Abstract
Aspergillus nidulans possesses three pmt genes encoding protein O-d-mannosyltransferases (Pmt). Previously, we reported that PmtA, a member of the PMT2 subfamily, is involved in the proper maintenance of fungal morphology and formation of conidia (T. Oka, T. Hamaguchi, Y. Sameshima, M. Goto, and K. Furukawa, Microbiology 150:1973-1982, 2004). In the present paper, we describe the characterization of the pmtA paralogues pmtB and pmtC. PmtB and PmtC were classified as members of the PMT1 and PMT4 subfamilies, respectively. A pmtB disruptant showed wild-type (wt) colony formation at 30°C but slightly repressed growth at 42°C. Conidiation of the pmtB disruptant was reduced to approximately 50% of that of the wt strain; in addition, hyperbranching of hyphae indicated that PmtB is involved in polarity maintenance. A pmtA and pmtB double disruptant was viable but very slow growing, with morphological characteristics that were cumulative with respect to either single disruptant. Of the three single pmt mutants, the pmtC disruptant showed the highest growth repression; the hyphae were swollen and frequently branched, and the ability to form conidia under normal growth conditions was lost. Recovery from the aberrant hyphal structures occurred in the presence of osmotic stabilizer, implying that PmtC is responsible for the maintenance of cell wall integrity. Osmotic stabilization at 42°C further enabled the pmtC disruptant to form conidiophores and conidia, but they were abnormal and much fewer than those of the wt strain. Apart from the different, abnormal phenotypes, the three pmt disruptants exhibited differences in their sensitivities to antifungal reagents, mannosylation activities, and glycoprotein profiles, indicating that PmtA, PmtB, and PmtC perform unique functions during cell growth.
Protein glycosylation, which is a major posttranslational modification, plays essential roles in eukaryotic cells from fungi to mammals (19). N-linked oligosaccharides in glycoproteins that share relatively common structures are structurally classified into high-mannose, complex, and hybrid types (3). O-linked oligosaccharides in glycoproteins are diverse with respect to their sugar components and the mode of sugar linkages among the eukaryotic organisms (8, 19). O mannosylation, which is commonly found in the glycoproteins of fungi, has been extensively studied in the budding yeast Saccharomyces cerevisiae (4, 21, 35). The initial reaction of mannose transfer to serine and threonine residues in proteins is catalyzed by protein O-d-mannosyltransferase (Pmt) in the endoplasmic reticulum (ER), where dolichyl phosphate-mannose is required as an immediate sugar donor (4). In the Golgi complex, O mannosylation in S. cerevisiae is linearly elongated by up to five mannose residues by mannosyltransferases (Mnt) that utilize GDP-mannose as the mannosyl donor. At least six Pmt-encoding genes (PMT1 to -6), three α-1,2-Mnt-encoding genes (KRE2, KTR1, and KTR3), and three α-1,3-Mnt-encoding genes (MNN1, MNT2, and MNT3) are known to be involved in O mannosylation in S. cerevisiae (21, 31, 45).
The Pmt family of proteins can be classified into the PMT1, PMT2, and PMT4 subfamilies based on phylogeny (6). Proteins of the PMT1 subfamily form a heteromeric complex with proteins belonging to the PMT2 subfamily, and PMT4 subfamily proteins form a homomeric complex (7). Simultaneous disruptions of three different types of PMT genes were lethal (4), suggesting that each class provided a unique function for O mannosylation. Yeasts other than S. cerevisiae, such as Schizosaccharomyces pombe (38, 41), Candida albicans (29), and Cryptococcus neoformans (28), possess three to five pmt genes, which have been characterized. Several studies provide evidence that protein O mannosylation modulates the functions and stability of secretory proteins and thereby affects the growth and morphology of these yeasts. O mannosylation by Pmt2 in S. cerevisiae (ScPmt2) provides protection from ER-associated degradation and also functions as a fail-safe mechanism for ER-associated degradation (11, 13, 23). Likewise, in C. albicans, CaPmt1- and CaPmt4-mediated O mannosylation specifically protects CaSec20 from proteolytic degradation in the ER (40). Cell wall integrity is maintained in S. cerevisiae by increased stabilization and correct localization of the sensor proteins ScWsc and ScMid2 due to O mannosylation by ScPmt2 and ScPmt4 (20). Similarly, the stability and localization to the plasma membrane of axial budding factor ScAxl2/Bud10 is enhanced by ScPmt4-mediated O mannosylation, increasing its activity (32). ScPmt4-mediated O glycosylation also functions as a sorting determinant for cell surface delivery of ScFus1 (30). CaPmt4-mediated O glycosylation is required for environment-specific morphogenetic signaling and for the full virulence of C. albicans (29).
With respect to filamentous fungi like Aspergillus that develop hyphae in a highly ordered manner, which then differentiate to form conidiospores, little is known about the function and synthetic pathway of the O-mannose-type oligosaccharides. O-Glycans in glycoproteins of Aspergillus include sugars other than mannose, and their structures have been determined (8). The initial mannosylation catalyzed by Pmts is found in Aspergillus and occurs as in yeasts (8).
We characterized the pmtA gene of Aspergillus nidulans (AnpmtA), belonging to the PMT2 subfamily, and found that the mutant exhibited a fragile cell wall phenotype and alteration in the carbohydrate composition, with a reduction in the amount of skeletal polysaccharides in the cell wall (26, 33). Recently, the Afpmt1 gene belonging to the PMT1 family of Aspergillus fumigatus, a human pathogen, was characterized. AfPmt1 is crucial for cell wall integrity and conidium morphology (46).
In this study, we characterize the pmtB and pmtC genes of A. nidulans to understand their contribution to the cell morphology of this filamentous fungus. We also demonstrate that the PmtA, PmtB, and PmtC proteins have distinct specificities for protein substrates and function differently during cell growth of filamentous fungi.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The A. nidulans strains (listed in Table 1) were grown on YG medium (0.5% [wt/vol] yeast extract, 2.5% [wt/vol] glucose) or minimal medium (MM) (1% [wt/vol] glucose, 0.6% [wt/vol] NaNO3, 0.052% [wt/vol] KCl, 0.052% [wt/vol] MgSO4·7H2O, 0.152% [wt/vol] KH2PO4, and Hunter's trace elements, pH 6.5). Liquid growth experiments to allow hyphal development in a submerged culture were done by inoculation of 2 × 108 conidia into 100 ml MM or YG medium in 500-ml shaking flasks. The flasks were reciprocally shaken at 120 rpm at 30°C. Standard transformation procedures for A. nidulans were used (44). Plasmids were propagated in Escherichia coli XL-1 Blue. Genomic DNA and total RNA of A. nidulans were prepared as previously described (26). Southern and Northern hybridizations were done using a DIG labeling kit (Roche) according to the manufacturer's protocols.
TABLE 1.
A. nidulans strains used in this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| A26 | biA1 | FGSCb |
| A89 | biA1 argB2 | FGSC |
| AKU89 | biA1 argB2 nkuB::aurA+ | This study |
| ΔAnpmtA (P3) | biA1 argB2 pmtA::argB+ | 26 |
| ΔAnpmtB | biA1 argB2 nkuB::aurA+pmtB::ptrA+ | This study |
| BΔAnpmtB | biA1 argB2 nkuB::aurA+pmtB::ptrA+pyrG::pmtB | This study |
| ΔAnpmtA-pmtB | biA1 argB2 nkuB::aurA+pmtA::argB+pmtB::ptrA+ | This study |
| ΔAnpmtC | biA1 argB2 nkuB::aurA+pmtC::ptrA+ | This study |
| CΔAnpmtC | biA1 argB2 nkuB::aurA+pmtC::ptrA+pyrG::pmtC | This study |
| AKU89-glaA | biA1 argB2 nkuB::aurA+glaA(P)-glaA-glaA(T) argB+ | This study |
| ΔAnpmtB-glaA | biA1 argB2 nkuB::aurA+pmtB::ptrA+glaA(P)-glaA-glaA(T) argB+ | This study |
| ΔAnpmtC-glaA | biA1 argB2 nkuB::aurA+pmtB::ptrA+glaA(P)-glaA-glaA(T) argB+ | This study |
| AKU89-AnwscA | biA1 argB2 nkuB::aurA+glaA(P)-wscA::3HA-glaA(T) argB+ | This study |
| ΔAnpmtA-AnwscA | biA1 argB2 pmtA::argB+glaA(P)-wscA::3HA-glaA(T) ptrA+ | This study |
| ΔAnpmtB-AnwscA | biA1 argB2 nkuB::aurA+pmtB::ptrA+glaA(P)-wscA::3HA-glaA(T) argB+ | This study |
| ΔAnpmtC-AnwscA | biA1 argB2 nkuB::aurA+pmtC::ptrA+glaA(P)-wscA::3HA-glaA(T) argB+ | This study |
glaA(P), promoter region of A. awamori glaA; glaA(T), terminator region of A. awamori glaA.
FGSC, Fungal Genetics Stock Center.
Isolation of the AnpmtB and AnpmtC genes.
All oligonucleotide primers used in this study are listed in Table S1 in the supplemental material. Based on a multiple-sequence alignment among Pmt proteins of A. nidulans, S. cerevisiae, and C. albicans, degenerate oligonucleotide primers for the amplification of A. nidulans pmtB and pmtC genes were synthesized. Using primers pmtB-F/pmtB-R and pmtC-F/pmtC-R, regions of AnpmtB and AnpmtC, respectively, were amplified from A. nidulans A26 genomic DNA and used as probes to screen an A. nidulans cosmid library (Fungal Genetics Stock Center) for entire AnpmtB and AnpmtC genes. Isolated AnpmtB and AnpmtC genes were sequenced using a LIC4200L DNA sequencer (Li-Cor). The cDNAs of pmtB and pmtC were amplified by reverse transcription-PCR using total RNA from strain A26 with primer pairs An-pmtB-RT-F/An-pmtB-RT-R and An-pmtC-RT-F/An-pmtC-RT-R, respectively. The amplified DNA fragments were inserted into pGEM-T Easy (Promega) and sequenced. The sequences were analyzed with Genetyx (Genetyx Corp., Japan). BLAST searches were done using the A. nidulans genome database at http://www.broad.mit.edu/annotation/genome/aspergillus_group/MultiHome.html.
Construction of AnpmtB, AnpmtC, and AnpmtA AnpmtB disruptants.
To facilitate gene disruption in A. nidulans, nkuB, encoding a protein (AN4552.3) involved in nonhomologous end joining (24, 25), was replaced in host strain A89 with the selectable marker aurA. For this purpose, a gene replacement cassette encompassing 5′ nkuB, aurA, and 3′ nkuB was constructed by recombinant PCR (18). First, the nkuB 5′- and 3′-flanking regions were amplified from A89 genomic DNA with primer pairs F1-PnkuB/R1-PnkuB and F3-TnkuB/R3-TnkuB, respectively, and aurA was amplified from pAUR316 (TakaraBio, Japan) with primers F2-aur and R2-aur. Then, after the three purified DNA fragments were combined as a template, the gene replacement cassette was amplified with primers F1-PnkuB and R3-TnkuB using LA-Taq DNA polymerase (TakaraBio) and inserted into pGEM-T Easy, yielding pGEM-nkuB::aurA. After digestion with EcoRV, pGEM-nkuB::aurA was transformed into strain A89. Transformants resistant to aureobasidin A were analyzed for correct gene replacement by PCR and Southern blot analysis, yielding strain AKU89 with a disrupted AnnkuB gene.
For disruption of AnpmtB with ptrA, conferring resistance to pyrithiamine, plasmid pGEM-ΔpmtB was constructed by insertion of a 2.0-kb KpnI fragment containing ptrA, amplified with primers ptrA-KpnI-F and ptrA-KpnI-R from pPTR I (TakaraBio), into the KpnI site of AnpmtB, cloned into pGEM-T Easy after PCR amplification with primers pmtB-around-F and pmtB-around-R. Strain AKU89 was transformed with pGEM-ΔpmtB linearized with NaeI. The disruption of AnpmtB in pyrithiamine-resistant transformants was confirmed by Southern blot analysis using a 1.1-kb region of AnpmtB amplified with primers pmtB-pr-F and pmtB-pr-R as a probe and by PCR using primers F2-AnpmtB and R2-AnpmtB.
For complementation of an AnpmtB disruptant with wild-type (wt) pmtB, a gene replacement cassette encompassing 1.3-kb 5′-pyrG, 4.2-kb wt pmtB, and 1.1-kb 3′-pyrG was constructed by recombinant PCR using three primer pairs, F1-AnpyrG/R1-AnpGpB, F1-AnpGpB/R2-AnpGpB, and F2-AnpGpB/R1- AnpyrG. The resultant DNA fragment amplified with primers F1-AnpyrG and R1-AnpyrG was used to transform the AnpmtB disruptant. Transformants were selected on MM with 10 mM arginine, 5 mM 5-fluoroorotic acid, 5 mM uridine, and 5 mM uracil. Introduction of the wt pmtB gene into the AnpmtB disruptant at the pyrG locus was confirmed by PCR using primer pairs F-pmtBPr/F-pmtBPr and F1-AnpyrG/R1-AnpyrG.
Similarly, AnpmtC was disrupted in AKU89 by ptrA insertion after transformation of SpeI-linearized pGEM-ΔpmtC, which was constructed analogously to pGEM-ΔpmtB with primers pmtC-around-F and pmtC-around-R to amplify pmtC. A 1.0-kb region of AnpmtC, amplified with primers pmtC-pr-F and pmtC-pr-R, was used as a probe in Southern blot analysis to verify correct disruption of AnpmtC. PCR with primers F3-AnpmtC and R3-AnpmtC was also done to verify disruption of AnpmtC.
For complementation of the AnpmtC disruptant with wt pmtC, plasmid pGTΔsal-pyrG::pmtC was constructed as follows. A 3.5-kb DNA fragment of pyrG was amplified by PCR with primers F1-AnpyrG and R1-AnpyrG and inserted into pGEM-T Easy, yielding pGT-pyrG. pGT-pyrG was digested with NsiI and self-ligated, yielding pGTΔsal-pyrG. A 3.5-kb DNA fragment of pmtC was replaced by part of the coding sequence of pyrG at the SalI site, yielding pGTΔsal-pyrG::pmtC. The 6.5-kb DNA fragment carrying 1.3-kb 5′-pyrG, 3.5-kb wt pmtC, and 1.7-kb 3′-pyrG from pGTΔsal-pyrG::pmtC was used to transform the AnpmtC disruptant. Introduction of the wt pmtC gene into the AnpmtC disruptant at the pyrG locus was confirmed by PCR using primer pairs F1-AnpyrG/R1-AnpyrG and F-AnpmtCPr/R-AnpmtCPr.
An AnpmtA and AnpmtB double disruptant was created from the ΔAnpmtB strain (AKU89 with pmtB::ptrA+) (Table 1) by transformation of BssHII-linearized pBS-ΔpmtA (26), targeting integration of argB into AnpmtA and complementing the strain's arginine auxotrophy. Southern blot analysis confirmed the disruption of both AnpmtA and AnpmtB in the selected transformants.
Analysis of the efficiency of conidiation.
About 105 conidia were spread onto an 84-mm minimum agar medium. After 3 days of incubation at 30°C or 42°C, the conidia formed were suspended in 5 ml 0.01% (wt/vol) Tween 20 solution and counted using a hemocytometer.
Microscopy.
Submerged hyphae of A. nidulans were observed as follows. Conidia were inoculated into liquid medium, and then the culture was poured into a petri dish containing glass coverslips. After incubation at 30°C or 42°C for 10 to 50 h, the submerged hyphae adhering to the coverslip were stained and fixed with Myco-Perm Blue (Scientific Device Laboratory). The aerial hyphae of A. nidulans were observed as follows. Conidia were inoculated on agar medium and incubated at 30°C or 42°C for 2 to 7 days. The adhesive side of Fungi-Tape (Scientific Device Laboratory) was gently pressed against the aerial hyphae. The tape with the aerial hyphae was mounted on a glass slide and then stained and fixed with Myco-Perm Blue. The hyphae were observed using a Nikon Eclipse E600 microscope.
Expression of GAI of A. awamori.
For expression of the gene encoding glucoamylase I (GAI) of Aspergilllus awamori (AaglaA), pGEM-glaAargB was constructed as follows. AaglaA amplified by PCR from pBR-glaA (9) with primers F1-PglaA and R1-TglaA was inserted into pGEM-T Easy to yield pGT-glaA. The argB gene was amplified from pDC1 (1) with primers argB-ApaI-F and argB-ApaI-R and inserted into the ApaI site of pGT-glaA to yield pGEM-glaAargB. Strains AKU89, ΔAnpmtB, and ΔAnpmtC were transformed with pGEM-glaAargB, and integration of the AaglaA expression cassette was confirmed by Southern blot analysis. The selected argB+ transformants carrying AaglaA were cultured in 100 ml culture A medium (27) with 2.0% (wt/vol) maltose and 0.05% (wt/vol) glucose to induce AaglaA expression for 36 h at 30°C. Culture filtrates of the transformants were concentrated by centrifugation through Microcon YM-10 (Millipore) filter units and loaded onto sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Detection of GAI was done by immunoblotting using an anti-GAI-antibody as described previously (27).
Expression of HA-tagged A. nidulans wscA.
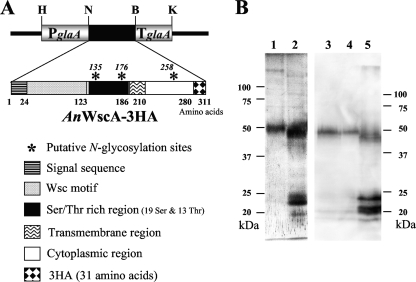
A C-terminally triple-hemagglutin (3HA)-tagged version of AN5660.3, the A. nidulans homolog of the highly O-glycosylated cell surface protein Wsc1 of S. cerevisiae, was expressed after integration of plasmids containing a KpnI/HindIII fragment with an AnwscA::3HA fusion in between the AaglaA promoter and terminator regions derived from pGEM-glaAargB (see Fig. 8A). pARG-PTglaAwscA3ha corresponds to pUC119 containing the above-mentioned fusion and a 1.7-kb EcoRI fragment carrying the argB gene from pDC1 (1), while pPTR-PTglaAwscA3ha corresponds to the fusion construct cloned into pPTR I (TakaraBio). The integrative plasmid pPTR-PTglaAwscA3ha or pARG-PTglaAwscA3ha was transformed into strain P3 (ΔAnpmtA argB+) or AKU89, ΔAnpmtB, and ΔAnpmtC, respectively. PtrA+ or argB+ transformants carrying AnwscA::3HA were cultured in 100 ml culture A medium with 2.0% (wt/vol) maltose and 0.05% (wt/vol) glucose for 36 h at 30°C, harvested by filtration, and lyophilized. The cells were mechanically broken with a Multi-beads shocker (Yasui Kikai, Japan), and proteins were extracted with 1% SDS, 9 M urea, 1 mM EDTA, 0.7 M β-mercaptoethanol, 50 mM Tris-Cl, pH 6.8. After separation of the proteins on 10% SDS-PAGE gels, WscA-3HA was detected by immunoblotting using anti-HA monoclonal antibody (Sigma).
FIG. 8.
AnWscA-3HA expression in the wt and the Anpmt disruptants. (A) Schematic representation of the integrated expression cassette consisting of the AaglaA promoter (P) and terminator (T) flanking an Anwsc open reading frame C-terminally tagged with 3HA. The protein domains and restriction sites used are indicated: H, HindIII; N, NotI; S, SalI; B, BglII; and K, KpnI. (B) AnWscA-3HA expressed in integrants derived from the AKU89 (lanes 1 and 3), ΔAnpmtA (lane 2), ΔAnpmtB (lane 4), and ΔAnpmtC (lane5) strains was detected by immunoblotting using monoclonal anti-HA antibody.
Triton X-100 membrane protein fraction preparation.
A. nidulans was grown in liquid MM and harvested. Freeze-dried cells were mechanically broken with the Multi-beads shocker with an equal volume of 0.5-mm glass beads, suspended in TM buffer (50 mM Tris-Cl, 5 mM MgCl2, Complete protease inhibitor [Roche], pH 7.5) and centrifuged at 17,530 × g for 10 min. The resultant pellet was washed two times with TM buffer, resuspended in extraction buffer (50 mM Tris-Cl, 5 mM MgCl2, 1% [vol/vol] Triton X-100, pH 7.5), and centrifuged at 17,530 × g for 10 min. The supernatant designated as a Triton X-100 fraction contained solubilized membrane proteins that were separated by SDS-PAGE and stained with Coomassie brilliant blue (CBB). Glycoproteins were detected by lectin blotting (10) using alkaline phosphatase-conjugated concanavalin A (EY Laboratory Inc.).
RESULTS
AnpmtB and AnpmtC genes encode Pmts.
We previously characterized AnpmtA encoding Pmt of A. nidulans (26). In the present paper, we describe the remaining Anpmt genes, termed AnpmtB and AnpmtC, which we cloned from a genomic cosmid library of A. nidulans using probes prepared with degenerate primers (see Materials and Methods). BLAST searches against the A. nidulans genome database with the obtained cDNA sequences of AnpmtB and AnpmtC identified the genes as AN4761.3 and AN1459.3, respectively. AnpmtB is a gene of 3,044 bp containing six exons and five introns and encodes the AnPmtB protein, which consists of 918 amino acids with a putative molecular mass of 103.3 kDa. AnpmtC is comprised of 2,424 bp with three exons and two introns and encodes a protein, AnPmtC, of 773 amino acids with a putative molecular mass of 88.2 kDa. AnPmtB and AnPmtC share relatively low (34.5%) amino acid sequence homology with each other and 37.5% and 32.8%, respectively, with AnPmtA. AnPmtB showed the highest sequence homology with Pmts from other sources, such as AfPmt1 (76.0%), CaPmt1 (41.9%), and ScPmt1 (39.6%). The highest sequence homology of AnPmtC was with ScPmt4 (48.4%). Thus, the AnPmtA, AnPmtB, and AnPmtC proteins belong to distinct families, namely, PMT2, PMT1, and PMT4, respectively, based on the phylogenetic tree constructed by the unweighted-pair group method using average linkages (34) (see Fig. S1 in the supplemental material).
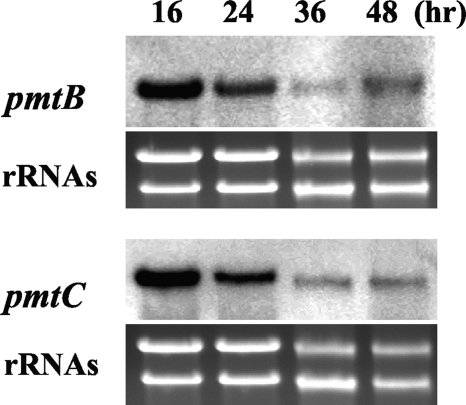
We previously demonstrated that AnpmtA was constitutively transcribed during incubation in liquid MM at 30°C. As determined by Northern blotting, AnpmtB and AnpmtC were expressed in liquid MM for 16 h to 48 h at 30°C (Fig. 1), indicating that both genes, as well as AnpmtA, as previously reported (26), are functional throughout hyphal development, whereas the expression levels of AnpmtB and AnpmtC decreased over time, suggesting that the genes are mainly required for early stages of hyphal development.
FIG. 1.
Transcriptional analysis of AnpmtB and AnpmtC. Total RNA of A. nidulans wt strain FGSC26 was prepared after growth in MM at 30°C, and 10 μg was loaded in each lane. Northern blot hybridization was performed using pmtB and pmtC as probes, and rRNAs were stained with ethidium bromide.
Disruptions of pmtB and pmtC genes.
To understand the effects of AnpmtB and AnpmtC on the growth of A. nidulans, we disrupted each gene in A. nidulans AKU89 by gene replacement with ptrA+ (see Fig. S2 in the supplemental material), yielding the ΔAnpmtB and ΔAnpmtC strains, respectively. Southern blot analysis using the 3′ region of AnpmtB or AnpmtC as a probe and PCRs with primer pairs F2-AnpmtB/R2-AnpmtB and F3-AnpmtC/R3-AnpmtC revealed that site-specific recombination had occurred at the AnpmtB or AnpmtC locus and that a single copy of ptrA+ had been integrated into the chromosomal DNA.
We introduced wt AnpmtB and wt AnpmtC genes into the ΔAnpmtB and ΔAnpmtCstrains, yielding BΔAnpmtB and CΔAnpmtC, respectively. PCRs with primer pairs F-pmtBPr/R-pmtBPr and F1-AnyrG/R1-AnpyrG, or F-pmtCPr/R-pmtCPr and F1-AnyrG/R1-AnpyrG, revealed that site-specific recombination of wt AnpmtB or wt AnpmtC had occurred at the pyrG locus and that a single copy of the wt pmt gene had been integrated into the chromosomal DNA (see Fig. S2 in the supplemental material).
Underglycosylation of heterologous GAI in Anpmt disruptants.
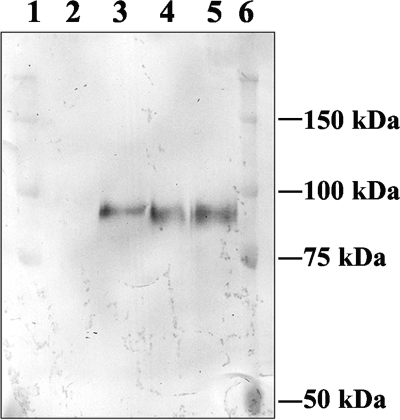
GAI from A. awamori is an extracellular protein, consisting of three domains, namely, the amino-terminal catalytic domain, a serine/threonine-rich region that is glycosylated, and the starch binding domain at the carboxy terminus (12). We previously used GAI as a reporter to measure glycosylation activity and demonstrated that AnpmtA disruption affected the O mannosylation of GAI (26). We integrated the GAI-encoding gene into the ΔAnpmtB and ΔAnpmtC strains (see Materials and Methods) and determined whether the absence of either PMT activity would have an effect on the glycosylation of GAI. The electrophoretic mobilities of GAI secreted from the ΔAnpmtB and ΔAnpmtC strains were similar to each other on 7% SDS-PAGE but slightly faster than that of GAI produced by the wt strain (Fig. 2). This result indicates that both AnpmtB and AnpmtC disruptions led to underglycosylation of GAI. Thus, in A. nidulans, complete glycosylation of GAI in vivo requires the presence of all three functionally active Pmts, AnPmtA (26), AnPmtB, and AnPmtC.
FIG. 2.
Protein O-d-mannosylation of GAI. Proteins in the culture filtrates of AKU89 (lane 2) or of strains expressing GAI—AKU89-glaA (lane 3) and the ΔAnpmtB-glaA (lane 4) and ΔAnpmtC-glaA strains (lane 5)—were separated on 7% SDS-PAGE and immunoblotted using anti-GAI antiserum. Precision plus protein standards (Bio-Rad) were used as molecular mass markers (lanes 1 and 5).
AnPmtB functions independently of AnPmtA.
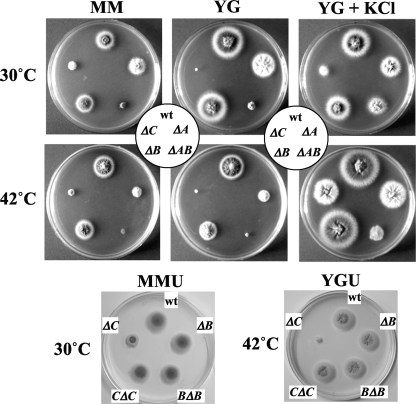
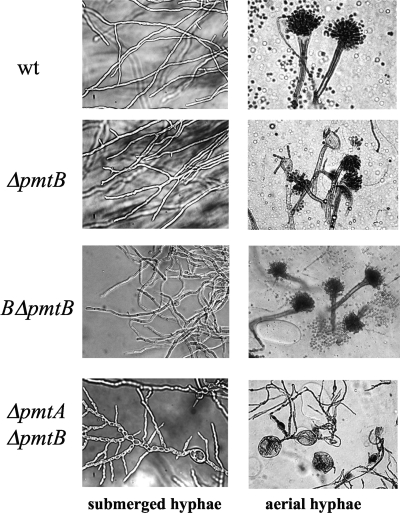
Since PMT1 and PMT2 subfamily proteins form heterodimers, whereas the PMT4 subfamily proteins homodimerize (7), we expected that the ΔAnpmtB strain would show a phenotype comparable to that of the ΔAnpmtA strain (26). However, in contrast to the ΔAnpmtA strain, the ΔAnpmtB strain showed colony phenotypes more similar to those of the wt strain at 30°C, and only at an elevated temperature of 42°C were slightly smaller colonies formed, which was remedied in the presence of 0.6 M KCl as an osmotic stabilizer (Fig. 3). Furthermore, the ΔAnpmtB strain formed submerged hyphae similar to those of the wt strain, although with more frequent hyphal branching (Fig. 4). Aerial hyphae of the ΔAnpmtB strain developed normal conidiophores with wt conidia, but some hyphae ended in abnormally swollen vesicles containing a few conidia. Accordingly, the number of conidia formed in the ΔAnpmtB strain was reduced to 56% of the wt hyphae cultivated under similar growth conditions (on MM at 30°C for 3 days). The BΔAnpmtB strain carrying wt pmtB at the pyrG locus in the ΔAnpmtB strain showed a phenotype identical to that of the wt strain with respect to colony and hyphal morphologies (Fig. 3 and 4), confirming that disruption of AnpmtB affects only the function of AnpmtB.
FIG. 3.
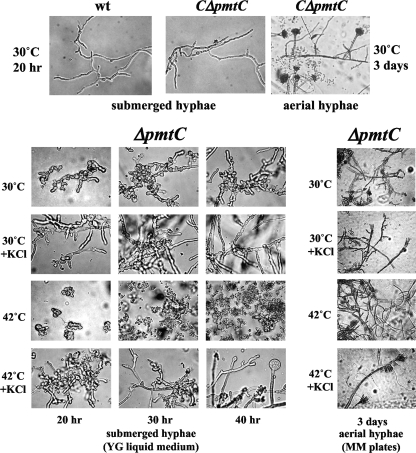
Colony formation by the wt strain and Anpmt disruptants. A. nidulans strains were grown in MM and YG medium with or without 0.6 M KCl at 30°C or 42°C for 3 days. MM and YG supplemented with 5 mM uracil and 5 mM uridine (MMU and YGU, respectively) were used for cultivation of the BΔAnpmtB and CΔAnpmtC strains.
FIG. 4.
Hyphal morphology of Anpmt disruptants. The wt strain AKU89 and the ΔAnpmtB, BΔAnpmtB, and ΔAnpmtA-pmtB strains were grown at 30°C in liquid MM or MMU (MM supplemented with 5 mM uracil and 5 mM uridine) (left) and on an MM or MMU plate (right), respectively. The AKU89, ΔAnpmtB, and BΔAnpmtB strains were cultivated at 30°C for 2 days and the ΔAnpmtA-pmtB strain for 3 days.
The fact that the ΔAnpmtB strain phenotypes were completely different from those of the ΔAnpmtA strain (26) suggested that the proteins have independent functions that do not rely on their heterodimerization, as reported for members of the PMT2 and PMT1 subfamilies (7). We further assessed the relative contributions of these proteins to the growth of A. nidulans by testing the ΔAnpmtA-AnpmtB double disruptant (see Fig. S2 in the supplemental material). The growth of the ΔAnpmtA-AnpmtB strain was severely impaired at 30 and 42°C, and although it slightly improved upon addition of the osmotic stabilizer, colony formation was under all conditions exceedingly more impaired than was observed in the case of the single disruptants, suggesting a synthetic defect (Fig. 3) (26). Also, abnormalities observed in the hyphal structure of the ΔAnpmtA-AnpmtB strain were cumulative with respect to the single disruptants (Fig. 4). The hyphae in the double disruptant were slightly swollen, with balloon structures characteristic of the ΔAnpmtA strain (26), and hyperbranching, as found in the ΔAnpmtB strain. These results confirmed that AnPmtA and AnPmtB have independent functions and that disruption of these genes caused divergent phenotypes in A. nidulans.
Disruption of AnpmtC impairs hyphal elongation and conidium formation.
Of the three Anpmt disruptions, removal of AnpmtC caused the most remarkable defect in colony formation, which was significantly recovered only at 42°C in the presence of 0.6 M KCl (Fig. 3), 0.8 M NaCl, or 1.2 M sorbitol as an osmotic stabilizer (data not shown). Interestingly, osmotic stabilization and high temperature restored wt-like extension of submerged ΔAnpmtC strain hyphae, which otherwise remained aberrantly swollen and branched out frequently in a random spatial pattern with shorter cells (Fig. 5). Stalks with a vesicle at the hyphal tip developed after 40 h at 42°C in liquid cultures under conditions of osmotic stabilization. Conidiophores were not observed in aerial hyphae of the ΔAnpmtC strain unless the cultures were grown on plates containing an osmotic stabilizer, and conidia were formed only at 42°C despite an aberrant conidiophore structure containing several clusters of sterigmata and conidia without vesicles. Conidium formation was reduced to 6% of wt levels under these conditions. Despite osmotic stabilization, conidia were not produced from sterigmata at 30°C. The CΔAnpmtC strain carrying wt pmtC at the pyrG locus in the ΔAnpmtC strain showed a phenotype identical to that of the wt strain with respect to colony formation, hyphal morphology, and conidiation (Fig. 3 and 5), confirming that disruption of AnpmtC affected only the function of pmtC.
FIG. 5.
(Bottom) Hyphal morphology of the ΔAnpmtC strain grown in YG liquid medium and on an MM plate with or without 0.6 M KCl. (Top) The wt and CΔAnpmtC strains were grown in liquid MMU (MM supplemented with 5 mM uracil and 5 mM uridine) (submerged hyphae) and on an MMU plate (aerial hyphae).
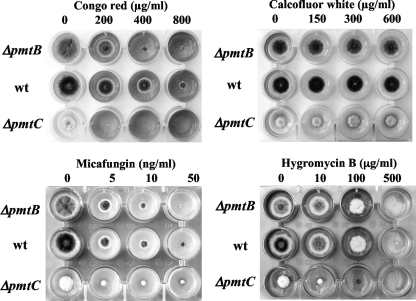
Sensitivity to antifungal reagents.
All Anpmt disruptants differed from each other and from the wt in their morphologies. Osmotic stabilization of the media helped to reduce many of these defects, suggesting that the cell wall no longer fully contributed to maintaining the proper architecture of these filamentous fungi. Since Congo red, micafungin, and calcofluor white (CFW) are known to inhibit cell wall synthesis, we determined the sensitivities of pmt disruptants to these compounds (Fig. 6). Compared to the wt strain, both the ΔAnpmtB and ΔAnpmtC strains were more sensitive to Congo red and micafungin, which inhibit the production of β-glucans and β-1,3-glucans. In contrast, and unlike the hypersensitive ΔAnpmtA strain (26), neither the ΔAnpmtB nor the ΔAnpmtC strain showed sensitivity to CFW, which inhibits chitin synthesis. Glycosylation mutants of yeast are hypersensitive to hygromycin B (HygB), probably due to increased permeability of the cell wall. Indeed, we previously found that the ΔAnpmtA strain was hypersensitive to HygB (data not shown). Of the other Anpmt disruptants, only the ΔAnpmtC strain was more sensitive to HygB than the wt strain. The ΔAnpmtB strain, however, did not show any sensitivity to HygB.
FIG. 6.
Effects of antifungal reagents on colony formation by the wt, ΔAnpmtB, and ΔAnpmtC strains. The strains were grown in MM in the presence of Congo red, CFW, micafungin, and HygB, as indicated, for 3 days at 30°C.
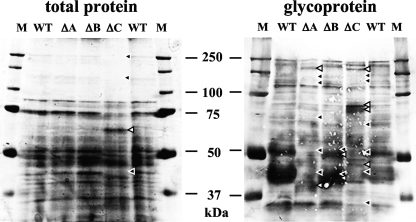
Glycoprotein profiles of Anpmt disruptants.
To obtain a better understanding of the substrate specificities of individual AnPmts, we compared the glycoprotein profiles from the three Anpmt disruptants on SDS-PAGE (Fig. 7). Secretory proteins that are subjected to protein glycosylation often localize to the plasma membrane. We therefore prepared membrane proteins extracted by Triton X-100 and analyzed them by staining and lectin blotting. Staining with CBB revealed comparable sets of membrane proteins from the wt, ΔAnpmtA, and ΔAnpmtB strains. In the protein set of the ΔAnpmtC strain, however, proteins larger than 100 kDa and with a mass of about 75 kDa were reduced, whereas proteins in the region of 65 kDa appeared to be more abundant. Lectin blotting with concanavalin A revealed an increase in mannose-containing glycoproteins of about 80 kDa in the ΔAnpmtC strain and a decrease in those of about 40 kDa in the ΔAnpmtC strain. The glycoprotein profile of the ΔAnpmtB strain was almost indistinguishable from the wt profile, suggesting a minor role in glycosylation of proteins localizing to the plasma membrane.
FIG. 7.
Comparison of soluble membrane proteins and glycoproteins. Proteins extracted with 1% Triton X-100 were loaded onto SDS-PAGE. Total proteins were stained with CBB, and the glycoproteins were detected by lectin blot analysis with concanavalin A as described in Materials and Methods. WT, ΔA, ΔB, and ΔC indicate proteins from wt, ΔAnpmtA, ΔAnpmtB, and ΔAnpmtC cells, respectively. Lanes M contained Precision plus protein standards used as molecular mass markers. Proteins that appeared in the wt but disappeared in the ΔAnpmt strain are indicated by black arrowheads; proteins that appeared in the ΔAnpmt strain but not in the wt are indicated by white arrowheads.
Substrate specificities of AnPmts.
In S. cerevisiae, ScPmt2 and ScPmt4 mannosylate the plasma membrane proteins ScWsc1 and ScMid2, which function as cell wall stress sensors (20). We tested whether the A. nidulans PMT2 (AnPmtA) and PMT4 (AnPmtC) proteins had comparable substrate specificities and therefore cloned the gene encoding AN5660.3 (termed AnWscA), which we identified in the A. nidulans genome as a homolog of ScWsc1. AnWscA is composed of 280 amino acids and shares 31.2% amino acid identity with ScWsc1. The protein has an N-terminal signal sequence of 24 amino acids, as predicted by the SignalIP program, and contains a Wsc motif (amino acids 25 to 123) rich in cysteine residues, a serine- and threonine-rich region (amino acids 124 to 186), a transmembrane region (amino acids 187 to 210), and a cytoplasmic domain at the C terminus (amino acids 211 to 280) (Fig. 8A). Three putative N-glycosylation sites are found at Asn135, Asn176, and Asn258, while 31 (19 serines and 13 threonines) out of the 63 residues of the serine/threonine-rich region can serve as O-glycosylation sites.
We expressed a 3HA-tagged version of AnwscA with the HA epitope tag attached to the C terminus in the wt and the Anpmt disruptants and assessed the extents of glycosylation by comparing the gel mobilities of the tagged proteins (Fig. 8B). The wt strain produced AnWscA-3HA proteins with an apparent molecular mass of 50 kDa, which is higher than the calculated molecular mass of 33.5 kDa due to N and O glycosylations. Deletion of the A. nidulans PMT1 protein in the ΔAnpmtB strain did not affect the mobility of the tagged protein on SDS-PAGE. However, absence of the PMT2- and PMT4-like proteins in the ΔAnpmtA and ΔAnpmtC strains caused slightly faster mobility of AnWscA-3HA, presumably due to underglycosylation of the tagged protein. In addition, several bands of around 20 to 25 kDa were detected in these disruptants, but not in the wt strain or when AnPmtB was absent, suggesting that glycosylation protects the protein from N-terminal degradation. These results indicate that AnWsc-3HA is a natural substrate of AnPmtA and AnPmtC, supporting their classification as PMT2 and PMT4 proteins.
DISCUSSION
Proteins going through the secretory pathway are posttranslationally modified by O glycosylation, which is generally protein O mannosylation in fungi. In Saccharomyces, Schizosaccharomyces, Candida, Cryptococcus, Trichoderma, and Aspergillus, initial protein O mannosylation is catalyzed by Pmts (9). Accordingly, all three pmt genes of A. nidulans contribute to normal hyphal development and are expressed throughout growth, implying that protein O mannosylation plays important roles for this fungal strain. We previously characterized AnpmtA and AapmtA genes encoding PmtA belonging to the PMT2 subfamily (26, 27). Here we characterized two Anpmt genes encoding AnPmtB and AnPmtC belonging to the PMT1 and PMT4 subfamilies, respectively. In A. nidulans, the phenotypes caused by gene disruption of AnpmtA and AnpmtB are different, and those of the double disruptant are cumulative with respect to each single pmt disruptant, suggesting that unlike most yeast PMT1 and PMT2 subfamily proteins, the corresponding AnPmtB and AnPmtA proteins in A. nidulans function in an independent manner. Interestingly, ScPmt6, the third PMT2 protein in S. cerevisiae, also does not behave as a canonical PMT protein in the sense that no interactions with other Pmts or with itself have been observed (7).
We attempted to determine the in vivo substrate specificities of AnPmt proteins by assessing the extent of glycosylation of GAI. Underglycosylation of GAI in the absence of AnPmtA was demonstrated previously (26). Strains in which either AnpmtB or AnpmtC was disrupted also secreted underglycosylated GAI, indicating that the three AnPmts share substrate specificity for GAI and are involved in the mannosylation of the high number of hydroxyamino acids.
In contrast, AnPmtB did not significantly contribute to the glycosylation of AnWscA-3HA. However, in the absence of either AnpmtA or AnPmtC, AnWscA mannosylation was affected, and two major protein bands with molecular masses between 20 and 25 kDa were formed, suggesting that proteolytic cleavage had occurred at around the middle of the protein, which corresponds to the serine/threonine-rich region. Thus, AnPmtB has a substrate specificity different from that of AnPmtA and AnPmtC. In S. cerevisiae, Wsc1, Wsc3, and Mid2, which act as the cell wall sensors, were determined to be substrates for ScPmt1-ScPmt2 complexes and ScPmt4 protein (20). In particular, ScPmt4 preferentially mannosylates the Ser/Thr-rich region flanked by a membrane anchor of secretory proteins (14). Thus, the substrate specificities toward Wsc proteins are conserved between the same PMT subfamily proteins of Saccharomyces and Aspergillus.
Disruption of pmt genes in Aspergillus led to phenotypes with pleiotropic abnormalities. As in the case of AnWscA, underglycosylation by a defect in Pmt activity may lead in general to proteolytic cleavage within the serine/threonine-rich regions of the Pmt substrates and subsequent underrepresentation of active versions of these proteins at their sites of action. In the absence of either the A. nidulans PMT2 protein AnPmtA or A. fumigatus PMT1, the mutant fungi lost their cell wall integrity, resulting in repressed colony formation (26, 46). AnpmtA and Afpmt1 disruptants were therefore also hypersensitive to high temperature, CFW, and HygB. In contrast, AnpmtB disruption did not significantly affect colony formation or make the fungus hypersensitive to CFW and HygB despite the high homology between AnPmtB and AfPmt1, which argues that they belong to the same PMT1 subfamily. While the AnpmtB disruptant was further sensitive toward high concentrations of Congo red or micafungin, the major phenotype characterizing this mutant was its highly branched hyphae. Therefore, presumably due to the absence of a protein normally glycosylated by AnPmtB, the mechanism by which the germination site for a new hypha is determined is no longer properly regulated. Thus, there is a possibility that AnPmtB is involved in polarity maintenance. Since the disruption of Afpmt1 does not affect the polarized growth of A. fumigatus, this strengthens our finding that PMT1 subfamily proteins of A. nidulans and A. fumigatus have different functions. Not only the disruption of AnpmtB, but also that of Angmt and AfmsdS genes involved in protein glycosylation, caused abnormal polarity (16, 17). Thus, some downstream, but unidentified, glycoproteins seem to control hyphal polarity.
Of the three analyzed AnPmts, AnPmtC appeared to be the most essential, as the absence of the protein caused the severest growth defect in terms of a repressed growth rate and aberrant morphology. The vital role AnPmtC plays for hyphal development and morphogenesis resembles that of AnChsB, a class III chitin synthase involved in the synthesis of cell wall chitin during hyphal growth and conidiation (2, 43). AnchsB mutants grow as minute colonies, form hyphae with a very high degree of branching, and cannot conidiate (15), a phenotype very similar to that of the AnPmtC disruptant. As AnChsB is a membrane protein of 916 amino acids that contains a total of 130 serine and threonine residues, it is tempting to speculate that the protein is a specific substrate for AnPmtC or interacts closely with one. Interestingly, a null mutant for another chitin synthase gene, AncsmB, forms abnormally branched conidiophores (37). Conidiophores generated in the absence of AnPmtC were also abnormal, with several clusters of sterigmata and conidia without vesicle formation.
Hyphal polarity, as well as cell wall integrity, is closely associated with the synthesis and degradation of α- and β-glucans and chitin. A. nidulans chiA encodes a class III chitinase with a Ser/Thr/Pro-rich region and a glycosylphosphatidylinositol anchor attachment motif. AnChiA is heavily O glycosylated and localizes at hyphal branching sites (42); however, disruption of the gene did not affect the hyphal and conidiophore morphology, as we observed in the absence of AnPmtA or AnPmtC, but decreased the hyphal growth rate (36). In S. cerevisiae, β-1,3-glucanosyltransferase (ScGas1), which is localized at the cell surface via a glycosylphosphatidylinositol anchor, is a substrate of ScPmt4 and ScPmt6 (5, 39). The Scgas1 null mutation resulted in defective cell wall architecture. A. fumigatus genes homologous to ScGAS1 were found to be Afgel1 and Afgel2. Disruption of Afgel1 did not cause a phenotype, but the Afgel2 disruptant exhibited slower growth than the wt, abnormal conidiogenesis, and altered cell wall composition (22). Both proteins contain a serine/threonine-rich region near the C terminus, and it will be interesting to know which AnPmts glycosylate these proteins.
An increasing number of genes responsible for synthesis and degradation of the cell wall and polarity establishment and maintenance have been identified in filamentous fungi. However, the localization and glycosylation of most of these proteins remain to be characterized. We are currently in the process of identifying these target proteins using the pmt disruptants, hoping to reveal how the glycoproteins maintain fungal morphology, hyphal development, and differentiation.
Supplementary Material
Acknowledgments
This work was supported in part by a Ministry of Education, Science, Sports, and Culture Grant-in-Aid for Scientific Research (C).
Footnotes
Published ahead of print on 31 July 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Aramayo, R., T. H. Adams, and W. E. Timberlake. 1989. A large cluster of highly expressed genes is dispensable for growth and development in Aspergillus nidulans. Genetics 122:65-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borgia, P. T., N. Iartchouk, P. J. Riggle, K. R. Winter, Y. Koltin, and C. E. Bulawa. 1996. The chsB gene of Aspergillus nidulans is necessary for normal hyphal growth and development. Fungal Genet. Biol. 20:193-203. [DOI] [PubMed] [Google Scholar]
- 3.Burda, P., and M. Aebi. 1999. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426:239-257. [DOI] [PubMed] [Google Scholar]
- 4.Gentzsch, M., and W. Tanner. 1996. The PMT gene family: protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J. 15:5752-5759. [PMC free article] [PubMed] [Google Scholar]
- 5.Gentzsch, M., and W. Tanner. 1997. Protein-O-glycosylation in yeast: protein-specific mannosyltransferases. Glycobiology 7:481-486. [DOI] [PubMed] [Google Scholar]
- 6.Girrbach, V., T. Zeller, M. Priesmeier, and S. Strahl-Bolsinger. 2000. Structure-function analysis of the dolichyl phosphate-mannose: protein O-mannosyltransferase ScPmt1p. J. Biol. Chem. 275:19288-19296. [DOI] [PubMed] [Google Scholar]
- 7.Girrbach, V., and S. Strahl. 2003. Members of the evolutionarily conserved PMT family of protein O-mannosyltransferases form distinct protein complexes among themselves. J. Biol. Chem. 278:12554-12562. [DOI] [PubMed] [Google Scholar]
- 8.Goto, M. 2007. Protein O-glycosylation in fungi: diverse structures and multiple functions. Biosci. Biotechnol. Biochem. 71:1415-1427. [DOI] [PubMed] [Google Scholar]
- 9.Goto, M., K. Ekino, and K. Furukawa. 1997. Expression and functional analysis of a hyperglycosylated glucoamylase in a parental host, Aspergillus awamori var. kawachi. Appl. Environ. Microbiol. 63:2940-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gravel, P. 2002. Identification of glycoproteins on nitrocellulose membranes using lectin blotting, p. 779-793. In J. M. Walker (ed.), The protein protocols handbook, 2nd ed. Humana Press Inc., Totowa, NJ.
- 11.Harty, C., S. Strahl, and K. Römisch. 2001. O-Mannosylation protects mutant alpha-factor precursor from endoplasmic reticulum-associated degradation. Mol. Biol. Cell 12:1093-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashida, S., K. Kuroda, K. Ohta, S. Kuhara, K. Fukuda, and Y. Sakaki. 1989. Molecular cloning of the glucoamylase I gene of Aspergillus awamori var. kawachi for localization of the raw-starch affinity site. Agric. Biol. Chem. 53:923-929. [Google Scholar]
- 13.Hirayama, H., M. Fujita, T. Yoko-o, and Y. Jigami. 2008. O-Mannosylation is required for degradation of the endoplasmic reticulum-associated degradation substrate Gas1*p via the ubiquitin/proteasome pathway in Saccharomyces cerevisiae. J. Biochem. 143:555-567. [DOI] [PubMed] [Google Scholar]
- 14.Hutzler, J., M. Schmid, T. Bernard, B. Henrissat, and S. Strahl. 2007. Membrane association is a determinant for substrate recognition by PMT4 protein O-mannosyltransferases. Proc. Natl. Acad. Sci. USA 104:7827-7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ichinomiya, M., T. Motoyama, M. Fujiwara, M. Takagi, H. Horiuchi, and A. Ohta. 2002. Repression of chsB expression reveals the functional importance of class IV chitin synthase gene chsD in hyphal growth and conidiation of Aspergillus nidulans. Microbiology 148:1335-1347. [DOI] [PubMed] [Google Scholar]
- 16.Jackson-Hayes, L., T. W. Hill, D. M. Loprete, L. M. Fay, B. S. Gordon, S. A. Nkashama, R. K. Patel, and C. V. Sartain. 2008. Two GDP-mannose transporters contribute to hyphal form and cell wall integrity in Aspergillus nidulans. Microbiology 154:2037-2047. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, H., H. Ouyang, H. Zhou, and C. Jin. 2008. GDP-mannose pyrophosphorylase is essential for cell wall integrity, morphogenesis and viability of Aspergillus fumigatus. Microbiology 154:2730-2739. [DOI] [PubMed] [Google Scholar]
- 18.Kuwayama, H., S. Obara, T. Morio, M. Katoh, H. Urushihara, and Y. Tanaka. 2002. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 30:E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehle, L., S. Strahl, and W. Tanner. 2006. Protein glycosylation, conserved from yeast to man: a model organism helps elucidate congenital human diseases. Angew. Chem. Int. Ed. Engl. 45:6802-6818. [DOI] [PubMed] [Google Scholar]
- 20.Lommel, M., M. Bagnat, and S. Strahl. 2004. Aberrant processing of the WSC family and Mid2p cell surface sensors results in cell death of Saccharomyces cerevisiae O-mannosylation mutants. Mol. Cell. Biol. 24:46-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lussier, M., A. M. Sdicu, and H. Bussey. 1999. The KTR and MNN1 mannosyltransferase families of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1426:323-334. [DOI] [PubMed] [Google Scholar]
- 22.Mouyna, I., W. Morelle, M. Vai, M. Monod, B. Léchenne, T. Fontaine, A. Beauvais, J. Sarfati, M. C. Prévost, C. Henry, and J. P. Latgé. 2005. Deletion of GEL2 encoding for a β(1-3)glucanosyltransferase affects morphogenesis and virulence in Aspergillus fumigatus. Mol. Microbiol. 56:1675-1688. [DOI] [PubMed] [Google Scholar]
- 23.Nakatsukasa, K., S. Okada, K. Umebayashi, R. Fukuda, S. Nishikawa, and T. Endo. 2004. Roles of O-mannosylation of aberrant proteins in reduction of the load for endoplasmic reticulum chaperones in yeast. J. Biol. Chem. 279:49762-49772. [DOI] [PubMed] [Google Scholar]
- 24.Nayak, T., E. Szewczyk, C. E. Oakley, A. Osmani, L. Ukil, S. L. Murray, M. J. Hynes, S. A. Osmani, and B. R. Oakley. 2006. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172:1557-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ninomiya, Y., K. Suzuki, C. Ishii, and H. Inoue. 2004. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 101:12248-12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oka, T., T. Hamaguchi, Y. Sameshima, M. Goto, and K. Furukawa. 2004. Molecular characterization of protein O-mannosyltransferase and its involvement in cell-wall synthesis in Aspergillus nidulans. Microbiology 150:1973-1982. [DOI] [PubMed] [Google Scholar]
- 27.Oka, T., Y. Sameshima, T. Koga, H. Kim, M. Goto, and K. Furukawa. 2005. Protein O-mannosyltransferase A of Aspergillus awamori is involved in O-mannosylation of glucoamylase I. Microbiology 151:3657-3667. [DOI] [PubMed] [Google Scholar]
- 28.Olson, G. M., D. S. Fox, P. Wang, J. A. Alspaugh, and K. L. Buchanan. 2007. Role of protein O-mannosyltransferase Pmt4 in the morphogenesis and virulence of Cryptococcus neoformans. Eukaryot. Cell 6:222-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prill, S. K., B. Klinkert, C. Timpel, C. A. Gale, K. Schroppel, and J. F. Ernst. 2005. PMT family of Candida albicans: five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol. Microbiol. 55:546-560. [DOI] [PubMed] [Google Scholar]
- 30.Proszynski, T. J., K. Simons, and M. Bagnat. 2004. O-Glycosylation as a sorting determinant for cell surface delivery in yeast. Mol. Biol. Cell 15:1533-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romero, P. A., M. Lussier, S. Veronneau, A. M. Sdicu, A. Herscovics, and H. Bussey. 1999. Mnt2p and Mnt3p of Saccharomyces cerevisiae are members of the Mnn1p family of α-1,3-mannosyltransferases responsible for adding the terminal mannose residues of O-linked oligosaccharides. Glycobiology 9:1045-1051. [DOI] [PubMed] [Google Scholar]
- 32.Sanders, S. L., M. Gentzsch, W. Tanner, and I. Herskowitz. 1999. O-Glycosylation of Axl2/Bud10p by Pmt4p is required for its stability, localization, and function in daughter cells. J. Cell Biol. 145:1177-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw, B. D., and M. Momany. 2002. Aspergillus nidulans polarity mutant swoA is complemented by protein O-mannosyltransferase pmtA. Fungal Genet. Biol. 37:263-270. [DOI] [PubMed] [Google Scholar]
- 34.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy. Freeman, San Francisco, CA.
- 35.Strahl-Bolsinger, S., M. Gentzsch, and W. Tanner. 1999. Protein O-mannosylation. Biochim. Biophys. Acta 1426:297-307. [DOI] [PubMed] [Google Scholar]
- 36.Takaya, N., D. Yamazaki, H. Horiuchi, A. Ohta, and M. Takagi. 1998. Cloning and characterization of a chitinase-encoding gene (chiA) from Aspergillus nidulans, disruption of which decreases germination frequency and hyphal growth. Biosci. Biotechnol. Biochem. 62:60-65. [DOI] [PubMed] [Google Scholar]
- 37.Takeshita, N., S. Yamashita, A. Ohta, and H. Horiuchi. 2006. Aspergillus nidulans class V and VI chitin synthases CsmA and CsmB, each with a myosin motor-like domain, perform compensatory functions that are essential for hyphal tip growth. Mol. Microbiol. 59:1380-1394. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka, N., Y. Fujita, S. Suzuki, M. Morishita, Y. Giga-Hama, C. Shimoda, and K. Takegawa. 2005. Characterization of O-mannosyltransferase family in Schizosaccharomyces pombe. Biochem. Biophys. Res. Commun. 330:813-820. [DOI] [PubMed] [Google Scholar]
- 39.Vai, M., E. Gatti, E. Lacanà, L. Popolo, and L. Alberghina. 1991. Isolation and deduced amino acid sequence of the gene encoding gp115, a yeast glycophospholipid-anchored protein containing a serine-rich region. J. Biol. Chem. 266:12242-12248. [PubMed] [Google Scholar]
- 40.Weber, Y., S. K. Prill, and J. F. Ernst. 2004. Pmt-mediated O mannosylation stabilizes an essential component of the secretory apparatus, Sec20p, in Candida albicans. Eukaryot. Cell 3:1164-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willer, T., M. Brandl, M. Sipiczki, and S. Strahl. 2005. Protein O-mannosylation is crucial for cell wall integrity, septation and viability in fission yeast. Mol. Microbiol. 57:156-170. [DOI] [PubMed] [Google Scholar]
- 42.Yamazaki, H., A. Tanaka, J. Kaneko, A. Ohta, and H. Horiuchi. 2008. Aspergillus nidulans ChiA is a glycosylphosphatidylinositol (GPI)-anchored chitinase specifically localized at polarized growth sites. Fungal Genet. Biol. 45:963-972. [DOI] [PubMed] [Google Scholar]
- 43.Yanai, K., N. Kojima, N. Takaya, H. Horiuchi, A. Ohta, and M. Takagi. 1994. Isolation and characterization of two chitin synthase genes from Aspergillus nidulans. Biosci. Biotechnol. Biochem. 58:1828-1835. [DOI] [PubMed] [Google Scholar]
- 44.Yelton, M. M., J. E. Hamer, and W. E. Timberlake. 1984. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc. Natl. Acad. Sci. USA 81:1470-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yip, C. L., S. K. Welch, F. Klebl, T. Gilbert, P. Seidel, F. J. Grant, P. J. O'Hara, and V. L. MacKay. 1994. Cloning and analysis of the Saccharomyces cerevisiae MNN9 and MNN1 genes required for complex glycosylation of secreted proteins. Proc. Natl. Acad. Sci. USA 91:2723-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou, H., H. Hu, L. Zhang, R. Li, H. Ouyang, J. Ming, and C. Jin. 2007. O-Mannosyltransferase 1 in Aspergillus fumigatus (AfPmt1p) is crucial for cell wall integrity and conidium morphology, especially at an elevated temperature. Eukaryot. Cell 6:2260-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.