Abstract
Iron acquisition is a critical aspect of the virulence of many pathogenic microbes, and iron limitation is an important defense mechanism for mammalian hosts. We are examining mechanisms of iron regulation and acquisition in the fungal pathogen Cryptococcus neoformans, and here, we characterize the roles of the ferroxidases Cfo1 and Cfo2. Cfo1 is required for the reductive iron uptake system that mediates the utilization of transferrin, an important iron source for C. neoformans during infection. The virulence of a cfo1 mutant was attenuated in a mouse model of cryptococcosis, and the mutant also displayed increased sensitivities to the antifungal drugs fluconazole and amphotericin B. Wild-type levels of drug sensitivity were restored by the addition of exogenous heme, which suggested that reduced levels of intracellular iron may curtail heme levels and interfere with ergosterol biosynthesis. We constructed green fluorescent protein (GFP) fusion proteins and found elevated expression of Cfo1-GFP upon iron limitation, as well as localization of the fusion to the plasma membrane. Trafficking to this location was disrupted by a defect in the catalytic subunit of cyclic AMP-dependent protein kinase. This result is consistent with findings from studies indicating an influence of the kinase on the expression of protein-trafficking functions in C. neoformans.
Pathogens possess iron acquisition mechanisms to compete with host systems that sequester and withhold iron during disease (5, 29). In mammals, the availability of iron is severely limited because the majority of extracellular iron is bound to iron carrier proteins such as transferrin, lactoferrin, and hemoglobin and intracellular iron is bound to the iron storage protein ferritin (33). Pathogenic fungi overcome the challenge of iron acquisition by three major mechanisms: reductive iron uptake, siderophore elaboration/uptake, and heme utilization. These systems, and others that likely remain to be discovered, are required individually or in combination in different pathogens.
Reductive iron uptake involves the reduction of ferric iron by cell surface reductases, followed by ferrous iron oxidation by a ferroxidase coupled to an iron permease for transport. The reductive system is particularly important in the pathogenesis of infections with Cryptococcus neoformans and Candida albicans because these fungi employ high-affinity iron permeases to acquire iron from transferrin in mammalian hosts (19, 21). Siderophores are low-molecular-weight iron chelators that specifically bind ferric iron with high affinity for subsequent uptake via specific transporters. Among fungal pathogens of humans, a role for siderophores in virulence has been clearly demonstrated for Aspergillus fumigatus. This fungus possesses siderophore transporters, and the inability to synthesize siderophores compromises virulence (12, 15, 16, 34). Heme and hemoglobin are also utilized by fungal pathogens as sources of iron. In C. albicans, the heme oxygenase Hmx1 and the cell wall mannoproteins Rbt5 and Rbt51 are involved in heme utilization (32, 44). Rbt5 has recently been shown to participate in the endocytosis of hemoglobin into the vacuole (45). C. neoformans is also able to utilize heme and hemoglobin as iron sources, although the mechanism(s) remains to be identified (19).
Iron availability influences the sensitivity of pathogenic fungi to antifungal drugs. For example, the combination of the iron-binding glycoprotein lactoferrin and antifungal drugs results in synergistic fungistatic effects against several clinical isolates of Candida spp. (22, 23, 43). Furthermore, iron chelation by bathophenanthroline disulfonic acid (BPS) and ferrozine increases the susceptibility of C. albicans to several antifungal drugs (30). The importance of iron is due likely to a requirement for heme in the ergosterol biosynthesis pathway that is targeted by azole antifungal drugs (3, 10). Specifically, these drugs target Erg11, a lanosterol demethylase that belongs to a cytochrome P450 protein family of heme-containing enzymes (14).
C. neoformans is a human fungal pathogen that can cause meningoencephalitis in immunocompromised individuals. We have investigated the iron acquisition systems and iron-related regulatory mechanisms in this fungus, and we recently discovered a central role for the iron-regulatory protein Cir1 in virulence (20). We have also characterized a siderophore transporter, Sit1, as well as the iron permeases Cft1 and Cft2. Sit1 participates in siderophore uptake, but the protein is not required for virulence (41). In contrast, mutants lacking the iron permease Cft1 are attenuated in virulence, show reduced accumulation in the central nervous systems of infected mice, and are defective in the utilization of iron from inorganic sources and transferrin. A role for Cft1 during cryptococcosis is also supported by the discovery of elevated transcript levels for the gene in cells from infected tissue and macrophages (9, 17, 39).
In this study, we continued our analysis of the reductive iron uptake system in C. neoformans by examining the roles of the CFO1 and CFO2 genes encoding candidate ferroxidases. Cfo1 and Cfo2 are similar to the Saccharomyces cerevisiae ferroxidase Fet3, which belongs to the multicopper oxidase family (1). In S. cerevisiae, Fet3 and the iron permease Ftr1 form a complex at the plasma membrane and mediate reductive iron uptake. The cfo1 and cfo2 mutants that we constructed revealed a role for Cfo1 in iron uptake and virulence. In addition, the cfo1 mutant and a CFO1-green fluorescent protein (GFP) gene fusion were used to examine the relationship between iron/heme homeostasis and antifungal drug sensitivity, as well as the role of cyclic AMP (cAMP)-dependent protein kinase A (PKA) in regulating the delivery of iron uptake functions to the plasma membrane.
MATERIALS AND METHODS
Strains and growth conditions.
C. neoformans var. grubii strain H99 (serotype A; MATα) was used in this study. The strain CDC1 (MATα ade2 pka1::ADE2) was used as a pka1 mutant derivative of strain H99 (6). Strains were maintained in yeast extract-Bacto peptone medium with 2.0% dextrose (YPD; Difco) or yeast nitrogen base (YNB; Difco) with 2.0% glucose. Defined low-iron medium was prepared as described previously, with slight modifications (25). Briefly, defined YNB medium was prepared and adjusted to pH 7.0 with 3-morpholinopropanesulfonic acid (MOPS), and the iron-limited condition was achieved by the addition of 100 μM BPS. Medium replete with iron was prepared by adding the iron source FeCl3, holotransferrin, heme, or feroxamine to low-iron medium at the final concentrations indicated below. To deplete intracellular iron, cells were precultured in low-iron medium at 30°C for 2 days for all experiments, as described previously (19). For antifungal sensitivity tests on plates, 10-fold serial dilutions of cells were spotted onto YPD plates containing fluconazole or amphotericin B with or without the supplemental iron sources indicated below. Plates were incubated at 30°C for 2 days before being photographed.
Construction of mutants.
The sequences of putative ferroxidases were obtained from the C. neoformans var. grubii serotype A genome database (http://www.broad.mit.edu/annotation/genome/cryptococcus_neoformans). The locus numbers for the corresponding genes are CNAG_06241.2 (CFO1) and CNAG_02958.2 (CFO2). The mutants were constructed using these sequences and primers listed in Table S1 in the supplemental material. The cfo1 mutant was constructed as follows. A gene-specific disruption cassette was constructed by PCR using primers CFO1-KO1, CFO1-KO22, CFO1-KO33, CFO1-KO4, CFO1-KO5, and CFO1-KO6, with genomic DNA and the plasmid pCH233 as templates (4, 46). The wild-type strain was biolistically transformed with the amplified construct as described previously (42). The genomic region of 2,499 bp that corresponds to the coding sequence of CFO1 was replaced with the nourseothricin acetyltransferase gene (NAT) using 5′ and 3′ flanking sequences of CFO1. Positive transformants were identified by PCR, and identification was confirmed by Southern blot analysis (see Fig. S1 in the supplemental material). For construction of the strain with CFO1 reintroduced, the CFO1 gene was amplified by PCR using wild-type genomic DNA and primers CFO1-KO1 and CFO1-KO4. The PCR fragment was digested with BglII and cloned into the BamHI site of the plasmid pJAF1 to construct pWH090, containing the neomycin resistance marker (NEO). The plasmid pWH090 was digested with DraI, and the cfo1 mutant was transformed with the digested plasmid. Positive transformants containing wild-type CFO1 at its authentic locus were identified by PCR. To construct the cfo2 mutant, the gene-specific disruption cassette was generated by overlap PCR using primers CFO2-KO1, CFO2-KO22, CFO2-KO3, CFO2-KO77, CFO2-KO5, and CFO2-KO6, with genomic DNA and the plasmid pCH233 as templates (4, 46). The genomic region of 2,500 bp that corresponds to the coding sequence of CFO2 was replaced by the nourseothricin acetyltransferase gene (NAT) using the 5′ and 3′ flanking sequences of CFO2. Positive transformants were selected by PCR, and identification was confirmed by Southern blot analysis (see Fig. S1 in the supplemental material). For construction of the strain with CFO2 reintroduced, the wild-type CFO2 gene was amplified by PCR using wild-type genomic DNA and primers CFO2-KO1 and CFO2-KO4. The PCR fragments were digested with BglII and cloned into the BamHI site of the plasmid pJAF1 to construct pWH089, containing the neomycin resistance marker (NEO). The plasmid pWH089 was digested with NdeI, and the cfo2 mutant was transformed with the digested plasmid. Positive transformants containing the wild-type CFO2 gene at its authentic locus were identified by PCR. The cfo1 cfo2 double mutants were constructed by transforming the cfo1 mutant with the CFO2 disruption cassette containing the NEO marker. The positive transformants were identified by PCR, and identification was confirmed by Southern blot analysis (see Fig. S1 in the supplemental material).
Construction of strains expressing GFP fusion proteins.
A plasmid that contained the GFP gene and had undergone codon optimization for C. neoformans (Cneo-GFP) was a generous gift from Peter Williamson (27). Cneo-GFP was cloned into pWH040, containing the Gal7 terminator and the NEO marker, to construct pWH091. To construct the Cfo1-GFP fusion protein, the wild-type CFO1 gene was amplified by PCR using wild-type genomic DNA and primers CFO1-KO4 and CFO1-GF-BglIIR. The PCR fragments were digested with BglII and cloned into the BamHI site of plasmid pWH091 to construct pWH097, containing the neomycin resistance marker (NEO). The plasmid pWH097 was digested with DraI, and the cfo1 mutant was transformed with the digested plasmid. Positive transformants containing the CFO1-GFP gene fusion at the CFO1 locus were identified by PCR. The pka1 and cir1 mutants were also transformed with the digested plasmid pWH097 to investigate the influence of the cAMP pathway and the loss of iron regulation, respectively. To generate the Cfo2-GFP fusion protein, the wild-type CFO2 gene was amplified by PCR using wild-type genomic DNA and primers CFO2-KO6 and CFO2-GF-BglIIR. The PCR fragments were digested with BglII and cloned into the BamHI site of the plasmid pWH091 to construct pWH098, containing the neomycin resistance marker (NEO). The plasmid pWH098 was digested with NdeI, and the cfo2 mutant was transformed with the digested plasmid. Positive transformants containing the CFO2-GFP gene fusion at the CFO2 locus were selected by PCR. All plasmids were checked by sequencing.
Microscopy.
Strains containing GFP-tagged proteins were grown in low-iron medium or high-iron medium at 30°C for 6 h. Cells were fixed with 4% paraformaldehyde for 20 min and washed twice with phosphate-buffered saline (PBS). Mounting medium (Prolong Gold antifade reagent with DAPI [4′,6-diamidino-2-phenylindole[) was added, and cells were visualized using an Axioplan 2 imaging microscope (Zeiss) or an LSM 510 META confocal microscope (Zeiss) with a magnification of ×1,000. The software Metamorph version 6.1r6 (Universal Imaging Corp.) or LSM Image Browser (Carl Zeiss Jena GmbH) was used to process images. The lipophilic dye FM4-64 [N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino)phenyl)hexatrienyl)pyridinium dibromide (catalog no. T-3166; Invitrogen Canada)] was used at a final concentration of 20 μM to stain internal membranes.
Virulence assays.
For the initial virulence assay, three female A/JCr mice obtained from Jackson Laboratories (Maine) were used for each cryptococcal strain. Fungal cells were cultured in 5 ml of YPD at 30°C overnight, washed twice with PBS, and resuspended in PBS. The A/JCr mice (4 to 6 weeks old) were anesthetized intraperitoneally with ketamine (80 mg/kg of body weight) and xylazine (5.5 mg/kg) in saline and suspended on a silk thread by the superior incisors. A suspension of 5 × 104 cells in 50 μl was slowly dripped into the nares of the anesthetized mice, and the mice were suspended for 10 min on the thread. The virulence of the strains was also tested with female BALB/c mice. Mice (4 to 6 weeks old) were obtained from Charles River Laboratories (Ontario, Canada), and groups of 10 mice were inoculated for each strain. The fungal cells were cultured and washed as described above. A suspension of 106 cells in 50 μl was used for inoculation by intranasal instillation. The status of the mice was monitored twice per day postinoculation. Statistical analyses of survival differences were performed by log rank tests using GraphPad Prism 4 for Mac (GraphPad Software, San Diego, CA). The protocol for the virulence assays (protocol A99-0252) was approved by the University of British Columbia Committee on Animal Care.
RESULTS
The ferroxidase encoded by CFO1 is required for iron acquisition.
The genome sequence of C. neoformans strain H99 encodes several candidate orthologs of the S. cerevisiae proteins responsible for high-affinity iron transport. These include the Cft1 and Cft2 proteins, with similarity to the S. cerevisiae iron permease Ftr1, and we previously demonstrated that Cft1 plays an important role in iron uptake and virulence in C. neoformans (19). The genome also contains two orthologs of the S. cerevisiae FET3 gene encoding the ferroxidase/multicopper oxidase that forms a complex with the Ftr1 iron permease for high-affinity transport; these genes were designated CFO1 and CFO2. CFO1 is adjacent to CFT1 on chromosome 12 and is divergently transcribed, and a similar arrangement is found for CFO2 and CFT2 on chromosome 3 (19).
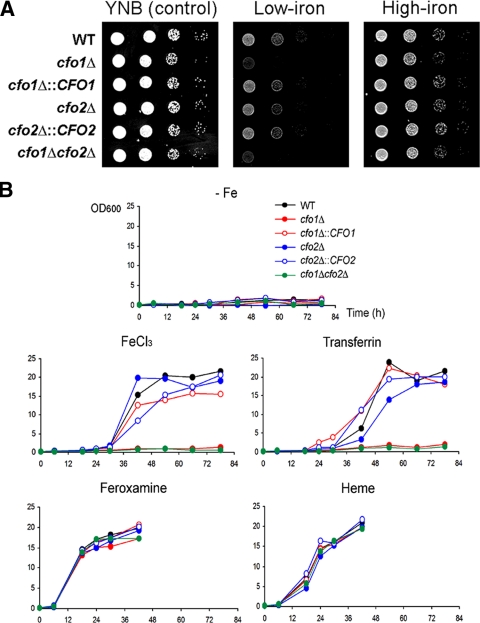
To investigate the roles of CFO1 and CFO2, we constructed mutants lacking one or both of the genes and tested the growth of the strains on solid medium (see Materials and Methods). Strains with reintroduced genes were also created by reintroducing the wild-type copy of CFO1 or CFO2 at the original locus, and these strains were included as controls in all experiments. While all strains grew well on the control medium YNB, the cfo1 mutant displayed a clear growth defect on low-iron medium. The reintroduction of CFO1 into the cfo1 mutant strain restored growth, thus confirming that the phenotype of the mutant was due to the deletion of CFO1. The growth of the cfo1 mutant was also restored in medium containing a higher concentration of iron (100 μM) (Fig. 1A). These results suggest that CFO1 is required for high-affinity iron transport and that an additional low-affinity uptake system is present to allow the mutants to grow under these conditions. The strains were also tested for their abilities to utilize other iron sources in liquid medium. The cfo1 and cfo1 cfo2 double mutants showed clear growth defects in liquid medium containing 10 μM FeCl3 or transferrin (Fig. 1B). These results further support the conclusion that CFO1 is required for high-affinity, reductive iron uptake, and they correspond to our previous findings for mutants defective in CFT1, the adjacent gene encoding the iron permease that is presumed to partner with Cfo1 (19). However, the cfo1 mutant and the cfo1 cfo2 double mutants grew as well as the wild-type strain in medium containing a siderophore (feroxamine) or heme as the sole iron source. Cfo1-mediated iron uptake therefore appears to be independent of siderophore or heme utilization. In contrast to the cfo1 mutant, the mutant lacking CFO2 displayed no visible growth defect under any of the conditions tested. This result is again consistent with our previous finding that the CFO2-linked gene CFT2 plays little role in iron uptake in vitro (19). Throughout the studies reported here, the cfo1 cfo2 double mutants presented the same phenotypes as the cfo1 single mutant.
FIG. 1.
CFO1 is required for high-affinity and reductive iron transport. (A) Tenfold serial dilutions of cells (starting at 104 cells) were spotted onto solid YNB medium (control), low-iron medium (YNB-100 μM BPS), and high-iron medium (YNB-100 μM BPS-100 μM FeCl3). Plates were incubated at 30°C for 2 days. Strains are indicated by genotypes. WT, wild type. (B) Cells were inoculated into liquid low-iron medium (YNB-100 μM BPS [−Fe]), low-iron medium containing FeCl3 (YNB-100 μM BPS-10 μM FeCl3), low-iron medium containing holotransferrin (YNB-100 μM BPS-10 μM transferrin), low-iron medium containing feroxamine (YNB-100 μM BPS-10 μM feroxamine), or low-iron medium containing 10 μM heme (YNB-100 μM BPS-10 μM heme). Cultures were incubated at 30°C, and optical densities at 600 nm (OD600) were measured. Note that the final optical density numbers on the y axes resulted from multiplication with a 10-fold dilution factor at each time point.
The loss of Cfo1 attenuates virulence.
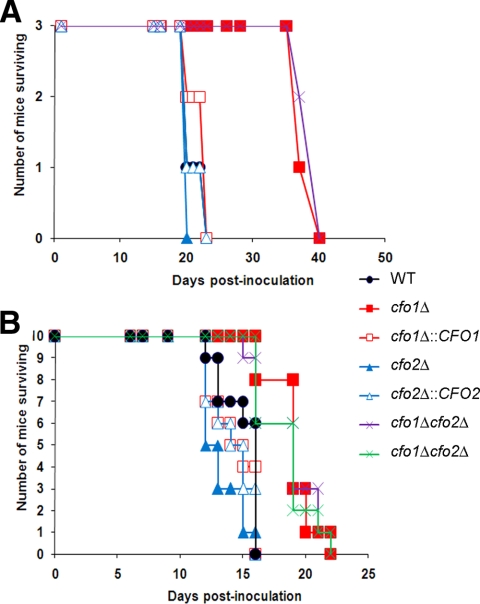
The virulence of the cfo1 and cfo2 mutants and the cfo1 cfo2 double mutants was tested using the mouse inhalation model of cryptococcosis. In the initial experiment, we employed three A/JCr mice for each cryptococcal strain and we observed significant attenuation of virulence in mice infected with the cfo1 single mutant or a cfo1 cfo2 double mutant (Fig. 2A). The loss of cfo2 did not influence virulence. These results are similar to our previous findings with the cft1 mutant in A/JCr mice (19). The attenuation of virulence of the cfo1 and cfo1 cfo2 mutants was also evident, although less prominent, when virulence was tested in a second experiment with 10 BALB/c mice per strain (Fig. 2B). The difference in the extent of attenuation of cfo1 mutant virulence between the two assays may reflect the different host strains, which may vary in immune responses or levels of available iron. We should note that the mutants grow as well as the wild-type strain at 37°C, thus ruling out temperature sensitivity as a reason for reduced virulence (data not shown). Overall, our data indicate that CFO1 but not CFO2 influences the virulence of C. neoformans.
FIG. 2.
The cfo1 mutant and the cfo1 cfo2 double mutants are attenuated in virulence. (A) Three female A/JCr mice were infected intranasally with each of the strains indicated, and the survival of the mice was monitored twice per day. The P values for results obtained with the wild-type strain (WT) versus those obtained with the cfo1Δ mutant or the cfo1Δ cfo2Δ mutants were each 0.0224. Strains are indicated by genotypes. (B) In a separate experiment, 10 female BALB/c mice were infected intranasally with each of the strains indicated and the survival of the mice was monitored twice per day. The results from both assays indicate that CFO1 is required for full virulence. The P values for the infections with BALB/c mice were as follows: results for the wild-type strain versus those for the cfo1Δ mutant, 0.001; results for the wild-type strain versus those for the first cfo1Δ cfo2Δ mutant, 0.0086; and results for the wild-type strain versus those for the second cfo1Δ cfo2Δ mutant, 0.0028. The two cfo1Δ cfo2Δ mutants were constructed independently, and their growth phenotypes were identical to each other and to that of the wild-type strain in culture (data not shown).
Deletion of CFO1 causes increased sensitivity to copper and antifungal drugs.
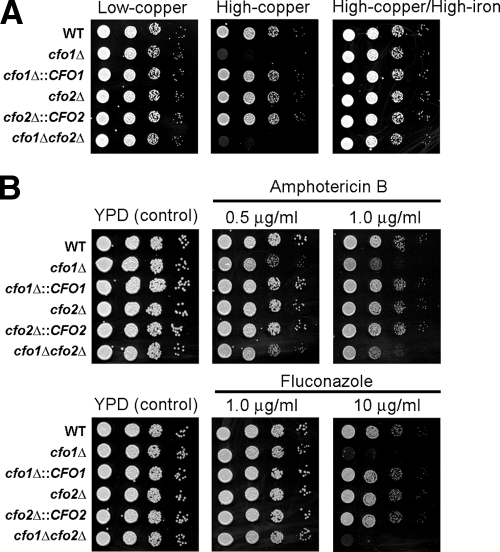
In S. cerevisiae, the deletion of FET3 causes poor growth under iron-deficient conditions and increases sensitivity to copper (26, 36, 40). Moreover, the growth defect of the fet3 mutant in the presence of elevated copper can be eliminated by the addition of iron, presumably due to Fe3+ inhibition of the ferric reductase (Fre1)-mediated reduction of Cu2+ to toxic Cu1+ (36). In this context, we tested our mutants for similar phenotypes and found that the cfo1 mutant, but not the cfo2 mutant, displayed increased sensitivity to copper. The copper sensitivity of the cfo1 mutant was restored to the wild-type level by the addition of iron, suggesting that Cfo1 may possess Cu1+ oxidation activity, as does its ortholog Fet3 in S. cerevisiae (Fig. 3A). In examining other phenotypes, we found that mutants lacking CFO1 showed increased sensitivities to the antifungal drugs amphotericin B and fluconazole (Fig. 3B). We speculate that iron deficiency resulting from the cfo1 mutation affects the biosynthesis of heme, which is an essential cofactor of several enzymes in the ergosterol biosynthesis pathway. These enzymes include Erg11, the cytochrome P450 lanosterol 14-α-demethylase that is the target of fluconazole inhibition. The decreased activity of this enzyme and the resulting change in the membrane ergosterol content would cause increased sensitivity to the antifungal drugs (30). Given the importance of these drugs in treating cryptococcosis, we examined the link between iron and drug sensitivity in more detail.
FIG. 3.
Deletion of CFO1 increases sensitivity to copper and antifungal drugs. (A) Tenfold serial dilutions of cells (starting at 104 cells) were spotted onto solid YNB medium containing the copper chelator bathocuproine disulfonate (BCS) at 100 μM (low copper), YNB medium containing 100 μM BCS and 100 μM CuSO4 (high copper), and YNB medium containing 100 μM BCS, 100 μM CuSO4, and 100 μM FeCl3 (high copper/high iron). Plates were incubated at 30°C for 2 days. Strains are indicated by genotypes. WT, wild type. (B) The growth of strains in media containing the antifungal drugs amphotericin B and fluconazole was monitored. Tenfold serial dilutions of cells (starting at 104 cells) were spotted onto YPD plates without or with the antifungal drug at the concentrations indicated. Plates were incubated at 30°C for 2 days.
Exogenous heme or an exogenous siderophore reduces the antifungal drug sensitivity of the cfo1 mutant.
As mentioned above, we hypothesized that diminished heme levels resulted in the increased sensitivities of the cfo1 mutant to the antifungal drugs. To test this idea, we analyzed the sensitivities of strains on media with fluconazole and different iron sources utilized by C. neoformans. Initially, we found that the addition of FeCl3 (or CuSO4) did not change the sensitivity of the cfo1 mutant to fluconazole (Fig. 4A). However, exogenous heme (Fig. 4B) or an exogenous siderophore (feroxamine) (Fig. 4C) restored the growth of the cfo1 mutant. We also tested the previously characterized cft1 iron permease mutant (19) and found that, like that of the cfo1 mutant, the fluconazole sensitivity of this strain was suppressed by heme addition (Fig. 4D). We previously reported the sensitivity of the cft1 mutant to miconazole and amphotericin B (19). Overall, these results support the idea that heme biosynthesis in the cfo1 and cft1 mutants is significantly affected by iron deficiency and that this effect may be the main cause of the increased sensitivity to fluconazole. The observation that the addition of heme or a siderophore, but not FeCl3, suppressed drug sensitivity suggests that low-affinity uptake of inorganic iron does not support sufficient heme synthesis to overcome the challenge imposed by fluconazole.
FIG. 4.
The addition of exogenous heme or an exogenous siderophore suppresses the antifungal drug sensitivities of the cfo1 and cft1 mutants. The growth of strains in medium containing the antifungal drug fluconazole with or without an iron source was monitored. Tenfold serial dilutions of cells (starting at 104 cells) were spotted onto YPD plates with fluconazole with or without an iron source as indicated. Plates were incubated at 30°C for 2 days. Strains are indicated by genotypes. WT, wild type.
A Cfo1-GFP fusion is more highly expressed in low-iron medium than in high-iron medium and is localized to the plasma membrane.
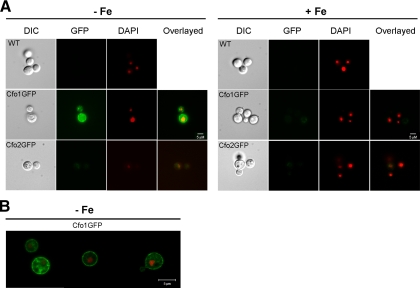
We previously showed that iron limitation leads to elevated CFT1 expression and that the iron-regulatory protein Cir1 negatively regulates CFT1 and CFO1 transcript levels (19, 20). To extend the analysis, we constructed fusions of CFO1 and CFO2 with the GFP gene and investigated the localization of the proteins and the influence of iron concentration on fluorescence levels. Cells containing the fusion proteins were grown in either low- or high-iron medium and visualized by fluorescence microscopy. We found that fluorescence from the Cfo1-GFP fusion protein was visible only in the cells grown in low-iron medium and that the levels of expression of the Cfo2-GFP fusion protein were substantially lower than those of Cfo1-GFP in cells grown in low- and high-iron media (Fig. 5A). The low level of Cfo2-GFP expression precluded a conclusion about the localization of Cfo2. Given that the cfo2 mutant did not show any mutant phenotypes, we were unable to test the fusion protein for its ability to complement the mutation; however, we did find that the Cfo1-GFP fusion functionally complemented the cfo1 mutation (see Fig. S2 in the supplemental material). In cells carrying the Cfo1-GFP fusion protein, fluorescence was detected mainly at the periphery of the cell, suggesting localization of the protein at the plasma membrane under iron-depleted conditions. Additional support for a plasma membrane location for the Cfo1-GFP fusion was obtained by confocal microscopy (Fig. 5B). Taken together, these results revealed that Cfo2 is expressed at a much lower level than Cfo1 and that the latter protein is expressed preferentially under low-iron conditions, with localization primarily to the plasma membrane.
FIG. 5.
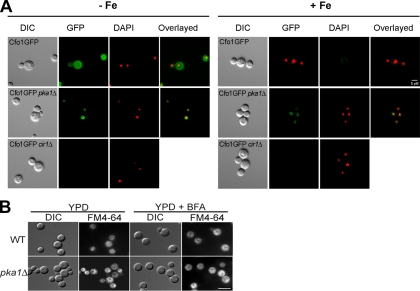
Expression and localization of Cfo1 and Cfo2. Iron-starved C. neoformans strains containing GFP-tagged proteins were grown in low-iron medium (YNB-100 μM BPS [−Fe]) or high-iron medium (YNB-100 μM BPS-100 μM FeCl3 [+Fe]) at 30°C for 6 h and visualized. The Cfo1-GFP fusion is more highly expressed in low-iron medium and is localized to the plasma membrane. At least 60 cells were analyzed for each experiment. DAPI was used to stain the DNA in the nuclei. The cells were observed with an Axioplan 2 imaging microscope (A) or an LSM 510 META confocal microscope (B), and the bars represent 5 μM as indicated. The images were processed as described in Materials and Methods such that the DAPI staining to reveal the DNA in the nuclei is shown in red. WT, wild type; DIC, differential interference contrast.
A defect in cAMP signaling leads to mislocalization of a Cfo1-GFP fusion protein.
We recently discovered that mutations in genes encoding the catalytic (Pka1) and regulatory (Pkr1) subunits of cAMP-dependent PKA influence the expression of genes for protein-trafficking functions (18). These observations led us to examine the location of the Cfo1-GFP and Cfo2-GFP fusion proteins in the pka1 mutant after growth in low- or high-iron medium (Fig. 6A). We found that the Cfo1-GFP fusion protein in the pka1 mutant was localized to an internal ring-like structure that is clearly distinct from the plasma membrane location in the wild-type strain (Fig. 5 and 6A). Additional work will be needed to determine whether the perinuclear location represents the endoplasmic reticulum. We also noted that the level of fluorescence from the Cfo1-GFP fusion protein was higher in the pka1 mutants than in the wild-type strain upon growth in medium replete with iron (Fig. 6A). In contrast, we did not observe differences in the localization or expression of the Cfo2-GFP fusion protein in the pka1 mutant, although the signal for this protein was weak (data not shown).
FIG. 6.
Influence of Pka1 or Cir1 on expression and localization of Cfo1. (A) Cfo1-GFP was expressed in the C. neoformans pka1Δ or cir1Δ strain in low-iron medium (YNB-100 μM BPS [−Fe]) or high-iron medium (YNB-100 μM BPS-100 μM FeCl3 [+Fe]) at 30°C for 6 h and visualized. Pka1 influences the localization of Cfo1 and negatively regulates its expression. Cir1 was shown to positively regulate the expression of Cfo1. DIC, differential interference contrast. (B) Internal membrane organization detected with FM4-64. The wild-type strain (WT) and the pka1 mutant were grown in YPD for 24 h at 30°C, stained with the lipophilic dye FM4-64 with or without brefeldin A (BFA) for 30 min, and observed for epifluorescence. The internal membrane structures are more prominent in the pka1 mutant than in the wild type, with some aggregation. At least 60 cells were analyzed for each experiment. DAPI was used to stain the DNA in the nuclei.
The influence of the pka1 mutation on Cfo1-GFP localization is consistent with our hypothesis that the cAMP pathway regulates intracellular trafficking. In further support of this idea, we found that FM4-64 staining revealed a marked difference in the organization of internal membrane structures between the pka1 mutant and the wild-type strain (Fig. 6B). Specifically, the mutant showed a more distinct pattern of accumulation of intracellular vesicles than the diffuse pattern seen in the wild-type strain. The results obtained with the pka1 mutation may therefore reflect a general influence on the trafficking machinery. Treatment with the secretion inhibitor brefeldin A did not substantially change the FM4-64 staining pattern for the pka1 mutant but did enhance the apparent accumulation of vesicles in the wild-type strain. We also tested whether the addition of cAMP or copper influenced the expression or localization of the Cfo1-GFP fusion protein in the pka1 mutant. Supplementation with copper ions was included because copper is necessary for proper membrane localization of the Cfo1 homolog protein Fet3 in S. cerevisiae. We found that neither cAMP nor copper addition restored the expression or localization of the Cfo1-GFP fusion protein in the pka1 mutant (data not shown). These results indicate that the cAMP pathway influences the localization of the Cfo1 protein and may also contribute to the regulation of expression under conditions of iron repletion. Finally, we found that the expression of the Cfo1-GFP fusion protein was significantly downregulated in the absence of the iron-regulatory protein Cir1 in both low- and high-iron media (Fig. 6A). These findings confirm the regulatory role of Cir1 in the expression of Cfo1 (21).
DISCUSSION
We have shown previously that iron plays a central role in the pathogenesis of C. neoformans. Specifically, we identified the iron-regulatory protein, Cir1, that is responsible for the transcriptional response to iron and that regulates both iron uptake functions and the expression of the major virulence factors (20). Our overall goals are to understand how C. neoformans acquires iron within mammalian hosts and to identify the key iron sources used by the fungus during infection. In the present study, we focused on the roles of the ferroxidases encoded by the CFO1 and CFO2 orthologs of the well-characterized FET3 gene of S. cerevisiae (1, 2, 37, 38).
The CFO1 and CFO2 genes are each paired with adjacent iron permease genes (CFT1 and CFT2, respectively), and the gene pairs are transcribed in a bidirectional fashion. Mutants with a deletion of CFO1 displayed growth defects when the cells were grown in low-iron medium, and they were unable to utilize FeCl3 or transferrin as a sole iron source in liquid culture. In contrast, the cfo1 mutants could utilize heme or the siderophore ferioxamine under the same conditions. These phenotypes are shared with mutants lacking CFT1, thus indicating that the CFO1-CFT1 gene cluster likely encodes proteins that together make up the reductive, high-affinity uptake system. The lack of mutant phenotypes for the cfo2 mutant mirrored the findings for the adjacent CFT2 gene, indicating that the CFO2-CFT2 gene cluster plays little role in iron uptake under the conditions tested. It is possible that Cft2 and Cfo2 function in intracellular iron transport.
We tested the virulence of the cfo1 and cfo2 mutants using A/JCr and BALB/c mice and found attenuation of virulence upon infection with the cfo1 single mutant and the cfo1 cfo2 double mutants. These results correspond with our finding that Cft1 contributes to virulence by a mechanism that likely involves the utilization of transferrin as an iron source during infection (19). In contrast, Eck et al. (7) found that a C. albicans mutant with a defect in the FET3-encoded ferroxidase did not show a virulence defect. However, this result may have been due to a high level of stored iron in the mutant and/or the presence of additional iron uptake mechanisms (7). The other C. neoformans ferroxidase, Cfo2, did not appear to make a contribution to virulence when the cfo1 cfo2 double mutants were tested, and this finding is in contrast to our previous demonstration that the loss of CFT2 further attenuated the virulence of a cft1 mutant (19). It is possible that other ferroxidase activities functionally replaced Cfo2 during infection. We noted that the attenuation of virulence was less pronounced in BALB/c mice than in A/JCr mice. There are undoubtedly complex differences between the mouse strains, but we speculate that the Nramp1 (natural resistance-associated macrophage protein) status of the mice may be particularly important in the context of iron acquisition. Nramp1 is a glycoprotein that is localized to endomembrane compartments. It has been shown previously that Nramp1 is recruited to phagolysosomes containing microbial pathogens (11, 35). BALB/c mice are Nramp1 deficient compared to A/JCr mice, and this difference may influence innate immunity to cryptococcal infections.
Our analysis of cfo1 mutants provided a foundation to investigate three additional aspects of iron acquisition/sensing in C. neoformans: the interplay between copper and ferroxidase activity, the relationship between heme and antifungal sensitivity, and the localization of ferroxidase proteins in wild-type and pka1 mutant cells. We found that the cfo1 mutants displayed hypersensitivity to copper, a phenotype which is similar to the copper sensitivity of the S. cerevisiae fet3 mutants (36). In S. cerevisiae, the membrane metalloreductase Fre1 reduces environmental Cu2+ and Fe3+ to Cu1+ and Fe2+, respectively, and then Cu1+ or Fe2+ becomes oxidized by the Fet3 protein. In the absence of Fet3, cytotoxic Cu1+ accumulation causes a growth defect for the mutant cells. Our analysis suggests that Cfo1 may also be required for the oxidation of Cu1+ to Cu2+ in C. neoformans. The deletion of CFO1 also caused increased sensitivity to the antifungal drugs amphotericin B and fluconazole. A relationship between iron and antifungal sensitivity in fungal pathogens has been established previously by Prasad et al. (30). Specifically, it was found that iron limitation increases the sensitivities of Candida spp. to azole antifungal drugs and that C. albicans mutants lacking the iron permeases Ftr1 and Ftr2 or the copper transporter Ccc2 display dramatically increased sensitivities to fluconazole (30). Moreover, it was shown that the increased sensitivities of the mutants to the drug were abated by the addition of inorganic iron and that the effect of iron limitation on drug sensitivity is independent of efflux pump proteins. Based on these findings, it was proposed that iron limitation causes a decrease in ergosterol content and increased membrane fluidity, resulting in enhanced passive diffusion of the drugs (30).
Although an influence on membrane fluidity may also be occurring in C. neoformans, we speculated that iron deficiency in the cfo1 mutants resulted in reduced levels of heme biosynthesis, which in turn affected the ergosterol biosynthesis pathway to alter sensitivity to antifungal drugs. This idea was supported by our observation that the addition of heme suppressed the antifungal drug sensitivities of the cfo1 and cft1 mutants. The influence of iron deprivation on heme biosynthesis may also be involved in the antifungal sensitivity observed in C. albicans and in other fungi (30). Reduced levels of Erg11, the heme-containing lanosterol 14-α-demethylase, are known to correlate with increased sensitivity to azole drugs (13, 28). For example, the S. cerevisiae strain lacking Dap1, which positively regulates Erg11, displays hypersensitivity to fluconazole and intraconazole (13, 28). We also found that the siderophore feroxamine could suppress the antifungal drug sensitivity of the cfo1 mutants, which suggests that iron from a siderophore can be utilized to synthesize heme in C. neoformans. Interestingly, the addition of FeCl3 was unable to suppress the sensitivity phenotypes of the cfo1 mutants, indicating that iron transported by low-affinity uptake mechanisms in the cfo1 mutants may not be sufficient for heme biosynthesis. This result differs from the finding for C. albicans that the addition of iron salts can reverse the enhanced sensitivities of the ftr1, ftr1 ftr2, and ccc2 mutants to fluconazole (30). It is possible that the patterns of iron involvement in drug sensitivity are different for C. albicans and C. neoformans at the level of mechanisms of iron acquisition or processing to generate heme.
Based on sequence and functional similarities to Fet3 in S. cerevisiae, we predicted that a Cfo1-GFP fusion protein would be localized at the plasma membrane (37). This was indeed the case, and we also found that Cfo1-GFP fluorescence was preferentially detected in cells grown in low-iron medium. This expression pattern was similar to that of the iron permease Cft1 and was consistent with our previous analysis of iron-regulated transcript levels in C. neoformans (20). The localization of Cfo1-GFP to the plasma membrane provided an opportunity to test a hypothesis arising from transcriptional profiling experiments that PKA regulates intracellular trafficking (18). We found that Cfo1-GFP in a pka1 mutant was localized to an internal ring structure surrounding the nucleus; this structure is likely to be the endoplasmic reticulum. The pattern of fluorescence resembled the localization seen for a Fet3-GFP fusion in S. cerevisiae in the absence of the Ftr1 permease; Ftr1 is known to form a complex with Fet3 and to be required for proper trafficking of the protein to the plasma membrane (37). These data, combined with the patterns of FM4-64 staining of internal membrane structures, support the conclusion that PKA regulates trafficking in C. neoformans.
It is also possible that cAMP signaling and PKA influence other aspects of iron acquisition. For example, in S. cerevisiae, one of three PKA catalytic subunits, Tpk2, negatively regulates the transcription of FET3 and CCC2, and the components of the cAMP pathway, Ras1 and Ras2, regulate ferric reductase activity (24, 31). In addition, the expression of the high-affinity FER2 iron permease is positively regulated by the regulatory subunit of PKA in Ustilago maydis (8). Moreover, we previously demonstrated that the cAMP pathway also influences the expression of the high-affinity iron uptake system in C. neoformans (18, 19). Specifically, the transcript level for the CFT1 gene was reduced in a pkr1 mutant lacking the regulatory subunit of PKA, and the CFT2 transcript level was elevated in a pka1 mutant lacking the catalytic subunit of the kinase (18, 19). We noted in our localization studies that the Cfo1-GFP fusion protein appeared to be expressed at a higher level in the pka1 mutant than in the wild-type strain under conditions of iron repletion. This result suggests that Pka1 negatively regulates the expression of Cfo1 in C. neoformans, although additional work is needed to determine the level at which this regulation occurs.
In summary, we examined the roles of the CFO1 and CFO2 genes, encoding candidate ferroxidases, and found that CFO1 is required for the utilization of inorganic iron and transferrin and for full virulence. In contrast, CFO2 was found not to play a demonstrable role. Together with the findings from our previous analyses of CFT1 and CFT2 (19), the results of these studies indicate that the iron permease and ferroxidase encoded in the same gene cluster (e.g., by CFT1 and CFO1 or by CFT2 and CFO2) likely have the same role in iron transport. This result is consistent with the observed specificities of iron permease-ferroxidase pairs in S. cerevisiae and Schizosaccharomyces pombe (2, 37, 38). Our analysis of Cfo1 also established a connection between heme and antifungal drug susceptibility and revealed a role for PKA in trafficking of the ferroxidase to the plasma membrane. This work lays the foundation for further characterization of iron uptake functions as possible targets for novel antifungal therapy, especially in combination with currently available azole drugs.
Supplementary Material
Acknowledgments
We thank Po-Yan Cheng and Guntram A. Grassl for advice and technical assistance, Kevin Hodgson for help with microscopy, Peter Williamson for the GFP gene, and Joseph Heitman for strains. This study was initiated during W.H.J.'s postdoctoral work at the University of British Columbia and completed at Chung-Ang University.
This work was supported by awards from the National Institutes of Health (grant no. R01 AI053721 to J.W.K.) and the Canadian Institutes of Health Research (to J.W.K.). J.W.K. is a Burroughs Wellcome Fund Scholar in molecular pathogenic mycology.
Footnotes
Published ahead of print on 21 August 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Askwith, C., D. Eide, A. Van Ho, P. S. Bernard, L. Li, S. Davis-Kaplan, D. M. Sipe, and J. Kaplan. 1994. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Askwith, C., and J. Kaplan. 1997. An oxidase-permease-based iron transport system in Schizosaccharomyces pombe and its expression in Saccharomyces cerevisiae. J. Biol. Chem. 272:401-405. [DOI] [PubMed] [Google Scholar]
- 3.Crisp, R. J., A. Pollington, C. Galea, S. Jaron, Y. Yamaguchi-Iwai, and J. Kaplan. 2003. Inhibition of heme biosynthesis prevents transcription of iron uptake genes in yeast. J. Biol. Chem. 278:45499-45506. [DOI] [PubMed] [Google Scholar]
- 4.Davidson, R. C., J. R. Blankenship, P. R. Kraus, M. de Jesus Berrios, C. M. Hull, C. D'Souza, P. Wang, and J. Heitman. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148:2607-2615. [DOI] [PubMed] [Google Scholar]
- 5.Doherty, C. P. 2007. Host-pathogen interactions: the role of iron. J. Nutr. 137:1341-1344. [DOI] [PubMed] [Google Scholar]
- 6.D'Souza, C. A., J. A. Alspaugh, C. Yue, T. Harashima, G. M. Cox, J. R. Perfect, and J. Heitman. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 21:3179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eck, R., S. Hundt, A. Hartl, E. Roemer, and W. Kunkel. 1999. A multicopper oxidase gene from Candida albicans: cloning, characterization and disruption. Microbiology 145(Pt. 9):2415-2422. [DOI] [PubMed] [Google Scholar]
- 8.Eichhorn, H., F. Lessing, B. Winterberg, J. Schirawski, J. Kamper, P. Muller, and R. Kahmann. 2006. A ferroxidation/permeation iron uptake system is required for virulence in Ustilago maydis. Plant Cell 18:3332-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan, W., P. R. Kraus, M. J. Boily, and J. Heitman. 2005. Cryptococcus neoformans gene expression during murine macrophage infection. Eukaryot. Cell 4:1420-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gollub, E. G., K. P. Liu, J. Dayan, M. Adlersberg, and D. B. Sprinson. 1977. Yeast mutants deficient in heme biosynthesis and a heme mutant additionally blocked in cyclization of 2,3-oxidosqualene. J. Biol. Chem. 252:2846-2854. [PubMed] [Google Scholar]
- 11.Gruenheid, S., E. Pinner, M. Desjardins, and P. Gros. 1997. Natural resistance to infection with intracellular pathogens: the Nramp1 protein is recruited to the membrane of the phagosome. J. Exp. Med. 185:717-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haas, H. 2003. Molecular genetics of fungal siderophore biosynthesis and uptake: the role of siderophores in iron uptake and storage. Appl. Microbiol. Biotechnol. 62:316-330. [DOI] [PubMed] [Google Scholar]
- 13.Hand, R. A., N. Jia, M. Bard, and R. J. Craven. 2003. Saccharomyces cerevisiae Dap1p, a novel DNA damage response protein related to the mammalian membrane-associated progesterone receptor. Eukaryot. Cell 2:306-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hannemann, F., A. Bichet, K. M. Ewen, and R. Bernhardt. 2007. Cytochrome P450 systems—biological variations of electron transport chains. Biochim. Biophys. Acta 1770:330-344. [DOI] [PubMed] [Google Scholar]
- 15.Hissen, A. H., J. M. Chow, L. J. Pinto, and M. M. Moore. 2004. Survival of Aspergillus fumigatus in serum involves removal of iron from transferrin: the role of siderophores. Infect. Immun. 72:1402-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hissen, A. H., A. N. Wan, M. L. Warwas, L. J. Pinto, and M. M. Moore. 2005. The Aspergillus fumigatus siderophore biosynthetic gene sidA, encoding l-ornithine N5-oxygenase, is required for virulence. Infect. Immun. 73:5493-5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, G., P. Y. Cheng, A. Sham, J. R. Perfect, and J. W. Kronstad. 2008. Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol. Microbiol. 69:1456-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu, G., B. R. Steen, T. Lian, A. P. Sham, N. Tam, K. L. Tangen, and J. W. Kronstad. 2007. Transcriptional regulation by protein kinase A in Cryptococcus neoformans. PLoS Pathog. 3:e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung, W. H., A. Sham, T. Lian, A. Singh, D. J. Kosman, and J. W. Kronstad. 2008. Iron source preference and regulation of iron uptake in Cryptococcus neoformans. PLoS Pathog. 4:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung, W. H., A. Sham, R. White, and J. W. Kronstad. 2006. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol. 4:e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knight, S. A., G. Vilaire, E. Lesuisse, and A. Dancis. 2005. Iron acquisition from transferrin by Candida albicans depends on the reductive pathway. Infect. Immun. 73:5482-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuipers, M. E., L. Beljaars, N. Van Beek, H. G. De Vries, J. Heegsma, J. J. Van Den Berg, D. K. Meijer, and P. J. Swart. 2002. Conditions influencing the in vitro antifungal activity of lactoferrin combined with antimycotics against clinical isolates of Candida. Impact on the development of buccal preparations of lactoferrin. APMIS 110:290-298. [DOI] [PubMed] [Google Scholar]
- 23.Kuipers, M. E., H. G. de Vries, M. C. Eikelboom, D. K. Meijer, and P. J. Swart. 1999. Synergistic fungistatic effects of lactoferrin in combination with antifungal drugs against clinical Candida isolates. Antimicrob. Agents Chemother. 43:2635-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lesuisse, E., B. Horion, P. Labbe, and F. Hilger. 1991. The plasma membrane ferrireductase activity of Saccharomyces cerevisiae is partially controlled by cyclic AMP. Biochem. J. 280(Pt. 2):545-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, L., D. Bagley, D. M. Ward, and J. Kaplan. 2008. Yap5 is an iron-responsive transcriptional activator that regulates vacuolar iron storage in yeast. Mol. Cell. Biol. 28:1326-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, L., and J. Kaplan. 1998. Defects in the yeast high affinity iron transport system result in increased metal sensitivity because of the increased expression of transporters with a broad transition metal specificity. J. Biol. Chem. 273:22181-22187. [DOI] [PubMed] [Google Scholar]
- 27.Liu, X., G. Hu, J. Panepinto, and P. R. Williamson. 2006. Role of a VPS41 homologue in starvation response, intracellular survival and virulence of Cryptococcus neoformans. Mol. Microbiol. 61:1132-1146. [DOI] [PubMed] [Google Scholar]
- 28.Mallory, J. C., G. Crudden, B. L. Johnson, C. Mo, C. A. Pierson, M. Bard, and R. J. Craven. 2005. Dap1p, a heme-binding protein that regulates the cytochrome P450 protein Erg11p/Cyp51p in Saccharomyces cerevisiae. Mol. Cell. Biol. 25:1669-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marquis, J. F., and P. Gros. 2007. Intracellular Leishmania: your iron or mine? Trends Microbiol. 15:93-95. [DOI] [PubMed] [Google Scholar]
- 30.Prasad, T., A. Chandra, C. K. Mukhopadhyay, and R. Prasad. 2006. Unexpected link between iron and drug resistance of Candida spp.: iron depletion enhances membrane fluidity and drug diffusion, leading to drug-susceptible cells. Antimicrob. Agents Chemother. 50:3597-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robertson, L. S., H. C. Causton, R. A. Young, and G. R. Fink. 2000. The yeast A kinases differentially regulate iron uptake and respiratory function. Proc. Natl. Acad. Sci. USA 97:5984-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santos, R., N. Buisson, S. Knight, A. Dancis, J. M. Camadro, and E. Lesuisse. 2003. Haemin uptake and use as an iron source by Candida albicans: role of CaHMX1-encoded haem oxygenase. Microbiology 149:579-588. [DOI] [PubMed] [Google Scholar]
- 33.Schaible, U. E., and S. H. Kaufmann. 2004. Iron and microbial infection. Nat. Rev. Microbiol. 2:946-953. [DOI] [PubMed] [Google Scholar]
- 34.Schrettl, M., E. Bignell, C. Kragl, C. Joechl, T. Rogers, H. N. Arst, Jr., K. Haynes, and H. Haas. 2004. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J. Exp. Med. 200:1213-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Searle, S., N. A. Bright, T. I. Roach, P. G. Atkinson, C. H. Barton, R. H. Meloen, and J. M. Blackwell. 1998. Localisation of Nramp1 in macrophages: modulation with activation and infection. J. Cell Sci. 111(Pt. 19):2855-2866. [DOI] [PubMed] [Google Scholar]
- 36.Shi, X., C. Stoj, A. Romeo, D. J. Kosman, and Z. Zhu. 2003. Fre1p Cu2+ reduction and Fet3p Cu1+ oxidation modulate copper toxicity in Saccharomyces cerevisiae. J. Biol. Chem. 278:50309-50315. [DOI] [PubMed] [Google Scholar]
- 37.Singh, A., S. Severance, N. Kaur, W. Wiltsie, and D. J. Kosman. 2006. Assembly, activation, and trafficking of the Fet3p · Ftr1p high affinity iron permease complex in Saccharomyces cerevisiae. J. Biol. Chem. 281:13355-13364. [DOI] [PubMed] [Google Scholar]
- 38.Spizzo, T., C. Byersdorfer, S. Duesterhoeft, and D. Eide. 1997. The yeast FET5 gene encodes a FET3-related multicopper oxidase implicated in iron transport. Mol. Gen. Genet. 256:547-556. [DOI] [PubMed] [Google Scholar]
- 39.Steen, B. R., S. Zuyderduyn, D. L. Toffaletti, M. Marra, S. J. Jones, J. R. Perfect, and J. Kronstad. 2003. Cryptococcus neoformans gene expression during experimental cryptococcal meningitis. Eukaryot. Cell 2:1336-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szczypka, M. S., Z. Zhu, P. Silar, and D. J. Thiele. 1997. Saccharomyces cerevisiae mutants altered in vacuole function are defective in copper detoxification and iron-responsive gene transcription. Yeast 13:1423-1435. [DOI] [PubMed] [Google Scholar]
- 41.Tangen, K. L., W. H. Jung, A. P. Sham, T. Lian, and J. W. Kronstad. 2007. The iron- and cAMP-regulated gene SIT1 influences ferrioxamine B utilization, melanization and cell wall structure in Cryptococcus neoformans. Microbiology 153:29-41. [DOI] [PubMed] [Google Scholar]
- 42.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venkatesh, M. P., and L. Rong. 2008. Human recombinant lactoferrin acts synergistically with antimicrobials commonly used in neonatal practice against coagulase-negative staphylococci and Candida albicans causing neonatal sepsis. J. Med. Microbiol. 57:1113-1121. [DOI] [PubMed] [Google Scholar]
- 44.Weissman, Z., and D. Kornitzer. 2004. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol. Microbiol. 53:1209-1220. [DOI] [PubMed] [Google Scholar]
- 45.Weissman, Z., R. Shemer, E. Conibear, and D. Kornitzer. 2008. An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Mol. Microbiol. 69:201-217. [DOI] [PubMed] [Google Scholar]
- 46.Yu, J. H., Z. Hamari, K. H. Han, J. A. Seo, Y. Reyes-Dominguez, and C. Scazzocchio. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41:973-981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.