-S-tag | This study |
Phylogenetic analysis.
Pmt sequences were obtained from the following databases: A. nidulans, http://www.broad.mit.edu/annotation/genome/aspergillus_group/MultiHome.html; C. albicans, http://www.candidagenome.org/; S. cerevisiae, http://www.yeastgenome.org/; S. pombe, http://www.genedb.org/genedb/pombe/index.jsp; Homo sapiens and Mycobacterium tuberculosis; http://www.ncbi.nlm.nih.gov/. The gene and accession numbers of the protein sequences are as follows: ScPmt1 (S. cerevisiae Pmt1), YDL095W; ScPmt2, YAL023C; ScPmt3, YOR321W; ScPmt4, YJR143C; ScPmt5, YDL093W; ScPmt6, YGR199W; AnPmtA (A. nidulans PmtA), AN5105; AnPmtB, AN4761; AnPmtC, AN1459; SpOgm1 (S. pombe Ogm1)/Oma1, SPAC22A12.07c; SpOgm2/Oma2, SPAPB1E7.09; SpOgm4/Oma4, SPBC16C6.09; HsPomt1 (H. sapiens Pomt1) isoform a, gi116517319; HsPomt2, gi32455271; MtPmt (M. tuberculosis Pmt), gi15608142; CaPmt1 (C. albicans Pmt1), orf19.5171; CaPmt2, orf19.6812; CaPmt4, orf19.4109; CaPmt5, orf19.7549; CaPmt6, orf19.3802. ClustalX2 was used for protein alignments, building neighbor-joining trees, and bootstrap analysis (17). Trees were viewed using TreeView 1.6.6 (25).
pmt gene replacements.
The pmt gene replacement cassettes were constructed by fusion PCR based on the method of Yang and colleagues (36). Approximately 2 kb upstream and downstream of pmt sequences was amplified using genomic DNA from strain A850 as the template (Table 1). The A. fumigatus marker genes were amplified from plasmids listed in Table 1. All primers used in this study are listed in Table 2. Each individual fragment was synthesized with the AccuPrime Pfx DNA polymerase in a total volume of 50 μl with adjusted annealing temperatures (Invitrogen Co., Carlsbad, CA). The amplicons were separated on an agarose gel, purified using the QIAquick gel extraction kit (Qiagen Inc., Valencia, CA), and transformed into TNO2A7, a ΔnkuA::argB strain. Transformants were tested for homologous integration by PCR, using a forward primer located upstream of the cassette and a reverse primer located inside the auxotrophic marker (Table 2), and Southern hybridization using standard protocols (2). Each Δpmt ΔnkuA strain was crossed with appropriately marked nkuA+ strains, and Δpmt nkuA+ progeny were selected. To facilitate double-mutant construction, we created an nkuA::Afpyro (A. fumigatus pyro) strain. Approximately 2 kb upstream and downstream of nkuA (AN7753), sequence was amplified using genomic DNA from strain A850 as the template. The pyro gene of A. fumigatus was amplified from plasmid pBS::Afpyro (Table 2). Fusion PCR and transformation were performed as described above. The replacement cassette was transformed into TNO2A7 (pyrG89 ΔnkuA::argB pyroA4 riboB2). Transformants were tested for homologous integration by PCR and Southern hybridization. A ΔnkuA::Afpyro transformant was crossed to ATK40 to obtain the argB marker, yielding strain ATK45. Random ascospore analysis was based on the method of Harris (11).
TABLE 2.
Primers
| Primer name | Sequence |
|---|---|
| pmtBupF | TAGGCGTTGTAGTATGTTGGAGTTTCCATG |
| pmtBupR | CAGGACTCCCAAGGAGAAACGAATCAATCTG |
| pmtB-AfpyrGF | GATTCGTTTCTCCTTGGGAGTCCTGGCCTCAAACAATGCTCTTCACCCTC |
| pmtB-AfpyrGR | GTTCAACAGACCGACATATTATCCTGTCTGAGAGGAGGCACTGATGCG |
| pmtBdnF | GATAATATGTCGGTCTGTTGAACTACCTGCC |
| pmtBdnR | ACTTTCACTTCTCACTGTCGTCGTATTCGC |
| CheckpmtBF | GAATATACCGGACCAACAGGTTGAGCG |
| CheckAfpyrGR | CAGAGCCCACAGAGCGCCTTGAG |
| PmtB-AfpyroF | GATTCGTTTCTCCTTGGGAGTCCTGGACATCAGATGCTGGATTACTAAG |
| PmtB-AfpyroR | GTTCAACAGACCGACATATTATCCACAATCAGCTTTTCAGAATTCGCG |
| PmtBdnF | GATAATATGTCGGTCTGTTGAACTACCTGCC |
| PmtBdnR | GCGAATACGACGACAGTGAGAAGTGAAAGT |
| CheckAfpyroR | CAGCGCTTGTGCCCCTCCATCTCCC |
| pmtAUFF | GGTACAACAGACGTGTCCTTATTGCAATGTC |
| pmtAUPR | GATGAGAGATAATAGGAGAAGTGGCCGGTC |
| pmtAdnF | GAACCGTGATAGAGCGTTACAGTTCCCGTTG |
| pmtAdnR | GCGATCCCAATCCTTCCTATCTCTGTCATC |
| pmtA-AfpyrGF | CCACTTCTCCTATTATCTCTCATCGCCTCAAACAATGCTCTTCACCCTC |
| pmtA-AfpyrGR | AACTGTAACGCTCTATCACGGTTCCTGTCTGAGAGGAGGCACTGATGCG |
| CheckpmtAF | TTACACGTCAAGAAGGGGAACGATCTTACG |
| checkpmtAR | GGCTGATATGGGTCGACCGATTTTCTATTC |
| PmtCupF | GCGCACCTCATATTAGGATAGGATGTGATC |
| PmtCupR2 | AATCACCAGGCCAAAACAACAGAGATG |
| PmtCdnF | GAGCATCTGATCTCATCTCGTTCTCTCCC |
| PmtCdnR | CTATCCACGGTATGAGCTGAGCGAGTAATG |
| pmtC-AfargBF | GTTGTTTTGGCCTGGTGATTAATGGCTGGGGGAGTGGGG |
| pmtC-AfargBR | AGAACGAGATGAGATCAGATGCTCCGATTTTCATAGGATTTTCCCCTTG |
| UpnKuF | CCATCCCACGAGTCCGAGAACTGATCATG |
| UPnKuR | GGCGTCTTGAATACAACTGGGGGTTCGATC |
| nKu-AfpyroF | AACCCCCAGTTGTATTCAAGACGCCGACATCAGATGCTGGATTACTAAG |
| nKu-AfpyroR | CCTAGATAACCGACAACAGAGTCACCACAATCAGCTTTTCAGAATTCGC |
| DNnKuF | GTGACTCTGTTGTCGGTTATCTAGGACTCG |
| DNnKuR | GTCTCTCGTTCCAGTCATCTACAGCGGTTC |
| CheckKuAF | GCAGTTGCCGCGCGGTTGGTGTGTC |
| AN5660F | CACCATGAGGTCGTTCACGCTATCCACAGTCTTC |
| AN5660StagR | GCGCCTGCACCAGCTCCGTGCCGGTCAGGATTCG |
| StagF | GGAGCTGGTGCAGGCGCTGGAGC |
| StagR | GCGCCAATTGCTGTTGCCAGGTGAGG |
Chemical sensitivity tests.
Strain A850 was used as a control for all experiments. Sensitivity to chemicals was tested by spotting 5 μl of a 10-fold serial dilution of conidia (106 to 102) on CM agar plates. The plates were incubated at 25°C, 30°C, 37°C, or 42°C. Stock solutions of chemicals were prepared as follows: Calcofluor, 1% in 25 mM KOH, and 10 mg/ml Congo red. All chemicals were filtered, sterilized, and added to autoclaved medium to give the following final concentrations: Calcofluor, 10 μg/ml; Congo red, 25 μg/ml.
Microscopic examination.
To examine germlings, 1 × 105 spores were grown in 10 ml of liquid CM in a petri dish containing a glass coverslip and incubated at 30°C or 42°C for 5 to 12 h. To examine conidiophores, 100 conidia in 5 μl of water were dropped on each side of a CM agar block, placed between two coverslips, and incubated for 3 days at different temperatures. To stain cell walls and nuclei, coverslips with adhering cultures were incubated in fixer (3.7% formaldehyde, 50 mM phosphate buffer [pH 7.0], and 0.2% Triton X-100) for 30 min. The coverslips were then briefly dipped in deionized water, incubated for 5 min in a staining solution (10 mg/ml Calcofluor and 100 ng/ml Hoechst), briefly dipped in deionized water, and mounted on a microscope slide with mounting solution (50% glycerol, 10% phosphate buffer, pH 7.0, 0.1% n-propyl gallate) (19). The slides were viewed with a Zeiss Axioplan epifluorescence microscope and imaged with Zeiss AxioCam Mrc software. Photoshop CS was used for micrograph organization and optimization.
Conidiation efficiency.
Freshly prepared conidia (104 spores) were spread on CM or CM with osmoticum plates and incubated at 30°C or 42°C for 4 days. Four milliliters of sterile water was added to the plate to harvest spores by using a glass rod, and the conidial concentration in the resulting solution was determined with a hemocytometer. Graphs and standard errors were generated using Microsoft Excel. Data were from three biological replicates.
Construction and detection of S-tagged An5660/WscA.
A strain carrying the Wsc ortholog An5660 with a C-terminal fusion to the S tag (a peptide epitope of 15 amino acids used for protein purification) was constructed in several steps. First, the GA4 S-tag fragment with a stop codon was amplified from pAO81 (Table 2) and the An5660 gene was amplified from the start codon to one codon before the stop codon. The two amplicons were then fused together by fusion PCR and ligated into the pENTR/D-TOPO vector using the pENTR/D-TOPO Cloning Kit (Invitrogen Co., Carlsbad, CA), yielding plasmid pTK59. The plasmid was transferred into the pMT-DV2 destination vector (Table 2) using the Gateway LR Clonase in vitro recombination kit (Invitrogen Corp., Carlsbad, CA). The resulting plasmid, pTK64, was transformed into the A850 and Δpmt strains (Table 1).
Western blot analysis.
For immunodetection, 1 ×107 conidia/ml were grown in 50 ml of CM and incubated on a rotary shaker at 220 rpm and 30°C for 8 h. Mycelia were filtered through cheese cloth, washed with cold stop buffer (0.9% NaCl, 1 mM NaN3, 10 mM EDTA, 50 mM NaF, pH 7.0), and ground in liquid nitrogen. Two milliliters of cold HK buffer (3) was added to 0.1 g of ground mycelia, vortexed for 1 min, and kept on ice for 1 min (four times). The cell suspension was then centrifuged at 500 × g for 5 min at 4°C. The supernatant was collected and centrifuged at 10,000 × g for 30 min at 4°C. The pellet containing crude membranes was resuspended in a small volume of HK buffer containing 15% glycerol. The protein contents of samples were quantified with an RC DC Protein Assay Kit (Bio-Rad Laboratories, Hercules, CA) using bovine serum albumin as a standard. An equal volume of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (2×) was added to the crude membrane fractions and heated for 5 min at 95°C. The solubilized proteins (10 μg/lane) were resolved by modified Laemmli peptide SDS-PAGE (16% gel) (2) and transferred to a nylon membrane. AN5660 S tag was detected by Rabbit anti-S-tag antibody (1:50,000), followed by anti-rabbit immunoglobulin G coupled to horseradish peroxidase (1:5,000) (Immunology Consultants Laboratory, Inc., Newberg, OR) and staining using an Amersham ECL Western blotting detection reagent and analysis system (GE Healthcare, United Kingdom).
RESULTS
A. nidulans possesses three putative Pmts.
In previous work, we identified the swoA mutant in a screen for temperature-sensitive polarity mutants and showed that the swoA gene encodes a protein with high homology to Pmt2, a Pmt in S. cerevisiae. swoA was renamed pmtA (21, 29). Fungi contain three to seven Pmts falling into three subfamilies named Pmt1, Pmt2, and Pmt4 based on the S. cerevisiae members (9).
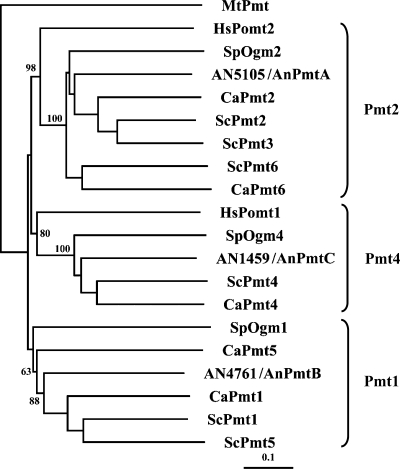
Using the S. cerevisiae Pmt2 protein sequence to query the A. nidulans genome database at the Broad Institute (http://www.broad.mit.edu), we found that A. nidulans possesses three hypothetical Pmts, AN4761, AN5105, and AN1459. A BLASTP search of the Saccharomyces Genome Database (http://www.yeastgenome.org) and phylogenetic analysis (Fig. 1) showed that AN4761 is 60% similar to ScPmt1 and is a member of subfamily 1, AN5105 is 67% similar to Pmt2 and is a member of subfamily 2, and AN1459 is 68% similar to ScPmt4 and is a member of subfamily 4. We refer to AN5105, AN4761, and AN1459 as pmtA, pmtB, and pmtC, respectively.
FIG. 1.
Phylogenetic tree of Pmts from A. nidulans (AN5105, AN4761, and AN1459), S. cerevisiae (Pmt1, Pmt 2, Pmt 3, Pmt 4, Pmt 5, and Pmt6), C. albicans (Pmt1, Pmt2, Pmt4, Pmt5, and Pmt6), S. pombe (Oma1, Oma2, and Oma4), H. sapiens (Pomt1 and Pomt2), and M. tuberculosis (Pmt). ClustalX2 was used for multiple-sequence alignments. The tree was created from a Bootstrap neighbor-joining tree. The numbers (percentages) are bootstrap values based on 1,000 trails. MtPmt was used as an outgroup. A branch length standard is indicated. The tree was drawn using the Tree View program.
Single pmt deletion mutants are viable, but not all double or triple pmt deletion mutants are viable.
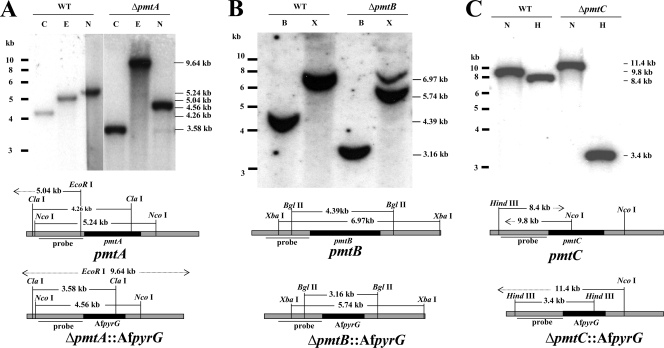
To understand the role of each Pmt, we constructed Δpmt strains by replacing the entire open reading frame with the pyrG auxotrophic marker from A. fumigatus. Strains carrying single correct gene replacements were determined by PCR (data not shown) and Southern blotting (Fig. 2). All three single Δpmt mutants were viable and showed a variety of phenotypes described in detail below. To confirm that the phenotypes were solely caused by deletion of pmt, each deletion mutant was complemented by expressing the corresponding pmt gene behind the constitutive gpd promoter. All transformed strains showed complemented phenotypes. Complementation ranged from partial to full restoration of the wild-type phenotype (data not shown), likely because of variation in the copy numbers and positions of integration of gpd-pmt cassettes.
FIG. 2.
Southern hybridization of pmt deletion strains. Genomic DNAs of the wild type (A850) (WT) and strains transformed with deletion cassettes for pmtA (ATK08) (A), pmtB (ATK16) (B), and pmtC (ATK38) (C) were digested with the restriction enzymes indicated and probed with a radiolabeled upstream fragment of the corresponding pmt. B, BglII; C, ClaI; E, EcoR I; H, HindIII; N, NcoI; X, XbaI. The approximately 7-kb band in ΔpmtB cut with XbaI is from the left flank of the probe. This band is obscured by the 6.97-kB band in the wild type.
Gentzsch and Tanner reported that S. cerevisiae pmt2 pmt3, pmt2 pmt4, and pmt1 pmt2 pmt3 mutants at elevated temperatures survived only in the presence of an osmoticum (7). To test whether A. nidulans strains with deletions in multiple pmt genes were viable, we crossed the single Δpmt mutants with each other and germinated progeny on medium supplemented with 1 M sorbitol. Crosses among single Δpmt mutants showed that the only double mutant that is viable is the ΔpmtA ΔpmtB mutant. To attempt to generate the triple-knockout mutant, the ΔpmtA ΔpmtB strain was crossed with the ΔpmtC strain. None of the 450 progeny from two crosses was identified as the triple mutant. Further, no ΔpmtB ΔpmtC or ΔpmtA ΔpmtC progeny were identified.
All Δpmt strains show reduced growth at high temperatures.
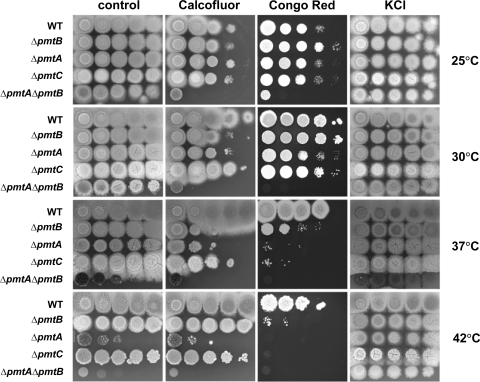
The phenotypes of the Δpmt mutants were examined by inoculating a series of 10-fold dilutions of conidia on solid CM and incubating them for 3 days at 25 to 42°C. Colonial growth of all Δpmt single mutants was comparable to that of the wild type at 25°C and 30°C; however, growth was reduced at 37°C and 42°C (Fig. 3). Growth of the ΔpmtA ΔpmtB double mutant was slightly retarded even at 25°C (Fig. 3). The ΔpmtA ΔpmtB double mutant was most sensitive to high temperatures, followed by the ΔpmtA, ΔpmtC, and ΔpmtB mutants. Adding osmoticum partially restored growth in all cases (Fig. 3).
FIG. 3.
Cell wall-perturbing agents alter the growth of Δpmt mutants. A 10-fold serial dilution of conidia (106 to 102) was spotted onto CM and CM containing 10 μg/ml Calcoflour, 25 μg/ml Congo red, or 0.6 M KCl as an osmotic stabilizer. The plates were incubated for 3 days at 25°C, 30°C, 37°C, or 42°C. WT, wild type.
Growth of Δpmt strains is altered by cell wall-perturbing agents.
To test the sensitivity of Δpmt mutants to cell wall-perturbing agents, serial dilutions of conidia were spotted on CM containing the cell wall-perturbing agent Calcofluor or Congo red (Fig. 3). The ΔpmtA and ΔpmtA ΔpmtB mutants were more sensitive than the wild type to these agents at all temperatures tested. The ΔpmtC strain was resistant to Calcofluor at all temperatures tested but was sensitive to Congo red at elevated temperatures. Surprisingly, the effects of cell wall-perturbing agents on the ΔpmtB mutant varied with temperature. At 25°C, the ΔpmtB mutant grew as well as the wild type. At 30°C and 37°C, the ΔpmtB mutant was hypersensitive to Calcofluor; however, at 42°C, the ΔpmtB mutant was resistant to Calcofluor. On the other hand, the ΔpmtB mutant was hypersensitive to Congo red at 37°C and 42°C and resistant to the chemical at 25°C and 30°C.
The Δpmt mutants have defects in early growth.
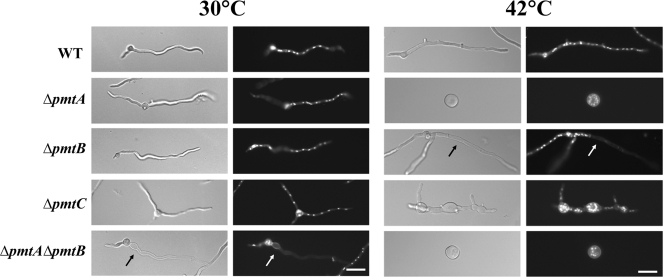
When an A. nidulans conidium breaks dormancy, it first expands isotropically. After approximately 6 h under standard conditions, the cell switches to grow asymmetrically, extending a tubular cell (germ tube) and continuously adding new material only at the tip. After approximately 8 h under standard conditions, cross walls (septa) are formed and partition the hypha into compartments. From these compartments, new tubular cells arise, forming branches (20). To determine the roles of Pmts in the growth and development of A. nidulans, we examined Δpmt mutants from conidial germination through early growth (Fig. 4). After 6 h of incubation at 30°C, 12% of the ΔpmtA and 15.5% of the ΔpmtA ΔpmtB mutants sent out germ tubes compared to 5% of the wild-type, ΔpmtB, and ΔpmtC strains (data not shown). After 12 h of incubation at 30°C, all germlings appeared normal, except that the ΔpmtC mutant sent out multiple germ tubes. Approximately 45% of the ΔpmtC mutant germlings had three or four germ tubes, while only 1.5% of the wild type and other deletion mutants had multiple germ tubes (n = 300).
FIG. 4.
Phenotypes of Δpmt mutants. Conidia of Δpmt mutants were inoculated onto CM, incubated for 12 h at 30°C or 42°C, fixed, and stained with Hoechst and Calcofluor white to label the nuclei and cell walls, respectively. (Left) Differential interference contrast images. (Right) Fluorescence images. The arrow indicates an empty apical compartment. Bar, 10 μm. WT, wild type.
All of the Δpmt mutants showed more severe mutant phenotypes at 42°C than at 30°C. After 12 h of incubation at 42°C, the ΔpmtA mutant was swollen, with diameters two to three times that of wild-type conidia, and did not send out germ tubes, a phenocopy of the original swoA temperature-sensitive mutant (Fig. 4) (29). After 12 h of incubation at 42°C, 20 to 30% of hyphal tips of ΔpmtB germlings grown in liquid medium lysed (n = 200) (Fig. 4). No similar lysis was seen in the wild type. The ΔpmtC mutant at restrictive temperature showed swollen germ tubes and hyperbranching. No similar swelling or hyperbranching was seen in the wild type. The ΔpmtA ΔpmtB mutant showed an additive phenotype. Hyphal tips lysed at permissive temperature and failed to switch to polar growth at restrictive temperature (Fig. 4). The hyphal-tip lysis of ΔpmtB and ΔpmtA ΔpmtB, the polar growth defect of ΔpmtA and ΔpmtA ΔpmtB, and the swollen hyphae of ΔpmtC mutants were corrected by adding osmoticum (KCl or sorbitol) to the medium (data not shown).
Δpmt mutants develop abnormal conidiophores.
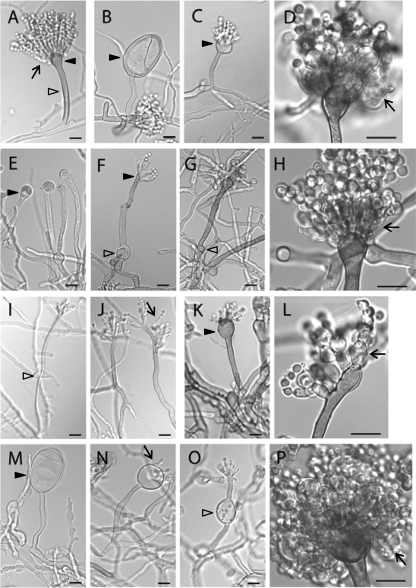
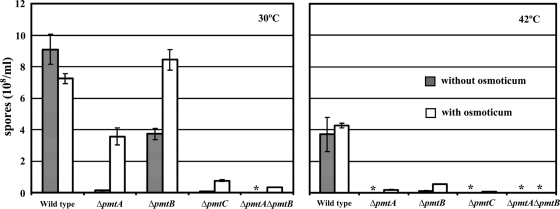
Asexual reproduction in A. nidulans relies on the development of a conidiophore structure that forms from an elongated aerial hypha (conidiophore stalk) with a swollen tip (vesicle). Primary and secondary sterigmata are produced from the vesicle surface to become conidiogenous layers that give rise to chains of conidia (1) (Fig. 5A). Such development requires temporal and spatial regulation of gene expression and coordination and remodeling of cell wall components. To examine the effects of pmt deletion on conidiation, mutants were inoculated on agar blocks on a coverslip and incubated for 3 days. Hyphae and conidiophores adhering to the coverslip were examined microscopically. At permissive temperature, the ΔpmtA vesicles and conidiogenous layers were swollen (Fig. 5B to D). Many vesicles made partial or no conidiogenous layers. At higher temperatures, the swelling was more severe. All conidiophores produced fewer conidia than the wild type (Fig. 6 and 7). At permissive temperature, the majority of ΔpmtB conidiophores were normal, though lysed vesicles were occasionally observed (Fig. 8D). Defects in ΔpmtB conidiophores were more severe at restrictive temperature, showing lysed vesicles and swollen conidiophore stalks (Fig. 5E to G). Although normal conidiophores were present (Fig. 5H), many conidiophores failed to form vesicles, resulting in fewer conidia being produced (Fig. 5F). The ΔpmtC mutant at 30°C had excessive aerial hyphae and produced fewer conidiophores (Fig. 7). Conidiophore stalks were elongated (Fig. 5I), and conidiogenous layers were misplaced (Fig. 5I and J). At 37°C, the vesicles and conidiogenous layers of the ΔpmtC strain were swollen (Fig. 5K and L). Conidiation yielded fewer spores at all temperatures tested (Fig. 6). The ΔpmtA ΔpmtB conidiophores showed an additive phenotype. Swollen conidiophore stalks, vesicles, and conidiogenous layers were visible at 25°C (Fig. 5M to P). At 37°C, the vegetative and aerial hyphae were badly swollen and conidiophores were not detectable (data not shown). Conidial production by all Δpmt mutants was reduced at all tested temperatures (Fig. 6). At permissive temperature, conidial production was partially restored in the presence of an osmostabilizer, except for ΔpmtC and ΔpmtA ΔpmtB mutants. Neither adding osmoticum nor reducing the temperature to 25°C significantly increased the spore numbers of these mutants (Fig. 6 and data not shown).
FIG. 5.
Conidiophores of Δpmt mutants are abnormal. (A) Wild type grown at 30°C. (B to D) ΔpmtA mutant grown at 30°C. (E to H) ΔpmtB mutant grown at 42°C. (I and J) ΔpmtC mutant grown at 30°C. (K and L) ΔpmtC mutant grown at 37°C. (M to P) ΔpmtA ΔpmtB mutant (ATK94) grown at 30°C. The arrows indicate conidiogenous layers, the empty arrowheads indicate conidiophore stalks, and the dark arrowheads indicate vesicles. Bar, in panel I is 5 μm; all others are 10 μm.
FIG. 6.
Conidiation efficiencies of Δpmt mutants are reduced. A total of 104 spores were spread on CM and CM containing 0.6 M KCl. The plates were incubated for 4 days at 30°C or 42°C and washed with 4 ml of sterile water. Conidia were counted using a hemocytometer. The asterisks indicate samples with too few conidia to count.
FIG. 7.
The ΔpmtC mutant produces excessive aerial hyphae. The wild type (A) and ΔpmtA (B), ΔpmtB (C), ΔpmtC (D), and ΔpmtA ΔpmtB (E) mutants were grown on solid medium at 30°C for 3 days. All panels are at the same magnification.
FIG. 8.
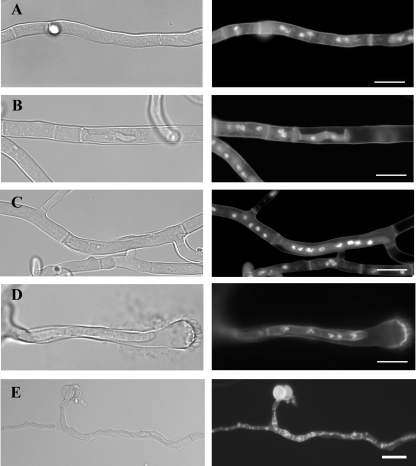
ΔpmtB and ΔpmtA ΔpmtB mutants make intrahyphal hyphae. The wild type (A) and the ΔpmtB (B to D) and ΔpmtA ΔpmtB (E) mutants were grown on solid medium at 30°C for 3 days and stained with Hoechst and Calcofluor white to label nuclei and cell walls, respectively. (Left) Differential interference contrast images. (Right) Fluorescence images. Bar, 10 μm.
The ΔpmtB and ΔpmtAΔpmtB mutants make intrahyphal hyphae.
In addition to the conidiophore defects seen in ΔpmtB and ΔpmtA ΔpmtB mutants after 3 days of incubation at 30°C, it appeared that hyphal compartments had frequently lysed and that hyphal tips from the adjacent compartments invaded these empty compartments, forming intrahyphal hyphae (Fig. 8). This “intrahyphal hyphae” phenotype is very similar to that previously reported for chitin synthase mutants (31).
PmtA and PmtC modify an ortholog of Wsc1, a known Pmt target.
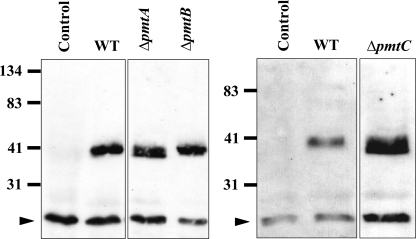
In S. cerevisiae, Wsc family proteins are modified by Pmts (18). Wsc proteins serve as sensors of stress, such as high temperature and cell wall-perturbing chemicals. Pmts add the first mannose residue to specific Ser or Thr residues of their target proteins. Without such modification, the oligo sidechain cannot be elongated, resulting in a protein of lower molecular mass. In order to determine if the A. nidulans Pmts are involved in modifying target proteins, we analyzed an ortholog of the S. cerevisiae Wsc1 protein. A BLASTP search of the A. nidulans genome database using ScWsc1 as the query returned two orthologs, AN6927 and AN5660, with 34% and 37% identity, respectively. The predicted amino acid sequence of AN5660 contains a long stretch of Ser and Thr residues, which is a characteristic of Pmt client proteins. We fused AN5660 with an S tag and expressed it behind the gpd promoter in ΔpmtA, ΔpmtB, and ΔpmtC strains of A. nidulans. Crude membrane fractions from each Δpmt mutant were separated on SDS-PAGE, transferred to membranes, and probed with anti-S-tag antibody. S-tagged AN5660 has a predicted mass of 32 kDa. As shown by Western blot analysis (Fig. 9), in wild-type and ΔpmtB strains, the S-tagged Wsc ortholog had an apparent molecular mass of approximately 40 kDa, while in the ΔpmtA and ΔpmtC strains, the S-tagged Wsc ortholog was slightly smaller, with an apparent molecular mass of approximately 38 kDa. The lower molecular mass of S-tagged AN5660 in ΔpmtA and ΔpmtC strains of A. nidulans is consistent with hypoglycosylation and is very similar to results for tagged Wsc1 expressed in pmt2Δ and pmt4Δ strains of S. cerevisiae (18).
FIG. 9.
PmtA and PmtC modify AN5660 in vivo. Crude membrane fractions from Δpmt mutants were separated on 16% SDS-PAGE. AN5660 S tag on a Western blot was probed with antibody against S tag. The arrowheads indicate nonspecific signals. WT, wild type.
DISCUSSION
A. nidulans has a single representative of each of the three Pmt subfamilies. Neither the Pmt1 representative (pmtB), the Pmt2 representative (pmtA), nor the Pmt4 representative (pmtC) is essential (Fig. 3). This makes A. nidulans the only fungus in which each subfamily has been individually deleted without loss of viability. Though deletion of single Pmt1 or Pmt2 representatives in S. cerevisiae is not lethal, it is impossible to say if these subfamilies are essential in S. cerevisiae because it contains at least two members of both subfamilies and deletion of all representatives of each subfamily has not been reported. Similarly, though deletion of a single Pmt1 representative is not lethal in C. albicans, deletion of the complete Pmt1 subfamily has not been reported. The C. albicans Pmt2 subfamily also contains two members, pmt2 and pmt6, and deletion of pmt2 is lethal. Like A. nidulans, S. pombe contains three Pmts, one from each subfamily. Also like A. nidulans the single Pmt1 subfamily representative is not essential. In contrast with A. nidulans, deletion of the single Pmt2 in S. pombe is lethal. A. nidulans, S. cerevisiae, C. albicans, and S. pombe all have a single Pmt4 representative that is not essential.
In S. cerevisiae and S. pombe, Pmt1 and Pmt2 subfamily representatives are thought to form heteromeric complexes based on immunoprecipitation assays (8, 34). In A. nidulans, ΔpmtA ΔpmtB strains show a synthetic phenotype that is more severe than that of either single mutant, suggesting that Pmt1 and Pmt2 subfamily members might also form heteromeric complexes in this filamentous fungus. The fact that A. nidulans ΔpmtA ΔpmtB mutants are viable with both Pmt1 and Pmt2 subfamilies completely eliminated suggests either that the targets modified by the Pmt1/Pmt2 complex are not needed for viability or that the remaining Pmt4 subfamily member is able to compensate, probably by promiscuous target modification. Consistent with the idea that the nonessential Pmt4 representative might be required to compensate for loss of Pmt1 or Pmt2, we were unable to recover either ΔpmtB ΔpmtC (subfamilies 1 and 4) or ΔpmtA ΔpmtC (subfamilies 2 and 4) mutant strains. Further, ΔpmtC showed much less effect from the cell wall-altering agent Calcofluor (Fig. 3), consistent with the idea that the Pmt4 representative might be most important when Pmt1 or Pmt2 is perturbed. Deletion of representatives of both the Pmt1 and Pmt4 subfamilies in combination appears to be lethal in C. albicans and S. pombe (26, 34).
One of our more intriguing observations was the increased severity of the Δpmt mutant phenotypes at elevated temperatures. Indeed, we first identified pmtA in a screen for temperature-sensitive polarity mutants (21). The pmtA allele (at the time called swoA) produced a dramatic phenotype at elevated temperature of highly swollen conidia from which germ tubes failed to extend. Later work showed that the swoA lesion was in a Pmt2 ortholog and was predicted to result in the truncation of the C-terminal 77 amino acids (29). Interestingly, the ΔpmtA strain described in this paper exactly phenocopies the original temperature-sensitive mutant, suggesting that the original swoA (pmtA) allele was a functional null and that the Pmt2 subfamily plays a role in response to elevated temperature, probably by virtue of the protein targets it modifies.
The more severe phenotypes at elevated temperatures that we observed in the A. nidulans Δpmt strains is a common feature of loss of Pmt activity in other fungi, as well. In S. cerevisiae, the temperature sensitivity of Δpmt2 and Δpmt3 strains (subfamily 2) is clearly connected to the cell wall integrity pathway. The cell wall integrity pathway allows S. cerevisiae to respond to stresses, including elevated temperature and cell wall-perturbing drugs, by upregulating cell wall synthesis (13, 38). In S. cerevisiae, Pmt2 and Pmt4 modify Wsc1p, one of the cell wall proteins that transmits the stress signal to the downstream Pkc1p and MAPK cascade signaling pathways that ultimately activate cell wall biosynthetic genes (18). Though the cell wall integrity pathway is not as well understood in A. nidulans, there are many orthologs of cell wall integrity pathway gene products, including the Wsc proteins (5). We found that a Wsc1 ortholog ran with an apparently reduced molecular mass in A. nidulans ΔpmtA and ΔpmtC strains (Fig. 9), showing that it is modified by these Pmt 2 and Pmt4 subfamily representatives in A. nidulans. It is likely that WscA plays a role in the cell wall integrity pathway in A. nidulans, since introduction of WscA on a high-copy-number plasmid partially suppressed the temperature sensitivity of the ΔpmtA mutant on agar plates (data not shown). However, overexpression of WscA in the ΔpmtA strain could not suppress polar growth in liquid culture at 42°C for 12 h (data not shown).
Most of the cell wall-related phenotypes of the A. nidulans Δpmt mutants, such as lysis, were visible only at high temperature or in the presence of cell wall-perturbing agents. These phenotypes were largely corrected by the addition of osmoticum, suggesting a weakened cell wall. Thus, many of the cell wall-related phenotypes of the A. nidulans Δpmt mutants could be explained by a requirement for modification of cell wall integrity pathway members by Pmts. In S. cerevisiae, several cell wall integrity pathway members are targets of Pmt modification, including Wsc proteins and Mid2 (18). It has been shown that Pmt modification is needed for stability of Wsc1 and Mid2 (18). Though most of the cell wall-related phenotypes of Δpmt mutants in A. nidulans are consistent with Pmt modification of cell wall integrity pathway proteins, it is possible that other cell wall-related proteins are also modified by Pmts, as is true for S. cerevisiae.
In addition to the cell wall-related phenotypes seen under the stress conditions of elevated temperature or drug treatment, all A. nidulans Δpmt mutants showed developmental-patterning defects under normal growth conditions. The ΔpmtA mutant sent out germ tubes earlier than the wild type, while the ΔpmtC mutant sent out more germ tubes and more branches than the wild type. The ΔpmtA ΔpmtB double mutant sent out germ tubes too early, and the ΔpmtB mutant made hyphae within hyphae. All A. nidulans Δpmt mutants made abnormal conidiophores and had reduced conidial numbers at normal growth temperatures (Fig. 6 and 7). Though a weakened cell wall could explain some of the swelling seen in conidiophores, we think that a developmental problem is the more likely cause because each mutant had a distinct pattern of enlarged areas. The ΔpmtA mutant vesicles and conidiogenous layers were swollen, the ΔpmtB mutant showed swellings of the stalk, and the ΔpmtC mutant made elongated stalks (Fig. 5). Further, these phenotypes were not fully rescued by adding osmoticum. These results suggest that different proteins serving as developmental cues are modified by each A. nidulans Pmt. Modification of developmental proteins by Pmts has a precedent in S. cerevisiae, where O mannosylation has been shown to be essential for the function, localization, and stability of Axl2, an axial-budding positional marker (28).
Clearly, the three A. nidulans Pmts play distinct roles in cell wall integrity and development. Though we cannot rule out direct roles of the Pmts, it is most likely that these roles are the result of Pmt modification of target proteins involved in these processes. Future work will focus on identifying targets of Pmt modification and their modes of action.
Acknowledgments
This work was supported by DOE Biosciences award DE-FG02-97ER20275 and the Office of the Vice President for Research at the University of Georgia. T.K. was supported by the Royal Thai Government.
We thank Yangsil Ha for construction of the Δpmt2::Afpyro strain.
Footnotes
Published ahead of print on 7 August 2009.
REFERENCES
- 1.Adams, T. H., J. K. Wieser, and J. H. Yu. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62:35-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2001. Current protocols in molecular biology. J. Wiley, New York, NY.
- 3.Booher, R. N., C. E. Alfa, J. S. Hyams, and D. H. Beach. 1989. The fission yeast cdc2/cdc13/suc1 protein kinase: regulation of catalytic activity and nuclear localization. Cell 58:485-497. [DOI] [PubMed] [Google Scholar]
- 4.Deshpande, N., M. R. Wilkins, N. Packer, and H. Nevalainen. 2008. Protein glycosylation pathways in filamentous fungi. Glycobiology 18:626-637.18504293 [Google Scholar]
- 5.Fujioka, T., O. Mizutani, K. Furukawa, N. Sato, A. Yoshimi, Y. Yamagata, T. Nakajima, and K. Abe. 2007. MpkA-dependent and -independent cell wall integrity signaling in Aspergillus nidulans. Eukaryot. Cell 6:1497-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gemmill, T. R., and R. B. Trimble. 1999. Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim. Biophys. Acta 1426:227-237. [DOI] [PubMed] [Google Scholar]
- 7.Gentzsch, M., and W. Tanner. 1996. The PMT gene family: protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J. 15:5752-5759. [PMC free article] [PubMed] [Google Scholar]
- 8.Girrbach, V., and S. Strahl. 2003. Members of the evolutionarily conserved PMT family of protein O-mannosyltransferases form distinct protein complexes among themselves. J. Biol. Chem. 278:12554-12562. [DOI] [PubMed] [Google Scholar]
- 9.Girrbach, V., T. Zeller, M. Priesmeier, and S. Strahl-Bolsinger. 2000. Structure-function analysis of the dolichyl phosphate-mannose: protein O-mannosyltransferase ScPmt1p. J. Biol. Chem. 275:19288-19296. [DOI] [PubMed] [Google Scholar]
- 10.Gorka-Niec, W., M. Pniewski, A. Kania, U. Perlinska-Lenart, G. Palamarczyk, and J. S. Kruszewska. 2008. Disruption of Trichoderma reesei gene encoding protein O-mannosyltransferase I results in a decrease of the enzyme activity and alteration of cell wall composition. Acta Biochim Pol. 55:251-259. [PubMed] [Google Scholar]
- 11.Harris, S. D. 2001. Genetic analysis of ascomycete fungi, p. 47-58. In N. Talbot (ed.), Molecular and cellular biolgy of filamentous fungi: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 12.Harrison, M. J., A. S. Nouwens, D. R. Jardine, N. E. Zachara, A. A. Gooley, H. Nevalainen, and N. H. Packer. 1998. Modified glycosylation of cellobiohydrolase I from a high cellulase-producing mutant strain of Trichoderma reesei. Eur. J. Biochem. 256:119-127. [DOI] [PubMed] [Google Scholar]
- 13.Heinisch, J. J., A. Lorberg, H. P. Schmitz, and J. J. Jacoby. 1999. The protein kinase C-mediated MAP kinase pathway involved in the maintenance of cellular integrity in Saccharomyces cerevisiae. Mol. Microbiol. 32:671-680. [DOI] [PubMed] [Google Scholar]
- 14.Hill, T., and E. Kafer. 2001. Improved protocols for Aspergillus minimal medium: trace element and minimal medium salt stock solutions. Fungal Genet. Newsl. 48:20-21. [Google Scholar]
- 15.Hutzler, J., M. Schmid, T. Bernard, B. Henrissat, and S. Strahl. 2007. Membrane association is a determinant for substrate recognition by PMT4 protein O-mannosyltransferases. Proc. Natl. Acad. Sci. USA 104:7827-7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kafer, E. 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19:33-131. [DOI] [PubMed] [Google Scholar]
- 17.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 18.Lommel, M., M. Bagnat, and S. Strahl. 2004. Aberrant processing of the WSC family and Mid2p cell surface sensors results in cell death of Saccharomyces cerevisiae O-mannosylation mutants. Mol. Cell. Biol. 24:46-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Momany, M. 2001. Using microscopy to explore the duplication cycle, p. 119-125. In N. J. Talbot (ed.), Molecular and cellular biology of filamentous fungi: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 20.Momany, M. 2002. Polarity in filamentous fungi: establishment, maintenance and new axes. Curr. Opin. Microbiol. 5:580-585. [DOI] [PubMed] [Google Scholar]
- 21.Momany, M., P. J. Westfall, and G. Abramowsky. 1999. Aspergillus nidulans swo mutants show defects in polarity establishment, polarity maintenance and hyphal morphogenesis. Genetics 151:557-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Nayak, T., et al. 2006. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172:1557-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oka, T., T. Hamaguchi, Y. Sameshima, M. Goto, and K. Furukawa. 2004. Molecular characterization of protein O-mannosyltransferase and its involvement in cell-wall synthesis in Aspergillus nidulans. Microbiology 150:1973-1982. [DOI] [PubMed] [Google Scholar]
- 23.Oka, T., Y. Sameshima, T. Koga, H. Kim, M. Goto, and K. Furukawa. 2005. Protein O-mannosyltransferase A of Aspergillus awamori is involved in O-mannosylation of glucoamylase I. Microbiology 151:3657-3667. [DOI] [PubMed] [Google Scholar]
- 24.Olson, G. M., D. S. Fox, P. Wang, J. A. Alspaugh, and K. L. Buchanan. 2007. Role of protein O-mannosyltransferase Pmt4 in the morphogenesis and virulence of Cryptococcus neoformans. Eukaryot. Cell 6:222-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 26.Prill, S. K., B. Klinkert, C. Timpel, C. A. Gale, K. Schroppel, and J. F. Ernst. 2005. PMT family of Candida albicans: five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol. Microbiol. 55:546-560. [DOI] [PubMed] [Google Scholar]
- 27.Rouabhia, M., M. Schaller, C. Corbucci, A. Vecchiarelli, S. K. Prill, L. Giasson, and J. F. Ernst. 2005. Virulence of the fungal pathogen Candida albicans requires the five isoforms of protein mannosyltransferases. Infect. Immun. 73:4571-4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanders, S. L., M. Gentzsch, W. Tanner, and I. Herskowitz. 1999. O-Glycosylation of Axl2/Bud10p by Pmt4p is required for its stability, localization, and function in daughter cells. J. Cell Biol. 145:1177-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaw, B. D., and M. Momany. 2002. Aspergillus nidulans polarity mutant swoA is complemented by protein O-mannosyltransferase pmtA. Fungal Genet. Biol. 37:263-270. [DOI] [PubMed] [Google Scholar]
- 30.Strahl-Bolsinger, S., M. Gentzsch, and W. Tanner. 1999. Protein O-mannosylation. Biochim. Biophys. Acta 1426:297-307. [DOI] [PubMed] [Google Scholar]
- 31.Takeshita, N., S. Yamashita, A. Ohta, and H. Horiuchi. 2006. Aspergillus nidulans class V and VI chitin synthases CsmA and CsmB, each with a myosin motor-like domain, perform compensatory functions that are essential for hyphal tip growth. Mol. Microbiol. 59:1380-1394. [DOI] [PubMed] [Google Scholar]
- 32.Timpel, C., S. Strahl-Bolsinger, K. Ziegelbauer, and J. F. Ernst. 1998. Multiple functions of Pmt1p-mediated protein O-mannosylation in the fungal pathogen Candida albicans. J. Biol. Chem. 273:20837-20846. [DOI] [PubMed] [Google Scholar]
- 32a.Toews, M. W., et al. 2004. Establishment of mRFP1 as a fluorescent marker in Aspergillus nidulans and construction of expression vectors for high-throughput protein tagging using recombination in vitro (GATEWAY). Curr. Genet. 45:383-389. [DOI] [PubMed] [Google Scholar]
- 33.VanderVen, B. C., J. D. Harder, D. C. Crick, and J. T. Belisle. 2005. Export-mediated assembly of mycobacterial glycoproteins parallels eukaryotic pathways. Science 309:941-943. [DOI] [PubMed] [Google Scholar]
- 34.Willer, T., M. Brandl, M. Sipiczki, and S. Strahl. 2005. Protein O-mannosylation is crucial for cell wall integrity, septation and viability in fission yeast. Mol. Microbiol. 57:156-170. [DOI] [PubMed] [Google Scholar]
- 35.Willer, T., M. C. Valero, W. Tanner, J. Cruces, and S. Strahl. 2003. O-Mannosyl glycans: from yeast to novel associations with human disease. Curr. Opin. Struct. Biol. 13:621-630. [DOI] [PubMed] [Google Scholar]
- 36.Yang, L., L. Ukil, A. Osmani, F. Nahm, J. Davies, C. P. De Souza, X. Dou, A. Perez-Balaguer, and S. A. Osmani. 2004. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell 3:1359-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou, H., H. Hu, L. Zhang, R. Li, H. Ouyang, J. Ming, and C. Jin. 2007. O-Mannosyltransferase 1 in Aspergillus fumigatus (AfPmt1p) is crucial for cell wall integrity and conidium morphology, especially at an elevated temperature. Eukaryot. Cell 6:2260-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zu, T., J. Verna, and R. Ballester. 2001. Mutations in WSC genes for putative stress receptors result in sensitivity to multiple stress conditions and impairment of Rlm1-dependent gene expression in Saccharomyces cerevisiae. Mol. Genet. Genomics 266:142-155. [DOI] [PubMed] [Google Scholar]