Abstract
Dense core granules (DCGs) in Tetrahymena thermophila contain two protein classes. Proteins in the first class, called granule lattice (Grl), coassemble to form a crystalline lattice within the granule lumen. Lattice expansion acts as a propulsive mechanism during DCG release, and Grl proteins are essential for efficient exocytosis. The second protein class, defined by a C-terminal β/γ-crystallin domain, is poorly understood. Here, we have analyzed the function and sorting of Grt1p (granule tip), which was previously identified as an abundant protein in this family. Cells lacking all copies of GRT1, together with the closely related GRT2, accumulate wild-type levels of docked DCGs. Unlike cells disrupted in any of the major GRL genes, ΔGRT1 ΔGRT2 cells show no defect in secretion, indicating that neither exocytic fusion nor core expansion depends on GRT1. These results suggest that Grl protein sorting to DCGs is independent of Grt proteins. Consistent with this, the granule core lattice in ΔGRT1 ΔGRT2 cells appears identical to that in wild-type cells by electron microscopy, and the only biochemical component visibly absent is Grt1p itself. Moreover, gel filtration showed that Grl and Grt proteins in cell homogenates exist in nonoverlapping complexes, and affinity-isolated Grt1p complexes do not contain Grl proteins. These data demonstrate that two major classes of proteins in Tetrahymena DCGs are likely to be independently transported during DCG biosynthesis and play distinct roles in granule function. The role of Grt1p may primarily be postexocytic; consistent with this idea, DCG contents from ΔGRT1 ΔGRT2 cells appear less adhesive than those from the wild type.
In eukaryotes, the directional transport of lumenal proteins throughout the network of membrane-bound organelles depends on reversible assembly of multisubunit protein complexes in the cytoplasm. For example, the assembly of a localized clathrin coat at a cell's surface facilitates both the concentration of specific transmembrane receptors together with their bound ligands at that site and the invagination and budding of the plasma membrane, resulting in endocytosis (18). Similarly, other cytosolic coats assemble and direct traffic at the endoplasmic reticulum (ER) and Golgi apparatus (4). For one protein trafficking pathway in eukaryotic cells, however, the determinative protein self-assembly occurs not in the cytoplasm but within the lumen of the secretory pathway itself. Dense core granules (DCGs) are secretory vesicles whose lumenal cargo consists of a condensed polypeptide aggregate. This cargo is secreted when the vesicles fuse with the plasma membrane in response to a specific extracellular stimulus, an event called regulated exocytosis. The aggregation of the cargo occurs progressively within the secretory pathway, beginning in the trans-Golgi network (TGN), and may be promoted by multiple factors including compartment-specific proton and calcium levels (23). Aggregation facilitates the vesicular storage of concentrated secretory proteins but also serves as a sorting mechanism to segregate DCG proteins from proteins that are secreted via other pathways. Evidence for this mechanism includes in vitro experiments showing that some proteins released via constitutive exocytosis remain soluble under TGN-like conditions that promote DCG protein aggregation (10). In vivo, sorting would result if aggregated and soluble proteins exit the TGN in different carriers. Importantly, there is no evidence that sorting of DCG proteins at the TGN requires assembly of cytosolic coat complexes.
While aggregative sorting represents an attractively simple mechanism, relatively little is known about the structure or dynamic properties of the aggregates themselves. This is an interesting issue, as illustrated by several phenomena. First, aggregates in some cell types, like those formed by proinsulin in pancreatic β cells, can become reordered as protein crystals during a multistage process called granule maturation (13). Second, Aplysia bag cells can sort different subsets of DCG proteins into distinct granules, suggesting that aggregation can be finely regulated and that different aggregates have different properties in vivo (20). Both of these phenomena have also been observed within the DCGs of unicellular ciliates (3, 14). In addition, ciliate DCGs demonstrate another degree of subtlety in DCG formation because the granule cores in many of these organisms are divided into distinct domains (25). The domain organization indicates that DCG proteins in these cells can segregate from one another even as they are sorted to the same vesicular destination. While the structures of DCGs in many ciliates have been captured by electron microscopy, molecular studies have advanced in two species, Tetrahymena thermophila and Paramecium tetraurelia (30, 33).
In many ciliates, the individual DCGs are organized in at least two distinct domains within the lumen. First, the bulk of the cargo is organized as a core crystal that expands, spring-like, upon exocytosis (28). This expansion can drive rapid extrusion of the DCG contents, which may be essential for hunting or defensive behaviors (17). In addition, many ciliate DCGs possess a single polarized tip structure that is involved in DCG docking to the plasma membrane and exocytic fusion (25). These tip structures are also filled with condensed, highly organized proteins, which appear by both genetic and morphological criteria to be different from proteins making up the expansible core (1, 21). The proteins that form the distinct domains are beginning to be identified and analyzed. Those that constitute the expansible springs are encoded by homologous families of genes named GRL (granule lattice) in Tetrahymena and tmp (trichocyst matrix) in Paramecium (11, 12, 15). Assembly of Grl proteins begins in the ER with formation of heterooligomers. This is an obligatory step, as shown by the fact that deletion of individual Grl proteins by targeted gene disruption resulted in the ER retention of remaining Grl proteins (12). Further assembly of Grl proteins to form a crystal occurs during DCG maturation and is accompanied by site-specific proprotein processing (34). Upon exocytosis, the expansion of the crystalline core is controlled by calcium binding to the fully processed Grl proteins (34).
In addition to the GRL family-encoded proteins, 13 other lumenal DCG proteins have been putatively or definitively identified in Tetrahymena, and homologous proteins are predicted in the Paramecium genome (6). The entire set belongs to a gene family that is defined by a carboxy-terminal β/γ-crystallin domain, which may function as a DCG-targeting motif (16). Studies of two different members of this family in Tetrahymena, IGR1 (induced during granule regeneration 1) and GRT1 (granule tip 1), suggested that these proteins are functionally distinct from the spring-forming Grl proteins. First, whereas gene disruption of any of the highly transcribed GRL genes resulted in grossly aberrant spring formation, no such defect was seen upon disruption of IGR1 (16). However, this could be explained by the fact that IGR1 encodes a relatively low-abundance protein in DCGs, and furthermore its function could be redundant with that of the highly related gene, IGR2.
The second protein in the β/γ-crystallin domain family that has been investigated is the 80-kDa product of the GRT1 gene. Grt1p was first detected as one of the most abundant DCG components released during exocytosis (32). Biochemical analysis showed that Grt1p differs in its solubility from the Grl proteins and also that it is packaged intact in DCGs rather than undergoing proteolytic processing (31). Since processing is essential for Grl protein assembly and function, this difference appears highly significant. Second, Grt1p accumulates at a single pole of each DCG, corresponding to the tip of the organelle that docks and then fuses with the plasma membrane (5). Two Mendelian mutants with defects in DCG maturation show delocalized Grt1p, and these mutant DCGs can dock but do not appear to undergo exocytosis (5). These results suggested that Grt1p might be involved in forming a DCG tip domain that interacted with the plasma membrane.
We have now investigated the trafficking and function of Grt1p. Our data provide both direct biochemical and cell-biological evidence that Grt1p and Grl proteins form distinct complexes during DCG biogenesis in Tetrahymena. Together with earlier results, our experiments provide genetic evidence that Grl and Grt complexes can be independently trafficked to DCGs. Cells lacking GRT1, together with the closely related GRT2, still show rapid and efficient release of DCG contents upon stimulation with secretagogues, but the released DCG contents are subtly different from those of the wild type, suggesting that Grt1p may primarily serve a postexocytic function.
MATERIALS AND METHODS
Tetrahymena strains and culture conditions.
Wild-type CU428.1 T. thermophila (from Peter Bruns, Cornell University) and the SB281 mutant (from Eduardo Orias, University of California—Santa Barbara) were grown at 30°C in SPP medium (1% proteose peptone [BD, Sparks, MD] 0.2% dextrose, 0.1% yeast extract, 0.009% ferric EDTA). All reagents were from Sigma Chemical Co. unless otherwise indicated. Cell densities were measured using a Coulter Counter (Coulter Electronics Limited, Luton, United Kingdom).
GRT1 and GRT2 disruption.
A fragment containing the entire coding region of GRT1 and part of GRT2 (residues 1 to 2467), together with the intergenic sequence, was replaced in a genomic clone with the neo3 cassett, using EcoRI sites added to primers on both the 5′ and 3′ ends. The neo3 cassette includes a gene encoding paromomycin resistance under the control of the cadmium-inducible MTT1 promoter (27). The resulting construct was linearized and introduced by biolistic bombardment into vegetative Tetrahymena by particle bombardment to replace the corresponding endogenous sequence by homologous recombination (9). The replacement was driven to completion to generate GRT1/GRT2 double-knockout cells (ΔGRT1 ΔGRT2) by growing the initial transformants in the presence of increasing concentrations of paromomycin (up to 1 mg/ml) with 0.5 μg/ml cadmium chloride.
Southern blotting.
Genomic DNA samples from wild-type and ΔGRT1 ΔGRT2 cells were digested with NcoI and probed with an oligonucleotide corresponding to the 5′ flank of GRT1 (nucleotide positions −1511 to −42, relative to the translation start).
Western blotting.
Protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to 0.45-μM-pore-size nitrocellulose (Osmonics, Westborough, MA), under previously described conditions (7). Individual blots were incubated with polyclonal antibodies against either Grt1p (1:500), Grl1p (1:1,000) (32), Grl3p(1:400), or Grl4p (1:250) (12); proteins were detected either using horseradish peroxidase-conjugated goat anti-rabbit antibody at 1:2,000 (Jackson ImmunoResearch Laboratories, West Grove, PA), followed by development with Pierce Supersignal (Pierce Chemical, Rockford, IL) and exposure in an α-Imager (Alpha Innotech, San Leandro, CA), or using Alexa Fluor 680 donkey anti-rabbit immunoglobulin G (IgG) at 1:5,000 (Invitrogen, Carlsbad, CA) with an Odyssey infrared imaging system (Li-Cor Biosciences, Lincoln, NE).
Immunofluorescence.
Cells were fixed and immunolabeled as described previously (7). Grt1p and Grl3p were visualized using monoclonal antibody (MAb) 4D11 (20%, vol/vol) (31) and MAb 5E9 (20%, vol/vol) (5), respectively, followed by 1% (vol/vol) Texas Red-conjugated goat anti-mouse antibody (Jackson ImmunoResearch, West Grove, Pennsylvania). Images were obtained and analyzed with a Zeiss LSM 510 confocal microscope and software.
Electron microscopy.
Cells were fixed for 30 min with 3% glutaraldehyde in 0.1 M Na cacodylate buffer, pH 7.2, and then washed three times for 10 min each time with 10% sucrose in 0.1 M Na cacodylate, pH 7.2, and postfixed with 1% osmium tetroxide in the same buffer. After samples were washed with water, they were dehydrated by serial passage through solutions of increasing ethanol concentration, from 20 to 100%, and embedded in Epon resin. After polymerization at 60°C for 24 to 48 h, the embedded samples were cut to yield 100-nm sections, which were collected on formvar-coated copper grids and poststained in 2% uranyl acetate and Reynold's lead citrate. Electron micrographs were collected on an FEI Tecnai F30 electron microscope interfaced with a high-performance charge-coupled-device camera.
Dibucaine stimulation and purification of DCG contents.
Wild-type or ΔGRT1 ΔGRT2 cells were grown to 4 × 105 to 6 × 105 cells/ml and washed once with 10 mM Na HEPES (pH 7.2) after being pelleted for 30 s at 400 × g in a clinical centrifuge. Loose cell pellets (concentrated ∼10-fold relative to the initial culture) were stimulated for 30 s by addition of 2.5 mM dibucaine (from a 25 mM stock). The cells were then diluted at least fivefold with buffer and centrifuged at 1,200 × g for 1 min. This resulted in a two-layer pellet, with cells overlaid by flocculent extruded DCG contents. For experiments requiring purification of the DCG contents, the flocculent layer was gently removed with a pipette and dispersed thoroughly, and the centrifugation process was then repeated four to six times until the flocculent contained no visible trapped cells. The final pelleting of flocculent to remove trapped cells was at 10,000 × g for 5 min. The final flocculent was resuspended in buffer containing 1% SDS for electrophoretic analysis. Samples were incubated for 3 min at 100°C, separated by SDS-PAGE, and stained with Coomassie blue G250, and images were captured using an α-imager (Alpha Innotech, San Leandro, CA).
Size exclusion chromatography.
Cultures of SB281 (200 ml) (24) were grown overnight to ∼106 cells/ml and washed and resuspended in 5 ml of ice-cold buffer A (20 mM K-HEPES, pH 7.0, 38 mM KCl, 0.3 M sucrose, 2 mM MgCl2, 2 mM EGTA), supplemented with protease inhibitors (10 μg/ml chymostatin, 5 μg/ml leupeptin, 12.5 μg/ml antipain, and 10 μg/ml E64). Cells were lysed by passage through a ball-bearing homogenizer (H&Y Enterprise, Redwood City, CA) with a nominal clearance of 0.0007 in. Lysates were cleared by centrifugation at 500 × g for 10 min, and the resulting supernatants were centrifuged at 150,000 × g for 45 min at 4°C in a TLA 100.4 rotor (Beckman, Fullerton, CA). The pellets were solubilized for 30 min on ice in 0.5% Triton X-100 in buffer A supplemented with protease inhibitors and then centrifuged at 200,000 × g for 45 min using the same rotor. The cleared homogenates were then applied to a Sephacryl S300 fast protein liquid chromatography (FPLC) column (120-ml bed volume) preequilibrated with buffer A and 0.05% Triton X-100. Columns were run under pressure at 0.5 ml/min by FPLC (Bio-Rad), and 1-ml fractions were collected.
Expression and purification of Grt1p-His6.
The coding sequence of GRT1 was amplified using a forward primer containing a PmeI restriction site and a reverse primer containing the coding sequence for His6, followed by a stop codon and an ApaI restriction enzyme site. The amplified product was cloned into the pNCVB vector predigested with these two enzymes. The pNCVB vector was designed to integrate a gene of interest at the MTT1 locus, and transformants can be selected based on the presence of a blasticidin resistance cassette in the 3′ untranslated region (5). The linearized insert was used to transform the SB281 Tetrahymena strain by biolistic bombardment. We used the SB281 line rather than the wild type because wild-type cell lysates have a large pool of insoluble or partially soluble DCG proteins released from DCGs. SB281 transformants were selected and passaged in the presence of 60 μg/ml blasticidin and 1.0 μg/ml CdCl2. To purify Grt1p-His6, we first grew 25-ml transformant cultures overnight in the presence of blasticidin and CdCl2. These were then used to inoculate 400-ml cultures grown overnight to stationary phase (∼106 cells/ml) in the absence of drug or CdCl2. Transgene expression was subsequently induced by treating the 400-ml cultures with 1.0 μg/ml CdCl2 for 6 h at 30°C. The cells were pelleted and disrupted by suspension in 20 ml of ice-cold lysis buffer (50 mM Na-phosphate, pH 7.0, 200 mM NaCl, 1% Triton X-100, 10 mM imidazole), supplemented with protease inhibitors, for 30 min. Lysates were clarified by centrifugation at 150,000 × g for 45 min in a TLA 100.4 rotor (Beckman, Fullerton, CA), and the supernatants were incubated with 200 μl of Talon resin (BD Biosciences, Palo Alto, CA) for 2 h at 4°C with end-to-end shaking. The resin was washed sequentially with 100 volumes of lysis buffer followed by 3 volumes each of lysis buffer containing 40 mm and 60 mM final concentrations of imidazole. Bound protein was eluted with lysis buffer containing 250 mM imidazole. This purification protocol essentially follows that of Cowan et al. (12).
Expression analysis of GRT1 and GRT2.
Transcript levels of GRT1, GRT2, and IGR1 in growing, starved, and conjugating Tetrahymena cells were determined by hybridization to whole-genome microarrays as part of a previous genome-wide expression study (22). The database arising from that study, which is searchable at http://tged.ihb.ac.cn/, was queried to determine the expression profiles of GRT1, GRT2, and IGR1.
RESULTS
Grt1p and Grl1p are found in nonoverlapping transport complexes.
Grl proteins appear to be transported through the Tetrahymena secretory pathway as heterooligomeric complexes (12). To ask whether Grt1p was similarly transported as part of a large complex, we used gel filtration chromatography to resolve Grl1p- and Grt1p-containing complexes present in detergent cell lysates. In order to focus on DCG proteins in transit rather than on the proteins that accumulate in DCGs, we exploited a mutant cell line called SB281 that lacks DCGs and in which newly synthesized DCG proteins are rapidly secreted instead of accumulating in a storage compartment (7). A bulk membrane fraction of SB281 cells was prepared and solubilized in Triton X-100, and the proteins were resolved by gel filtration followed by SDS-PAGE and Western blotting. In these samples Grl1p appears in its proprotein form (pro-Grl1p) since there is no detectible enzymatic or morphological DCG maturation in this mutant (32).
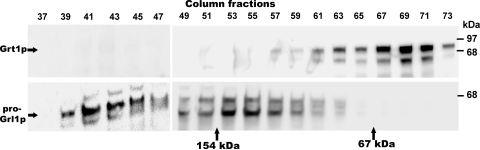
When SB281 lysates were analyzed by gel filtration chromatography, pro-Grl1p was found predominantly in fractions at or near the column void volume (Fig. 1). This is consistent with previous genetic evidence that pro-Grl1p is transported as part of a large oligomeric complex. On the same columns, however, Grt1p was predominantly found in fractions corresponding to much smaller proteins. Indeed, Grt1p, with a monomeric molecular mass of ∼80 kDa, eluted shortly after the bovine serum albumin (BSA) marker (67 kDa). There was no significant cofractionation of Grt1p and Grl1p. This suggests that most of the Grt1p is in a monomeric form not associated with other DCG proteins and that the monomer is very compact.
FIG. 1.
Grt1p and Grl1p are present in distinct transport complexes. A crude membrane fraction from SB281 cells was solubilized in Triton X-100, as described in Materials and Methods, and the cleared solute was applied to a Sephacryl S300 column in the same buffer. A void volume marker, blue dextran, eluted at fraction 39. Beginning with eluted fraction 37, the proteins in alternate fractions were resolved using SDS-PAGE, transferred to nitrocellulose, and Western blotted with polyclonal antibodies against Grt1p (upper panel) or Grl1p (lower panel) on parallel nitrocellulose filters. Fractions 37 to 47 were fitted on one blot, and fractions 49 to 73 were fitted on a separate blot and developed under identical conditions. Grl1p was found predominantly near the column void volume, consistent with formation of large heterooligomeric Grl-based complexes. Grt1p eluted in later column fractions, peaking near fraction 69. Two molecular mass markers were run in parallel, and their peak elution positions are shown. The bacteriophage N4 gp65 monomer (154 kDa) peaked in fraction 52, and BSA (67 kDa) peaked in fraction 66.
Purified Grt1p is not associated with Grl proteins.
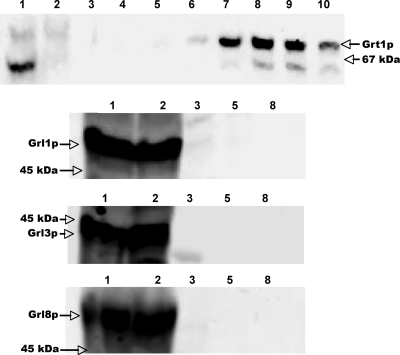
To corroborate these results using a different approach, we fused full-length Grt1p at its C terminus to a His6 tag. The GRT1-His6 construct was introduced on a multicopy plasmid, under the control of an inducible promoter, into the Tetrahymena strain SB281, and the His-tagged protein was purified from induced cell lysates. We previously used this approach to show that His-tagged Grl1p was associated with several other Grl proteins (12). As shown in Fig. 2, the His-tagged Grt1p from SB281 lysates could be readily bound and then eluted from nickel affinity columns (Fig. 2, top panel). In contrast, none of three different Grl proteins bound to the column or coeluted with Grt1p-His6. The results for Grt1p-His6 are consistent with the gel filtration profiles in Fig. 1 and suggest that Grt1p does not interact strongly with Grl proteins under these conditions.
FIG. 2.
Grt1p does not copurify with Grl proteins. Grt1p-His6 was expressed in SB281 cells and purified from detergent lysates by nickel-affinity chromatography as described in Materials and Methods. (Top panel) Nickel column fractions were subjected to SDS-PAGE and analyzed by Western blotting using anti-Grt1p antibodies. The upper band corresponds to Grt1p; the lower band is a cross-reactive species seen in SB281 lysates. Lane 1, lysate applied to column. lane 2, column flowthrough; lanes 3 to 10, elution with increasing concentrations of imidazole, with each fraction representing 1 column volume (fractions 3 and 4, 40 mM; 5 to 7, 60 mM; 8 to 10, 250 mM). Grt1p-His6 elutes primarily in fractions 7 to 9. Arrows on the right indicate the band corresponding to Grt1p and the position of the 67-kDa SDS-PAGE standard, BSA. The remaining three panels show the identical column fractions analyzed by Western blotting with antibodies against Grl1p, Grl3p, and Grl8p. Column fractions correspond to those in the top panel. In all cases, the Grl proteins are found in the column flowthrough and do not copurify with Grt1p-His6.
GRT1 has a closely related paralog, GRT2.
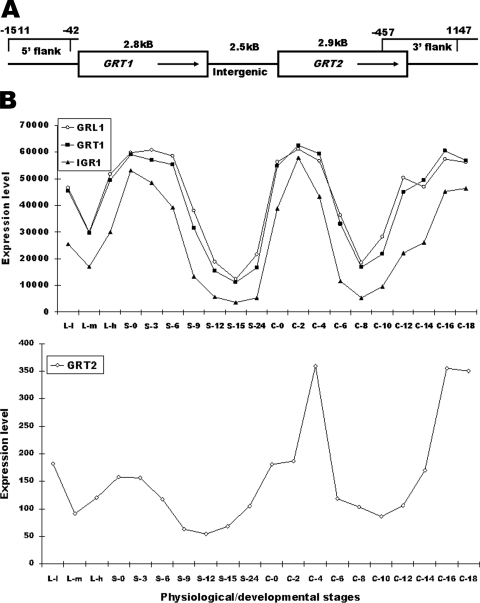
If Grt1p is not associated with Grl proteins, the proteins may be independently sorted to DCGs and play distinct roles within the DCG core. To test the function and sorting properties of Grt1p, we used homologous recombination to disrupt the endogenous gene. However, a BLAST search with the GRT1 coding sequence revealed that a second, very similar gene was present in T. thermophila, which we named GRT2. GRT1 and GRT2 are adjacent to one another on macronuclear chromosome CH445588, with 2.5 kb of intergenic sequence (Fig. 3A) (Tetrahymena genome database at http://ciliate.org). The predicted translation products of GRT1 and GRT2 are 83% identical and 92% similar (see Fig. S1 in the supplemental material).
FIG. 3.
Organization and expression of GRT1 and GRT2. (A) The organization of GRT1 and GRT2 on macronuclear chromosome CH445588. (B) The expression profiles of GRT1 and GRT2 at successive time points in growing (L-l, L-m, and L-h), starved (S-0, S-3, S-6, S-9, S-12, S-15, and S-24), and conjugating (C-0, C-2, C-4, C-6, C-8, C-10, C-12, C-14, C-16, and C-18) cultures, as determined by hybridization of stage-specific cDNAs to whole-genome microarrays. Details on the sampling times are found in Miao et al. (22). Also shown are the profiles for two known DCG protein genes, GRL1 and IGR1.
Based on whole-genome microarray analysis of mRNA isolated from Tetrahymena under a variety of conditions, GRT1 is highly expressed in growing, starved, and conjugating cells (22) (Fig. 3B). The profile of GRT1 expression is very similar to that of other DCG protein genes, including GRL1 and IGR1 for which the profiles are shown in Fig. 3. The microarray data suggest that GRT2 is transcribed at a much lower level than GRT1 under all conditions.
Gene disruption of GRT1 and GRT2.
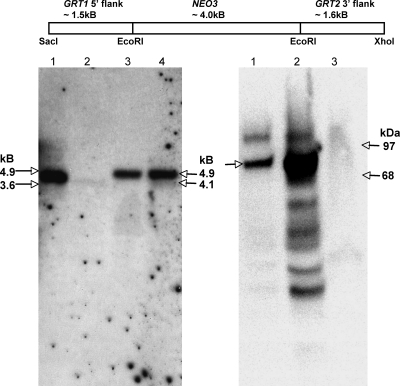
Since GRT1 and GRT2 may under some circumstances have redundant activities, we designed a single replacement construct to disrupt both genes. We transformed vegetative cells to integrate one or a small number of copies of the knockout construct at the macronuclear GRT1/GRT2 locus and passaged the cells in the presence of paromomycin. Under these conditions, the knockout construct replaced all endogenous alleles in the macronucleus, leaving intact only the two micronuclear alleles of GRT1/GRT2 (Fig. 4, left blot). Because there is no vegetative transcription in the micronucleus, the resulting strains have functional deletions of GRT1 and GRT2 (ΔGRT1 ΔGRT2) under the conditions in which the cells were characterized. As expected, these cells accumulated no detectible Grt1p (Fig. 4, right blot). The ability to derive such cells indicates that GRT1 and GRT2 are nonessential, and indeed ΔGRT1 ΔGRT2 cells grew comparably to wild-type cells (data not shown).
FIG. 4.
Macronuclear disruption of GRT1 and GRT2. At top is shown the organization of the construct used to disrupt GRT1 and GRT2 in the macronucleus. The NEO3 cassette confers resistance to paromomycin. Southern blot analysis (left panel) of wild-type cells and two independent ΔGRT1 ΔGRT2 cell lines (designated A and B) is shown. The probe used is shown in Fig. 3. The positions of the expected NcoI fragments for wild-type DNA (4.1 kb) and ΔGRT1 ΔGRT2 DNA (4.9 kb) are indicated on the right; the positions of electrophoresis standards (3.6 and 4.9 kb) are shown on the left. Lane 1, wild type; lane 2, 1/20th loading of wild-type cells to reveal the signal intensity expected for the two micronuclear copies; lane 3, ΔGRT1 ΔGRT2 line A; lane 4, ΔGRT1 ΔGRT2 line B. Western blot analysis of SDS lysates of wild-type and ΔGRT1 ΔGRT2 cells using a polyclonal antibody against Grt1p is shown at right. The position of Grt1p (80 kDa) is shown on the left; the positions of molecular mass standards (68 and 97 kDa) are shown on the right. Lane 1, wild-type lysate; lane 2, wild-type lysate, with 20× loading relative to lane 1; lane 3, ΔGRT1 ΔGRT2 (line A) lysate at a loading equivalent to that of lane 2.
ΔGRT1 ΔGRT2 cells are distinct from cells with disruptions in GRL genes.
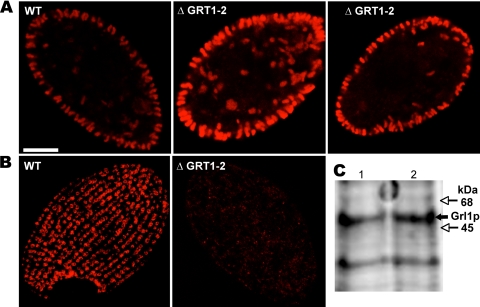
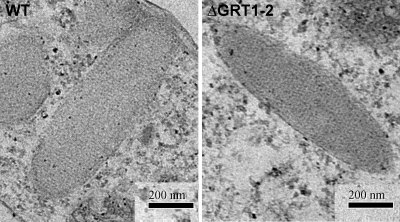
The disruption of any of the genes encoding the six abundant Grl proteins results in Tetrahymena cells whose DCG cores are no longer organized as a lattice (12). Such mutant cores are spherical rather than elongated, a difference that can be seen by indirect immunofluorescence using either of two MAbs. The MAb 5E9 recognizes Grl3p while MAb 4D11 recognizes Grt1p (5). In wild-type cells, both antibodies reveal large numbers of DCGs, most of which are docked in linearly arranged sites at the plasma membrane (Fig. 5A and B). As expected, immunofluorescence with MAb 4D11 did not reveal any DCGs in ΔGRT1 ΔGRT2 cells but showed only a diffuse background signal since the protein target of the antibody is absent (Fig. 5B). In contrast, MAb 5E9 labeled DCGs in the ΔGRT1 ΔGRT2 cells that were indistinguishable in both abundance and morphology from wild-type cells (Fig. 5A). The similar accumulation of DCGs in wild-type and ΔGRT1 ΔGRT2 cells was also demonstrated by Western blotting of whole-cell lysates with an antibody against the granule marker Grl1p, which showed that equivalent amounts of this protein accumulated in the wild-type and mutant cells (Fig. 5C). The DCGs in wild-type and ΔGRT1 ΔGRT2 cells were also indistinguishable at the level of electron microscopy (Fig. 6).
FIG. 5.
GRT1 and GRT2 are not essential for DCG formation and accumulation. (A) Visualization of docked DCGs by indirect immunofluorescence using a MAb (5E9) directed against the DCG lattice protein Grl3p. Shown are mid-cell sections of one wild-type (WT) and two ΔGRT1 ΔGRT2 (ΔGRT1-2) (cell line A) cells. Similar numbers of elongated DCGs are present, predominantly docked at the plasma membrane. (B) Visualization of docked DCGs by indirect immunofluorescence using a MAb (4D11) directed against Grt1p. Shown are tangential sections of one wild-type and one ΔGRT1 ΔGRT2 cell. The DCGs, seen en face, are docked at linearly arranged docking sites in the wild-type cell. No DCGs are seen in the ΔGRT1 ΔGRT2 cell since the MAb is directed against Grt1p. (C) Western blot analysis of SDS lysates of wild-type and ΔGRT1 ΔGRT2 (line A) cells using a polyclonal antibody against the granule lattice protein Grl1p. A total of 2 × 105 cell equivalents were loaded for each sample, and the bound antibody was detected using the Odyssey system (see Materials and Methods). Lane 1, wild type; lane 2, ΔGRT1 ΔGRT2. Wild-type and ΔGRT1 ΔGRT2 cells accumulate similar levels of Grl1p.
FIG. 6.
ΔGRT1 ΔGRT2 DCGs have lattice-organized cores. Wild-type and ΔGRT1 ΔGRT2 (line A) cells were analyzed by thin-section electron microscopy, as described in Materials and Methods. Both images show elongated DCG profiles in which the central core is visibly organized.
To test whether the DCGs lacking Grt1p and Grt2p could undergo exocytosis, we stimulated ΔGRT1 ΔGRT2 cells with two compounds that provoke rapid global DCG secretion in wild-type Tetrahymena. These are Alcian blue and dibucaine. When wild-type cells are treated with the polycationic dye Alcian blue, they become entrapped in robust capsules formed by the dye-dependent cross-linking of the exocytosed DCG contents (29). When we treated wild-type and ΔGRT1 ΔGRT2 cells in parallel with Alcian blue, virtually 100% of cells from both cultures were encapsulated (data not shown). The DCGs in ΔGRT1 ΔGRT2 cells are therefore capable of undergoing regulated exocytosis under these conditions. However, the Alcian blue assay does not allow one to assess the extent of exocytosis, that is, the fraction of DCGs that have released their contents.
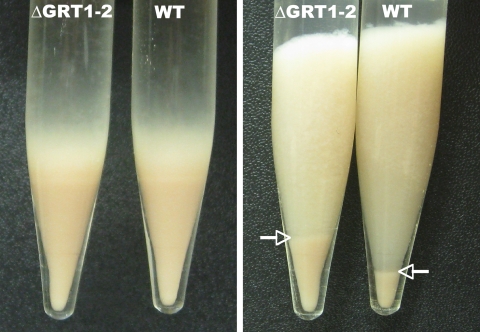
A second compound that acts as a potent secretagogue in Tetrahymena is the local anesthetic dibucaine (26). When treated with dibucaine, wild-type cells release their DCG contents within seconds. The DCG cores expand during release but remain highly insoluble, and they can be readily pelleted to form a flocculent layer. Measuring the volume of this flocculent layer provides a simple and quantitative assay for exocytosis (12). When wild-type and ΔGRT1 ΔGRT2 cultures were treated in parallel with dibucaine, both produced very similar volumes of flocculent (Fig. 7, left panel). Taken together, our results indicate that that ΔGRT1 ΔGRT2 cells accumulate wild-type levels of DCGs, that those DCGs undergo rapid exocytosis in response to dibucaine, and that the expansion of the ΔGRT1 ΔGRT2 cores is comparable to wild type.
FIG. 7.
Comparison of DCG secretion from wild-type and ΔGRT1 ΔGRT2 cells. Stationary overnight cultures (50 ml) of wild-type (WT) and ΔGRT1 ΔGRT2 (ΔGRT1-2; line A) cells were pelleted, resulting in similarly sized cell pellets (left panel). Cells were resuspended, stimulated with dibucaine for 20 s, and recentrifuged to produce a pellet of cells with an overlying flocculent (right panel). The flocculent corresponds to the DCG contents that were released from the cells. The boundary between the pelleted cells and the flocculent is indicated by arrows. The postdibucaine cell pellet from ΔGRT1 ΔGRT2 cultures is larger because a smaller proportion of cells is trapped in the ΔGRT1 ΔGRT2 flocculent (right panel).
During these experiments, we noted a subtle but reproducible difference between the exocytosed material in wild-type and ΔGRT1 ΔGRT2 cells. When Tetrahymena cultures are pelleted shortly after dibucaine addition, a large fraction of wild-type cells remains trapped in the expanded DCG cores that form the flocculent layer. This can be seen in the greatly reduced size of the cell pellet in poststimulation, compared to prestimulation, samples (Fig. 7, right panel). The reduction in cell pellet size represents trapping of cells in the flocculent layer and not cell mortality or lysis since the original volume of cells could be recovered by repeated dilution, dispersion, and repelleting of the flocculent layer (data not shown). ΔGRT1 ΔGRT2 cells were also trapped in the flocculent layer after stimulation, but the cell pellet was always larger than in parallel wild-type cultures; i.e., fewer ΔGRT1 ΔGRT2 cells were trapped (Fig. 7, right panel). One possibility is that the expanded DCG cores in wild-type cells are more adhesive than those from ΔGRT1 ΔGRT2 cells, suggesting a potential postexocytic function for Grt1p and Grt2p.
Grl protein sorting to DCGs is independent of Grt proteins.
Since Grt1p does not appear to be cotransported or to interact biochemically with Grl proteins under the conditions tested (Fig. 1 and 2) and since ΔGRT1 ΔGRT2 DCG cores still maintain the highly structured core that depends upon Grl proteins (Fig. 6), it appeared likely that the sorting of Grl proteins to DCGs was not perturbed by the absence of Grt1p and Grt2p.
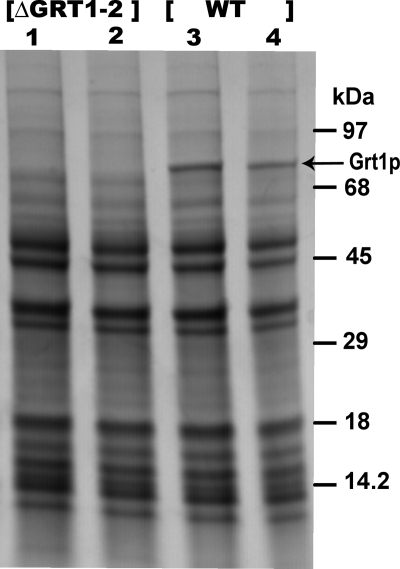
To obtain direct evidence, we analyzed the released DCG contents from ΔGRT1 ΔGRT2 cells. As previously demonstrated, all of the major polypeptide species in Tetrahymena DCGs correspond to Grl proteins with the exception of Grt1p itself (11, 34). Analysis by SDS-PAGE and Coomassie blue staining of wild-type versus ΔGRT1 ΔGRT2 flocculent showed that the only species visibly missing from the latter was Grt1p (Fig. 8). Therefore, the absence of Grt1p and Grt2p does not preclude normal sorting and accumulation in DCGs of the set of Grl proteins.
FIG. 8.
Targeting of DCG lattice proteins is independent of ΔGRT1 ΔGRT2. Wild-type (WT) and ΔGRT1 ΔGRT2 (ΔGRT1-2) cultures were stimulated with dibucaine, and the released flocculent was depleted of trapped cells by several rounds of pelleting and resuspension, as described in Materials and Methods. The final flocculents were precipitated by trichloroacetic acid, suspended in SDS sample buffer, and electrophoresed on a 5 to 15% polyacrylamide gel. Proteins were visualized by Coomassie blue staining. All lanes contain independent samples of the indicated cell type.
DISCUSSION
The elaborate DCGs found in ciliates reflect complex sorting and assembly within the eukaryotic secretory pathway. Although substantial progress has been made in identifying DCG constituents in Tetrahymena and Paramecium, we do not yet understand how proteins are targeted to DCGs in ciliates, and indeed the targeting mechanisms are not yet clearly understood even in the much better studied animal systems (23). The multilayered DCG structures formed in some ciliate species raise the additional question of how protein sorting and organization within the DCGs are coordinated. Previous studies in Tetrahymena and Paramecium have focused on one family of proteins, the Tmps/Grls, and have demonstrated the importance of proteolytic processing for control of assembly although the details (including the proteases themselves) are not yet known (2, 8). However, the Grls/Tmps appear to constitute only the central expansible crystal in these DCGs, so other proteins must be responsible for the other structures. In this paper we report the functional analysis of an abundant non-Grl protein in Tetrahymena DCGs, called Grt1p, and analysis of potential interactions between Grl proteins and Grt1p.
The major conclusion of the data, combined with results from previous studies, is that the Grl and Grt proteins, though targeted to the same DCGs, are likely to act independently with regard to assembly and sorting and to serve distinct functions for Tetrahymena. The assembly of Grl proteins involves obligate heterooligomer formation in the ER, transport through the secretory pathway in the form of large complexes, and extensive proteolytic processing during DCG maturation (12, 34). None of these appears to be relevant for Grt1p. Previous analysis showed that Grt1p did not undergo processing, and data in the current paper show that it also differs from the Grls in the other properties (5, 31). Gel filtration analysis indicated that Grt1p in the secretory pathway is primarily a monomer, and none of the protein fractionates with the much larger complexes that contain pro-Grl1p. These large complexes are known, from genetic experiments, to contain other pro-Grl proteins (12). Consistent with the absence of Grt1p from these complexes, we found that Grt1p, when affinity purified as a His-tagged variant from Tetrahymena cell lysates, did not copurify with Grl proteins. These biochemical experiments were all done using the SB281 strain, a mutant that fails to synthesize DCGs, in order to focus on protein in transit in the secretory pathway. In wild-type cell lysates, only a small fraction of the Grl or Grt proteins are derived from protein in transit while the vast majority reflect the pool released from DCGs. We previously used SB281 lysates to identify heterooligomeric Grl protein complexes, and genetic results strongly supported the existence of the same complexes in wild-type cells (12). We therefore expect that the characterization of Grt1p in SB281 lysates is fully relevant to wild-type cells in which DCGs are formed although we cannot rule out the possibility that there are additional complexes that form in the wild type but are absent in SB281. We do not yet know the genetic lesion in SB281 but a cell biological defect is clearly manifested at the level of the late Golgi compartment/TGN, where there is no sorting of constitutive versus regulated secretory cargo in the mutant (5, 7). If there is a difference between the TGN conditions in the wild-type and SB281 strains, these may have an effect on protein oligomerization in that compartment. Our data, in which protein-protein interactions are analyzed at neutral pH, cannot rule out the possibility that Grt1p and Grl proteins associate in the conditions that exist in the lumen of the ciliate TGN, about which nothing is currently known. Interactions could also be promoted in the TGN by compartment-specific posttranslational modification of Grl proteins or Grt1p though previous work suggests there is no detectible difference in the electrophoretic mobilities of any of these proteins when they are isolated from wild-type and SB281 cells (31, 32). This is an important issue because aggregation is believed to underlie DCG protein sorting in mammalian cells (23).
Importantly, even if there are Grt1p-containing complexes that we cannot detect for technical or biological reasons in SB281 lysates, our composite data in this system argue that such complexes cannot be essential for efficient sorting of either Grt1p or Grl proteins to DCGs in wild-type cells. First, in a thorough study of Grl protein function, we found that the absence of Grl1p had no effect on Grt1p accumulation in DCGs (11). In subsequent studies each of the GRL genes was disrupted, and in every case the resulting DCGs could still be visualized using a MAb directed against Grt1p (12). These results strongly suggested that Grt1p does not depend for sorting on any individual Grl protein. A remaining possibility is that Grt1p depends on Grl proteins for sorting but that any Grl protein is sufficient, i.e., there is extensive redundancy for this function. The data in the current paper provide the important complementary evidence that cells lacking Grt1p and the closely related Grt2p still sort all of the major Grl proteins to DCGs. This is clear because the ΔGRT1 ΔGRT2 cells synthesized DCGs with no exocytosis defects. These DCGs cannot lack any of the Grl proteins since such deficiencies have previously been shown to result in dramatic exocytosis defects (12). Moreover, we found by direct biochemical analysis that the amount of the Grl proteins released from DCGs in ΔGRT1 ΔGRT2 cells, as judged by Coomassie staining, was indistinguishable from amounts in the wild type.
The analysis of the ΔGRT1 ΔGRT2 cells allowed us to test ideas about potential roles of Grt1p. Prior models for Grt1p function were based on the localization of Grt1p at the tip of DCGs, where docking and fusion occur (5). This suggested that Tetrahymena DCGs might have a tip domain that was specialized for these functions. This model was also inspired by analogy with the DCGs in P. tetraurelia, which are both structurally and biochemically more complex than those in Tetrahymena. Paramecium DCGs have a morphologically distinct protrusion at the tips of the DCGs whose formation is essential for docking and fusion (1). The proteins that compose the Paramecium tip have not been identified, but one strong possibility is that they are encoded by the many Paramecium homologs of GRT1, whose roles have not yet been analyzed. Based on this analogy, we hypothesized that Tetrahymena DCGs possess a functionally important though morphologically undetectable tip domain, comprised in part by Grt1p. However, the ΔGRT1 ΔGRT2 cells showed no apparent defects in DCG docking or fusion and so do not support a model in which Grt1p is a determinant of a docking or fusion-specialized tip domain. It remains to be tested whether other members of the crystallin domain family in Tetrahymena, distantly related to GRT1 and GRT2, may play such roles.
GRT1 is the second non-GRL gene in Tetrahymena DCGs to be analyzed, and in both cases gene disruption had no effect on DCG core formation or exocytosis (16). In the case of IGR1 the lack of phenotype could be potentially explained by the low abundance of the protein and also by the fact that a second, closely related gene was subsequently found to be encoded in the genome. In contrast, Grt1p is an abundant protein in Tetrahymena DCGs, and we preempted any potential functional redundancy with the related GRT2 gene by generating the ΔGRT1 ΔGRT2 double knockout strain. The absence of exocytosis phenotypes for both IGR1 and GRT1 is therefore consistent with the idea that both Igr1p and Grt1p primarily serve postexocytic functions as secreted proteins, for example, by binding proteins or other molecules present in the cell's environment. Since Grt1p remains associated after exocytosis with the Grl-based expanded core, as judged by its pelleting in the flocculent layer, the dense cores of ciliate DCGs may serve not only as extrusion machines during exocytosis but also, by virtue of associated proteins like Grt1p, as biologically active complexes that are released into the medium. Since Grt1p is localized at the docking tip of the DCG, it is positioned to be among the first DCG contents to become externalized upon exocytosis. The function of DCGs in Tetrahymena is not yet known, but DCG exocytosis can protect Paramecium from predators wielding paralyzing toxins (17, 19). This could involve the rapid externalization of toxin-binding proteins at the tips of DCGs during a defense response. We have suggestive evidence that Grt1p is a sticky protein since the released contents of DCGs lacking Grt1/Grt2 appeared to be less adhesive than the contents of the wild-type DCGs. This conclusion is based on the observation that fewer cells were trapped in the extruded ΔGRT1 ΔGRT2 DCG contents than in the wild-type DCG contents in a pelleting assay. This model of DCG function in Tetrahymena can be contrasted with a second possibility, in which the role of expansible Grl-based cores is to rapidly disperse soluble molecules, such as peptides derived from large stretches of the Grl proproteins that are proteolytically degraded during DCG maturation. The models are not mutually exclusive, and the two mechanisms may contribute to multiple distinct functions of DCGs in these cells.
Supplementary Material
Acknowledgments
We thank Martin Gorovsky (University of Rochester), as well as members of the Turkewitz laboratory, including Lydia Bright, Sennep Herde, and Grant Bowman, for help and encouragement. Grant Bowman did important preliminary work on characterizing granule protein complexes in SB281 lysates. Jennifer McPartland (University of Chicago) provided the N4 gp65 gel filtration standard, and Kajari Dhar helped with FPLC while Yimei Chen performed electron microscopy. Marlo Nelson and Joseph Frankel (University of Iowa) generously donated monoclonal antibodies 5E9 and 4D11, and the SB281 cell line was the gift of Ed Orias (University of California—Santa Barbara).
This work was supported by National Institutes of Health grant GM077607 to A.P.T.
Footnotes
Published ahead of print on 14 August 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Adoutte, A. 1988. Exocytosis: biogenesis, transport and secretion of trichocysts, p. 325-362. In H. D. Gortz (ed.), Paramecium. Springer-Verlag, Berlin, Germany.
- 2.Adoutte, A., N. Garreau de Loubresse, and J. Beisson. 1984. Proteolytic cleavage and maturation of the crystalline secretion products of Paramecium. J. Mol. Biol. 180:1065-1081. [DOI] [PubMed] [Google Scholar]
- 3.Anderer, R., and K. Hausmann. 1977. Properties and structure of isolated extrusive organelles. J. Ultrastruc. Res. 60:21-26. [DOI] [PubMed] [Google Scholar]
- 4.Bonifacino, J. S., and J. Lippincott-Schwartz. 2003. Coat proteins: shaping membrane transport. Nat. Rev. Mol. Cell Biol. 4:409-414. [DOI] [PubMed] [Google Scholar]
- 5.Bowman, G. R., N. C. Elde, G. Morgan, M. Winey, and A. P. Turkewitz. 2005. Core formation and the acquisition of fusion competence are linked during secretory granule maturation in Tetrahymena. Traffic 6:303-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowman, G. R., D. G. Smith, K. W. Michael Siu, R. E. Pearlman, and A. P. Turkewitz. 2005. Genomic and proteomic evidence for a second family of dense core granule cargo proteins in Tetrahymena thermophila. J. Eukaryot. Microbiol. 52:291-297. [DOI] [PubMed] [Google Scholar]
- 7.Bowman, G. R., and A. P. Turkewitz. 2001. Analysis of a mutant exhibiting conditional sorting to dense core secretory granules in Tetrahymena thermophila. Genetics 159:1605-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradshaw, N. R., N. D. Chilcoat, J. W. Verbsky, and A. P. Turkewitz. 2003. Proprotein processing within secretory dense core granules of Tetrahymena thermophila. J. Biol. Chem. 278:4087-4095. [DOI] [PubMed] [Google Scholar]
- 9.Cassidy-Hanley, D., J. Bowen, J. H. Lee, E. Cole, L. A. VerPlank, J. Gaertig, M. A. Gorovsky, and P. J. Bruns. 1997. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics 146:135-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chanat, E., and W. B. Huttner. 1991. Milieu-induced, selective aggregation of regulated secretory proteins in the trans-Golgi network. J. Cell Biol. 115:1505-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chilcoat, N. D., S. M. Melia, A. Haddad, and A. P. Turkewitz. 1996. Granule lattice protein 1 (Grl1p), an acidic, calcium-binding protein in Tetrahymena thermophila dense-core secretory granules, influences granule size, shape, content organization, and release but not protein sorting or condensation. J. Cell Biol. 135:1775-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cowan, A. T., G. R. Bowman, K. F. Edwards, J. J. Emerson, and A. P. Turkewitz. 2005. Genetic, genomic, and functional analysis of the granule lattice proteins in Tetrahymena secretory granules. Mol. Biol. Cell 16:4046-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodson, G., and D. Steiner. 1998. The role of assembly in insulin's biosynthesis. Curr. Opin. Struct. Biol. 8:189-194. [DOI] [PubMed] [Google Scholar]
- 14.Dumont, J. N. 1961. Observations on the fine structure of the ciliate Dileptus anser. J. Eukaryot. Microbiol. 8:392-402. [Google Scholar]
- 15.Gautier, M. C., L. Sperling, and L. Madeddu. 1996. Cloning and sequence analysis of genes coding for Paramecium secretory granule (trichocyst) proteins. J. Biol. Chem. 271:10247-10255. [DOI] [PubMed] [Google Scholar]
- 16.Haddad, A., G. R. Bowman, and A. P. Turkewitz. 2002. New class of cargo protein in Tetrahymena thermophila dense core secretory granules. Eukaryot. Cell 1:583-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harumoto, T., and A. Miyake. 1991. Defensive function of trichocysts in Paramecium. J. Exp. Zool. 260:84-92. [Google Scholar]
- 18.Kirchhausen, T. 2000. Clathrin. Annu. Rev. Biochem. 69:699-727. [DOI] [PubMed] [Google Scholar]
- 19.Knoll, G., B. Haacke-Bell, and H. Plattner. 1991. Local trichocyst exocytosis provides an efficient escape mechanism for Paramecium cells. Eur. J. Protistol. 27:381-385. [DOI] [PubMed] [Google Scholar]
- 20.Kreiner, T., W. Sossin, and R. H. Scheller. 1986. Localization of Aplysia neurosecretory peptides to multiple populations of dense core vesicles. J. Cell Biol. 102:769-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lefort-Tran, M., K. Aufderheide, M. Pouphile, M. Rossignol, and J. Beisson. 1981. Control of exocytic processes: cytological and physiological studies of trichocyst mutants in Paramecium tetraurelia. J. Cell Biol. 88:301-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miao, W., J. Xiong, J. Bowen, W. Wang, Y. Liu, O. Braguinets, J. Grigull, R. E. Pearlman, E. Orias, and M. A. Gorovsky. 2009. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS ONE 4:e4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morvan, J., and S. A. Tooze. 2008. Discovery and progress in our understanding of the regulated secretory pathway in neuroendocrine cells. Histochem. Cell Biol. 129:243-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orias, E., M. Flacks, and B. H. Satir. 1983. Isolation and ultrastructural characterization of secretory mutants of Tetrahymena thermophila. J. Cell Sci. 64:49-67. [DOI] [PubMed] [Google Scholar]
- 25.Rosati, G., and L. Modeo. 2003. Extrusomes in ciliates: diversification, distribution, and phylogenetic implications. J. Eukaryot. Microbiol. 50:383-402. [DOI] [PubMed] [Google Scholar]
- 26.Satir, B. 1977. Dibucaine-induced synchronous mucocyst secretion in Tetrahymena. Cell Biol. Int. Rep. 1:69-73. [DOI] [PubMed] [Google Scholar]
- 27.Shang, Y., X. Song, J. Bowen, R. Corstanje, Y. Gao, J. Gaertig, and M. A. Gorovsky. 2002. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 99:3734-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sperling, L., A. Tardieu, and T. Gulik-Krzywicki. 1987. The crystal lattice of Paramecium trichocysts before and after exocytosis by X-ray diffraction and freeze-fracture electron microscopy. J. Cell Biol. 105:1649-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tiedtke, A. 1976. Capsule shedding in Tetrahymena. Naturwissenschaften 63:93. [DOI] [PubMed] [Google Scholar]
- 30.Turkewitz, A. P. 2004. Out with a bang! Tetrahymena as a model system to study secretory granule biogenesis. Traffic 5:63-68. [DOI] [PubMed] [Google Scholar]
- 31.Turkewitz, A. P., and R. B. Kelly. 1992. Immunocytochemical analysis of secretion mutants of Tetrahymena using a mucocyst-specific monoclonal antibody. Dev. Genet. 13:151-159. [DOI] [PubMed] [Google Scholar]
- 32.Turkewitz, A. P., L. Madeddu, and R. B. Kelly. 1991. Maturation of dense core granules in wild type and mutant Tetrahymena thermophila. EMBO J. 10:1979-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vayssie, L., F. Skouri, L. Sperling, and J. Cohen. 2000. Molecular genetics of regulated secretion in Paramecium. Biochimie 82:269-288. [DOI] [PubMed] [Google Scholar]
- 34.Verbsky, J. W., and A. P. Turkewitz. 1998. Proteolytic processing and Ca2+-binding activity of dense-core vesicle polypeptides in Tetrahymena. Mol. Biol. Cell 9:497-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.