Abstract
Candida albicans strains tolerate aneuploidy, historically detected as karyotype alterations by pulsed-field gel electrophoresis and more recently revealed by array comparative genome hybridization, which provides a comprehensive and detailed description of gene copy number. Here, we first retrospectively analyzed 411 expression array experiments to predict the frequency of aneuploidy in different strains. As expected, significant levels of aneuploidy were seen in strains exposed to stress conditions, including UV light and/or sorbose treatment, as well as in strains that are resistant to antifungal drugs. More surprisingly, strains that underwent transformation with DNA displayed the highest frequency of chromosome copy number changes, with strains that were initially aneuploid exhibiting ∼3-fold more copy number changes than strains that were initially diploid. We then prospectively analyzed the effect of lithium acetate (LiOAc) transformation protocols on the stability of trisomic chromosomes. Consistent with the retrospective analysis, the proportion of karyotype changes was highly elevated in strains carrying aneuploid chromosomes. We then tested the hypothesis that stresses conferred by heat and/or LiOAc exposure promote chromosome number changes during DNA transformation procedures. Indeed, a short pulse of very high temperature caused frequent gains and losses of multiple chromosomes or chromosome segments. Furthermore, milder heat exposure over longer periods caused increased levels of loss of heterozygosity. Nonetheless, aneuploid chromosomes were also unstable when strains were transformed by electroporation, which does not include a heat shock step. Thus, aneuploid strains are particularly prone to undergo changes in chromosome number during the stresses of DNA transformation protocols.
Candida albicans is the most prevalent fungal pathogen of humans and is found in nature as a diploid that usually carries both mating type genes. Population studies suggest that most isolates are clonal and that genetic exchange rarely occurs in the human host. In the laboratory, C. albicans can undergo a parasexual cycle in which diploid strains homozygous for the mating type-like locus (MTLhom) mate with one another to form tetraploids (50, 66). No bona fide meiosis has been detected; however, when these tetraploid cells are stressed by nitrogen starvation or growth on sorbose, a poor carbon source, chromosome numbers return to near-diploid levels by a process termed concerted chromosome loss (6, 32). Presumably, these tetraploid cells undergo nondisjunction events during mitosis, although the mechanism that generates concerted chromosome loss is not understood. Analysis of the near-diploid strains revealed that chromosome segregation is random (non-Mendelian) and that the majority of the near-diploid strains carry at least one trisomic chromosome (32).
The C. albicans genome has long been known to tolerate aneuploidy (reviewed in reference 78). Aneuploidy is seen in clinical isolates, often in those with colony morphology mutations, which appear more frequently in strains from deep-seated sites of infection (82; reviewed in reference 78). Recent studies show that the frequency of aneuploidy and loss of heterozygosity is also higher in strains propagated in vivo than in those maintained in vitro (34). Early studies of C. albicans strains used UV treatment to induce mutations and chromosome loss events in order to generate genetic linkage maps (98, 99). The demonstration of a linkage between genotype and phenotype was first demonstrated convincingly by Rustchenko and coworkers. They found that loss of chromosome 5 (Chr5) permitted growth on sorbose (53) and that subsequent reduplication of Chr5 under nonselective conditions reversed this phenotype. This phenotype is apparently due to multiple genes on the right arm of Chr5 that negatively regulate SOU1 (sorbose utilization 1), which is found on Chr4 (44, 54).
Aneuploidy is also prevalent in C. albicans strains that have acquired resistance to fluconazole (FluR). A survey of resistant and sensitive strains found that ∼50% of resistant strains carried at least one aneuploid chromosome and that ∼20% of these strains included two extra copies of Chr5L organized as an isochromosome [i(5L)] (81). FluR in these strains is due to the increase in the number of copies of two genes on Chr5: ERG11, the drug target, and TAC1, encoding a transcription factor that upregulates expression of the CDR1 and CDR2 efflux pumps (82).
Aneuploidies can be detected by several methods. Classic studies followed cosegregation of multiple mutant alleles (79). Pulsed-field gel electrophoresis by contour-clamped homogeneous electric field (CHEF) of the C. albicans karyotype can detect chromosome sizes and the relative intensity of different chromosome bands, but because of genome rearrangements, including translocations (68, 90), it requires Southern hybridization to verify assumptions about which gel bands correspond to which genome segments. Quantitative Southern blotting, although laborious, also can reveal alterations in relative copy number of different chromosomes (16). Loss of heterozygosity (LOH) of markers is detectable by single nucleotide polymorphism (SNP) analysis, using either a microarray format (33, 35) or analysis of SNPs that include restriction fragment length polymorphisms (36). Chromosome loss from a diploid strain results in LOH of all markers on both chromosome arms (62), although it is not possible to distinguish homozygous diploids from haploids by SNP analysis alone. Gain of a chromosome (from heterozygous disomy [a:b] to heterozygous trisomy [a:a:b]) results in skewed allelic ratios (1:2 instead of 1:1) (62) for multiple markers on both chromosome arms. Comparative genome hybridization (CGH), in which genomic DNA from a test strain is hybridized competitively with a strain of known chromosome copy number, provides a comprehensive analysis of relative copy number when performed with whole-genome microarrays (array CGH [aCGH]). When aCGH is combined with Southern analysis of chromosomes on CHEF gels, genome copy number and the chromosome size can be used to infer chromosome organization (for example, see reference 81).
In Saccharomyces cerevisiae, 8% of strains that underwent systematic transformation with DNA to delete whole open reading frames became aneuploid, usually due to segmental duplications (49). These copy number variations were initially revealed when gene expression levels, generated using whole-genome microarrays, were plotted as a function of chromosome position. aCGH demonstrated that increased transcript levels across a DNA region correlated well with increased DNA copy levels in the same DNA region (45, 49, 73). It is important to note that, while aCGH readily detects imbalances in the number of chromosomes in a strain, it cannot distinguish between genomes that are of different ploidies (completely haploid, diploid, triploid, or tetraploid).
Transformation with DNA has also been suspected of causing genome changes in C. albicans lab strains. Transformation of strain SC5314 to delete URA3 on Chr3 resulted in strain CAI-4 (31). CAI-4 isolates from different labs have been shown to carry an extra copy of Chr1 and/or Chr2 (16, 80). Subsequent transformation of CAI-4 to generate histidine auxotrophs resulted in homozygosis of the left arm of Chr2 (A. Forche, unpublished data), the loss of the Chr1 and -2 trisomies, and the loss of ∼38 kb from one Chr5 right-arm telomere in some transformants (33, 80). This small segmental aneuploidy arose near the HIS1 locus (80) and includes at least one essential gene because the intact copy of Chr5 cannot be lost (2). While these studies revealed the possibility that transformation protocols result in increased levels of whole-chromosome aneuploidy as well as specific segmental aneuploidies, the possibility has not been analyzed systematically.
Here we used whole-genome transcript profiles from over 400 microarray experiments to predict aneuploidy in C. albicans strains. In addition to the high levels of aneuploidy expected in UV/sorbose- and drug-treated strains, we found that DNA transformation was associated with changes in chromosome copy number, especially in strains that were initially aneuploid. We then tested this hypothesis and found that aneuploid chromosomes frequently change from trisomic to disomic upon DNA transformation with lithium acetate (LiOAc) using either a heat shock or an electroporation protocol. Nonetheless, short-term exposure to high temperature alone results in high levels of chromosome gains and losses and may explain why some strains become aneuploid during transfer between labs.
MATERIALS AND METHODS
Prediction of aneuploidy.
Transcription profiles (Table 1) were collected as previously described (52). The microarray data were prepared for analysis as follows. If the data were already normalized and in (log) ratio format, we used the ratios as provided. If the data were raw intensities, they were background corrected via the Normexp algorithm (75) and normalized using print-tip LOESS normalization (86) (if information on print-tip groups was available) or global LOESS normalization (86). Single-channel data were normalized using quantile normalization (9). The background correction and normalization were done in the R limma package from www.bioconductor.org. All log2 ratios were averaged over dye swaps and replicates for the same gene.
TABLE 1.
Summary of expression array data sets, experiment type, and aneuploid chromosomes
| Topic | Reference | No. of experiments
|
No. of arrays predicting aneuploidyb | Aneuploid chromosome(s) | ||||
|---|---|---|---|---|---|---|---|---|
| Totala | Time course | UV/sorbose | Drug | Transformation | ||||
| Azole resistance | 20 | 4 | 1 | 3 | ||||
| Cell wall | 39 | 1 | 1 | |||||
| Morphogenesis | 67 | 12 | 10 | 2 | 1 | 5, 7 | ||
| Mating | 95 | 12 | 12 | |||||
| Mating | 94 | 50 | 50 | 46 | 1, 2, 6 | |||
| Stress response | 28 | 8 | 8 | |||||
| Stress response | 48 | 32 | 32 | |||||
| Quorum sensing | 30 | 12 | 12 | |||||
| Stress response | 29 | 6 | 6 | |||||
| Cell cycle | 5 | 6 | 5 | 1 | ||||
| Morphogenesis | 60 | 8 | 6 | 2 | ||||
| Host interaction | 37 | 2 | 2 | |||||
| Cell wall | 70 | 1 | 1 | |||||
| Host interaction | 4 | 1 | 1 | |||||
| Azole resistance | 3 | 1 | 1 | |||||
| Mating | 22 | 13 | 7 | 6 | 6 | R, 1, 2, 3, 4, 5, 6, 7 | ||
| Morphogenesis | 55 | 19 | 16 | 3 | ||||
| Morphogenesis | 46 | 10 | 7 | 3 | 2 | 2 | ||
| Stress response | 83 | 3 | 2 | 1 | 1 | 2, 3 | ||
| Stress response | 8 | 4 | 3 | 1 | ||||
| Morphogenesis | 12 | 1 | 1 | |||||
| Signaling | 17 | 16 | 15 | 1 | ||||
| Drug response | 84 | 1 | 1 | |||||
| Quorum sensing | 15 | 2 | 2 | |||||
| Starvation | 93 | 6 | 4 | 2 | ||||
| Host interaction | 77 | 5 | 5 | |||||
| Azole resistance | 21 | 7 | 1 | 6 | 5 | 4, 5, 7 | ||
| Cell wall | 13 | 6 | 6 | |||||
| Azole resistance | 56 | 4 | 2 | 2 | 2 | 5, 6, 7 | ||
| Azole resistance | 57 | 15 | 13 | 2 | ||||
| Host interaction | 65 | 27 | 27 | |||||
| Azole resistance | 76 | 3 | 3 | 3 | 3 | |||
| Mating | 6 | 14 | 14 | |||||
| Mating | 7 | 17 | 14 | 3 | ||||
| Drug response | 61 | 1 | 1 | |||||
| Mating | 101 | 5 | 5 | |||||
| Host interaction | 89 | 10 | 9 | 1 | 1 | 5 | ||
| Stress response | 69 | 12 | 11 | 1 | 10 | 7 | ||
| Morphogenesis | 40 | 14 | 9 | 5 | ||||
| Drug response | 61 | 3 | 3 | |||||
| Morphogenesis | 23 | 4 | 1 | 3 | ||||
| Cytoskeleton | 71 | 18 | 11 | 7 | 1 | R, 1, 2, 3, 4, 5, 6, 7 | ||
| Drug response | 19 | 1 | 1 | |||||
| Morphogenesis | 85 | 1 | 1 | |||||
| Drug response | 11 | 5 | 1 | 1 | 3 | 1 | R | |
| Cell wall | 10 | 4 | 1 | 3 | ||||
| Adhesion | 101 | 2 | 2 | |||||
| Stress response | 97 | 2 | 2 | |||||
| Total | 48 | 411 | 290 | 9 | 17 | 95 | 79 | |
Only experiments with significant levels of expression were analyzed.
Whole chromosome or large segmental aneuploidies.
To predict aneuploid chromosomes from the expression data, a rank sum test was used to detect chromosomes whose median expression values differed significantly from the median expression levels of the nonaneuploid chromosomes of the rest of the genome. For each experiment, the median log2 ratio for the entire genome was initially set to 0. Then, expression values for each chromosome were compared to those of the rest of the genome. If the rank sum P value was less than 10−12, the chromosome was predicted to be aneuploid.
If a strain has multiple aneuploidies, the presence of one aneuploid chromosome can obscure prediction of other aneuploid chromosomes, because during the comparison of each chromosome to the rest of the genome, the overall expression level of the rest of the genome includes the effect of the other aneuploid chromosome(s). To account for this, the rank sum test was applied iteratively, and on each iteration the systematic up- or down-expression of earlier predicted aneuploidies was subtracted. On the first iteration of the rank sum test, the chromosome with the smallest P value was predicted to be aneuploid. To search for a possible second aneuploid chromosome, the first predicted aneuploid chromosome's median log2 ratio was reset to 0, the median log2 ratio for the remaining genome was also reset to 0, and the rank sum test was rerun. A P value cutoff was set at 10−12 to predict aneuploid chromosomes. To identify multiple chromosome aneuploidies within the same experiment, the rank sum test was repeated, with effects of aneuploid chromosomes predicted on earlier iterations being subtracted each time, until there were no longer any chromosomes identified with a P value of <10−12.
Visualization of expression array data.
Collected expression array data were plotted to the C. albicans genetic map using an updated version of the Chromosome_Map (80), based on Assembly 21 coordinates (96) (http://www.candidagenome.org/).
Strain maintenance and growth.
Strains were stored as freezer stocks in 50% glycerol at −80°C and grown on YPAD medium (1% yeast extract, 1% peptone, 2% glucose, 1.5% agar) at 30°C.
Strain construction.
Transformants of strains RM1000#2 (YJB7617), CAI-4 F2 (YJB8653), and CAI-4 F3 (YJB8654) were constructed using PCR-mediated gene deletion with 70 nucleotides of homology to the genomic locus with gene disruption primers (Table 2) and a lithium acetate-heat shock protocol (100) or electroporation protocol (58). BUD7 was deleted by amplification of URA3 from pGEM-URA3 (100). C. albicans transformants were isolated after incubation on SDC-uridine at 30°C.
TABLE 2.
List of primers used in this study
| Forward primer | Sequence | Reverse primer | Sequence | Probea | Purpose |
|---|---|---|---|---|---|
| 1045 | ATGCTACCTCCAACATTATCATCGATTCCTGAAGTTAAAGAAGAATATCCAGGTGTCTGTTTAATTGAAAGTTTTCCCAGTCACGACGTT | 1046 | TTATTCATCAGCATTATGAAGATTATACAATTTGATATAATCATTGTCCAAATCTGTCATCATTTCAATATGTGGAATTGTGAGCGGATA | bud7::URA3 disruption | |
| 3709 | TCAGATGTTCCAGCTACTGAATCT | 3710 | TTCAGTACCAGCTGGAGTCATTT | 15 | qRT-PCR for HWP1 |
| 3711 | TCAAGCTGATTGTGCTATTTTGA | 3712 | AATACCGGCTTCGAATTCAC | 131 | qRT-PCR for TEF1 |
| 3713 | TTGTTAAAGGGAATGGTTTGTTC | 3714 | CCCCTCTAATTTGATTTTTAACCA | 119 | qRT-PCR for CHA1 |
| 3715 | TTGTTAGTACCCATTTGATACCTTCA | 3716 | TCAATGTTTTCATCGGGACTC | 161 | qRT-PCR for RPB7 |
| 3717 | TGATGTTGGCTGCTATGACC | 3718 | AACACCGGAGCCAATAACC | 140 | qRT-PCR for CTR1 |
| 3719 | TTCTGATGGCACTTGTTTTGTC | 3720 | AGTGGTTTCTTCTGCATCATCA | 59 | qRT-PCR for FGR41 |
Roche Universal ProbeLibrary probe.
Quantitative real-time PCR.
The copy numbers of Chr1 and -2 were predicted by quantitative real-time PCR using Roche LightCycler 480 Probes Master and Universal ProbeLibrary probes. Primers and probes used in the reactions were designed by the Universal ProbeLibrary Assay Design Center (Roche Applied Science) and are listed in Table 2. Reactions were run on the Roche LightCycler 480 Real-Time PCR system. Analysis was performed using LightCycler 480 software release 1.5.0 sp3 and applying efficiency corrections to each primer set. Copy numbers of Chr1 and -2 were predicted as a normalized ratio of target to reference, where the target is Chr1 or -2 and the reference is Chr6. To validate the system, two primer sets from Chr1 and -2, as well as a primer set from Chr4 and -6, were used to analyze the parental strains (RM1000 2, CAI-4 F2, and CAI-4 F3), which have known karyotypes. Sets of transformants were then screened using one primer set each from Chr1, -2, and -6.
Extraction of genomic DNA.
Genomic DNA was prepared as previously described (80).
Heat shock at 50°C.
Single colonies were isolated on YPAD medium after 2 days of growth at 30°C. Three single colonies of strain SC5314 were inoculated into 2 ml YPAD and incubated at 30°C for 5 h to obtain actively dividing log-phase cultures. From each culture, 100 μl was diluted and plated onto YPAD to determine the total CFU prior to heat shock. The cultures were spun down and taken up in 100 μl YPAD, which was then transferred to 5-ml YPAD cultures prewarmed to 50°C. One milliliter of culture was taken out at 60, 90, and 120 s, and serial dilutions were generated and plated onto YPAD to determine the number of cells that survived the heat shock. From these YPAD plates, several small and large colonies were restreaked on YPAD to ensure that the colony phenotype was heritable.
Karyotype analysis by CHEF gel.
To determine whether strains subjected to heat shock had undergone gross chromosomal rearrangements, CHEF plugs were prepared for three independent small and large colonies per strain as previously described (82). CHEF plugs were run on a DRIII CHEF machine on a 1% Megabase agarose gel in 0.5× Tris-borate-EDTA under the following conditions: block 1, 60- to 120-s switch for 36 h at 6 V/cm with a 120° angle; block 2, 120- to 300-s switch for 12 h at 4.5 V/cm with a 120° angle. Strains with changes in CHEF karyotype were subjected to aCGH following established protocols (80).
Fluctuation analysis.
To determine whether rates of LOH are altered when cells are subjected to heat shock, we carried out fluctuation analysis using strain YJB9834, a Ura+ heterozygote derived from strain YJB7617 (RM1000). Strain YJB9834 was streaked for single colonies onto SDC-uri (to maintain the URA3 selection), and grown for 2 days at 30°C. Independent single colonies were each inoculated into 5 ml liquid YPAD cultures. All cultures were grown for 16 h, with 5 to 20 cultures each at 30°C, 39°C, and 42°C. Cells were pelleted, washed once with distilled water, and resuspended in 1 ml distilled water. Dilution series were made for each culture, and appropriate dilutions were plated onto YPAD for total cell count and onto 5-fluoroorotic acid (5-FOA) to obtain cells that lost the URA3 marker (5-FOAR). YPAD CFU were counted on day 2 and 5-FOAR colonies on day 3. URA3 loss rates were determined using the method of the median by Lea and Coulson (59) as described by Spell and Jinks-Robertson (87).
RESULTS
Prediction of aneuploidy using expression profiles of known drug-resistant strains.
Aneuploidy is a common feature of C. albicans strains that have undergone stresses such as exposure to, and survival of, antifungal drug treatment (81), as well as laboratory strains that have undergone series of transformations with DNA (16, 80; reviewed in reference 78). Because gene copy number correlates well with transcript level in many organisms (45, 49, 73, 92), including C. albicans (81), we first analyzed expression data from FluR strains that contained known aneuploidies. Indeed, known aneuploidies in several FluR strains were evident in published expression profiles when they were plotted as a function of chromosome position (for example, see reference 81).
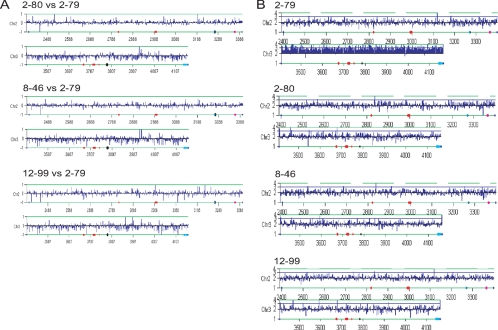
We then asked if aneuploidies predicted from published expression profiles were always detectable by aCGH. aCGH analysis of several fluconazole-treated strains confirmed all but one predicted aneuploidy (81) (data not shown). In this case, when expression data (76) from a series of drug-resistant strains isolated from a single human immunodeficiency virus-positive patient (74) were plotted as a function of chromosome position (Fig. 1A), the results suggested that Chr3 was monosomic in three resistant strains (2-80, 8-46, 12-99, corresponding to isolates 3, 15, and 17 in the patient [74]) because Chr3 expression levels were consistently low relative to expression levels in a less FluR isolate from the same patient (2-79, corresponding to isolate 2) (Fig. 1A). This result was particularly curious since several genes encoding drug efflux pumps (CDR1 and CDR2), as well as a regulator (MRR1) of the gene encoding the Mdr1p efflux pump, map to Chr3 (http://www.candidagenome.org/), and an earlier report suggested that Chr3 trisomy was associated with FluR (72). aCGH of all four strains used for the expression analysis was analyzed relative to that of an SC5314 reference control strain, and the analysis revealed that none of the three FluR isolates (isolates 3, 15, and 17) exhibited Chr3 monosomy (Fig. 1B). Rather, the less FluR isolate (isolate 2) that had been used as the reference control exhibited Chr3 trisomy (Fig. 1B). Thus, the relative expression of the strains did reflect a difference in copy number between the test and reference control strains, but the aneuploid strain was used as the reference.
FIG. 1.
RNA expression levels correlate with DNA copy number. (A) RNA expression (log2 ratios) analysis for Chr2 and -3 of fluconazole-resistant isolates (2-80, 8-46, and 12-99) compared to a drug-sensitive isolate (2-79). Lower relative expression of Chr3 suggests monosomy in all three fluconazole-resistant isolates. (B) aCGH analysis (DNA copy number) for Chr2 and -3 of the isolates used for panel A relative to the known diploid SC5314. Higher levels of Chr3 DNA in the drug-sensitive isolate (2-79) indicate trisomy in this isolate, while the fluconazole-resistant isolates (2-80, 8-46, and 12-99) are disomic for Chr3.
Predicting aneuploidy from a large collection of microarray data sets.
Since uniformly up- or down-expressed genes over an entire chromosome (or chromosome arm) are indicative of aneuploidy, we determined how frequently predicted aneuploidies occur in a broad range of C. albicans strains by analyzing expression data from a large set of published expression array data sets (Table 1). We collected data from over 400 expression array experiments, normalized the data when needed (see Materials and Methods), and analyzed them as a function of chromosome position (80). The presence of a predicted aneuploidy was defined by the likelihood (P < 10−12) that the median expression level of genes on a given chromosome was significantly increased or decreased compared to that of the genes on the nonaneuploid chromosomes.
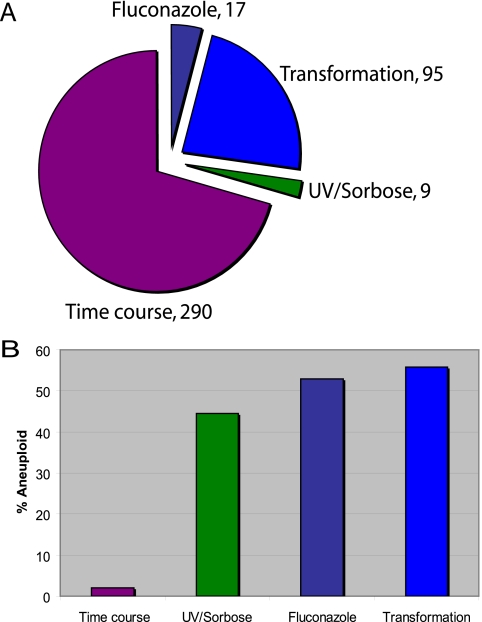
To determine what types of manipulations might cause these predicted aneuploidies, we then categorized the predicted aneuploidies based on the types of experiments performed: those involving treatment with UV and/or sorbose to select for genome changes, those involving treatment with the antifungal drug fluconazole, those in which strains differed from the reference control because they had been transformed with DNA, and those in which the same strain had been followed over time under different conditions. The proportions of experiments of each type are illustrated in Fig. 2A and are detailed in Table 1, and the proportion of the strains carrying at least one large aneuploidy is shown in Fig. 2B.
FIG. 2.
Proportion of aneuploid strains based on experiment type. (A) Four hundred eleven published expression experiments were analyzed to predict the frequency of aneuploidy in C. albicans strains. Experiment type included DNA transformation, treatment with fluconazole, exposure to UV and/or sorbose, and time course experiments. Numbers indicate the number of each experiment type included in this analysis. (B) The proportion of aneuploid strains from each experiment type was predicted by plotting expression data as a function of chromosome position. High levels of aneuploidy are predicted for UV and/or sorbose exposure, treatment with fluconazole, and DNA transformation, while very few aneuploids are predicted from time course experiments.
Experiments that were performed as time courses usually compared a specific strain as it was treated with an altered growth condition over time. The reference control was usually the same strain that did not undergo treatment or a mixture of strains from the experiment. Thus, it is not surprising that these time course experiments generally did not reveal predicted aneuploidies (Fig. 2B), since dramatic changes in chromosome composition are not expected to appear within the few division cycles monitored during these experiments.
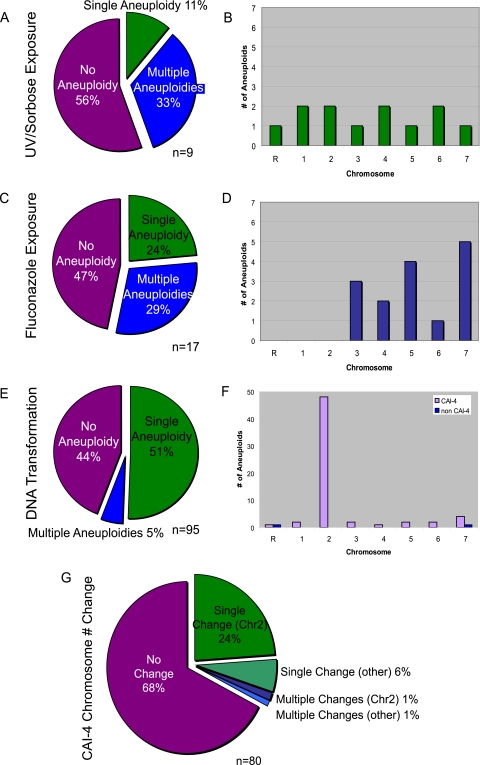
We also were not surprised to detect a high level (44%) of aneuploidy in strains exposed to UV and/or sorbose (Fig. 2B, 3A, and 3B), treatments that are used to deliberately induce genome changes (6, 53). These changes affected all eight C. albicans chromosomes with roughly equal frequencies. Interestingly, 75% of the aneuploid strains involved multiple chromosomes (Fig. 3A). Thus, sorbose and UV treatment promoted loss of chromosomes including, but not limited to or even particularly biased for, Chr5. This is consistent with the observations of others (1, 2, 32) and suggests that the stress of growing on a suboptimal carbon source may affect chromosome stability through multiple mechanisms.
FIG. 3.
Aneuploidy is common in strains treated with UV/sorbose or fluconazole and in strains transformed with DNA. The proportions of strains from each experiment type (UV/sorbose exposure, fluconazole exposure, DNA transformation) that exhibit no aneuploidy, a single aneuploid chromosome, and multiple aneuploid chromosomes (A, C, and E) and of chromosomes that became aneuploid (B, D, and F) are shown. Because CAI-4 was aneuploid prior to transformation, genome instability is indicated as a change in chromosome copy number, and this change frequently involved the aneuploid chromosome Chr2 (G).
Consistent with previous studies, approximately half of the strains that had been treated with fluconazole contained aneuploidies (81). Interestingly, there was a strong bias for aneuploidy of the smaller chromosomes (Chr3 to -7) in drug-treated strains (Fig. 3C and D). While many genes affecting drug efflux pumps are found on Chr3, -4, -5, and -6, and the drug target ERG11 locus is found on Chr5, it does not explain why Chr7 aneuploidy is high, or why we did not detect aneuploidy of ChrR, -1, and -2 in these strains. We note that in a clinical isolate used for experimental evolution studies (T118), appearance of i(5L) was usually accompanied by trisomy of Chr7 (A. Selmecki, K. Dulmage, L. E. Cowen, J. B. Anderson, and J. Berman, submitted).
Surprisingly, the expression array data predicted the highest frequencies of aneuploidy (56%) in strains that had undergone transformation with DNA (Fig. 2B). The high frequency of aneuploidy raises a serious concern about the integrity of strains following standard methods of molecular manipulation. Strains in this group included those that had been transformed with plasmid DNA or with PCR-amplified fragments used for gene disruption, deletion (100), or generation of fusion proteins (41-43). Less than 10% of the aneuploidies involved multiple chromosomes, indicating that the mechanism giving rise to genome changes is likely to be different from that induced during UV and/or sorbose treatment (Fig. 3E).
Curiously, while aneuploidies in the transformant strains were detected for all eight C. albicans chromosomes, 75% of the 64 aneuploid events detected were aneuploidy of Chr2 (Fig. 3E). Importantly, all cases of Chr2 aneuploidy appeared among strains derived from CAI-4, a strain that is trisomic for Chr1 or for Chr1 and -2 (16, 80) (Fig. 3F). Indeed, 84% (80/95 strains) of transformant strains here were CAI-4 derivatives. Of the CAI-4 derivatives that were predicted to be aneuploid, 94% (48 of 51 strains) exhibited Chr2 trisomy, which also was present in the parental strain CAI-4. In contrast to CAI-4 derivatives, 64% of which were aneuploid, only 13% of the non-CAI-4-derived transformants were aneuploid. The non-CAI-4-derived strains that became aneuploid usually became aneuploid for a single chromosome, and that chromosome was not targeted for modification by transforming DNA.
If a strain generated by transformation of the CAI-4 parent strain is compared to its parent strain and if both retain the same aneuploidies, comparative hybridization will not detect the aneuploidy in either of them. Since we are interested in genome changes due to transformation, we compared the frequency with which chromosome copy number changed, including a return to the diploid, nonaneuploid state, for all of the transformed strains. Of the CAI-4-derived transformant strains, 32% (26/80) exhibited a change in chromosome copy number (Fig. 3G). The majority of these (24/26) exhibited a change in the copy number of one chromosome relative to its CAI-4 parent. In 79% (19/24) of these, the changes were due to loss of one copy of Chr2, such that a disomic transformant was derived from the trisomic parent. In addition, a small number of strains (2% of CAI-4 derivatives) exhibited multiple chromosome changes, including a return to disomy of Chr2 along with other aneuploidies.
Aneuploid strains are more likely to undergo chromosome number changes during transformation.
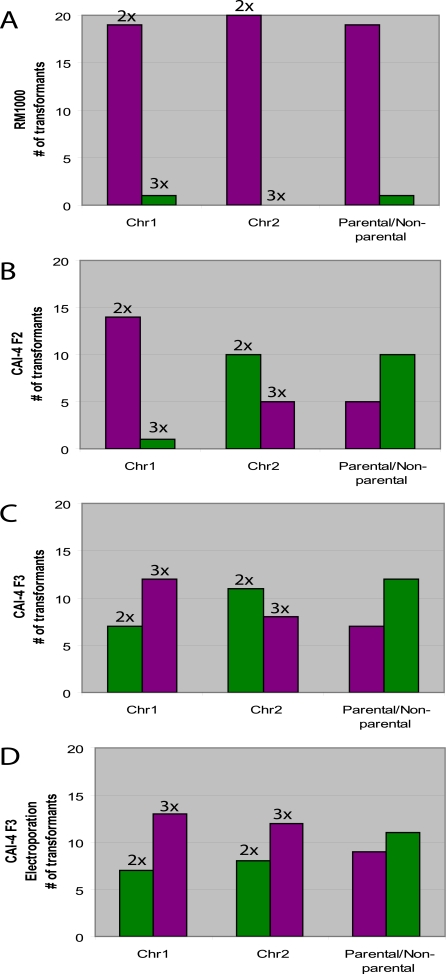
Because all the data above were retrospective, we next performed a prospective analysis to directly test the hypothesis that aneuploid strains are more unstable than isogenic diploid strains. For this experiment we used the sister strains CAI-4 F2 and CAI-4 F3, which carry one or two large trisomic chromosomes, and compared them to control strain RM1000#2, which was derived from CAI-4 and is disomic for all chromosomes (80). All three strains were transformed by the LiOAc-heat shock treatment method to delete one copy of bud7 by replacing it with URA3. We then analyzed up to 20 randomly selected transformants per strain using quantitative real-time PCR to predict copy numbers of Chr1 and -2, using Chr6 copy number as an internal control. For RM1000#2, we detected one transformant (5%) that had become trisomic for Chr1; all other transformants remained disomic for Chr1 and -2 (Fig. 4A). In contrast, for CAI-4 F2, which is trisomic for Chr2, 67% (10/15) of the transformants had lost the trisomic copy of Chr2, including one that also became trisomic for Chr1 (Fig. 4B); in CAI-4 F3, only 37% (7/19) retained both trisomic chromosomes, with 6 transformants losing a copy of both Chr1 and Chr2, 1 losing a copy Chr1, and 5 losing a copy of Chr2 (Fig. 4C). Thus, aneuploidy in a parental strain greatly increases the frequency with which that strain loses trisomic chromosomes during DNA transformation.
FIG. 4.
Chromosome copy number changes are frequent in aneuploid strains. Strains RM1000 2, CAI-4 F2, and CAI-4 F3 were transformed by LiOAc-heat shock transformation (A to C), and CAI-4 F3 was transformed by electroporation transformation (D). Chr1 and -2 copy numbers were predicted by quantitative real-time PCR. Because the different strains have different numbers of Chr1 and Chr2 (e.g., CAI-4 F2 parental chromosome copy numbers are two for Chr1 and three for Chr2), purple bars indicate that the transformants have not changed relative to the parental chromosome copy number; green bars indicate that transformants did have a change in chromosome copy number relative to the parent. Over half of the CAI-4 F2 and CAI-4 F3 transformants analyzed had a chromosome copy number that differed from that of the parent, while for nearly all RM1000 transformants, chromosome copy number resembled that of the parent (rightmost columns). CAI-4 F3 transformed by electroporation protocol also exhibited a large number of isolates that had an altered chromosome copy number relative to the parent.
Elevated temperature promotes changes in chromosome copy number.
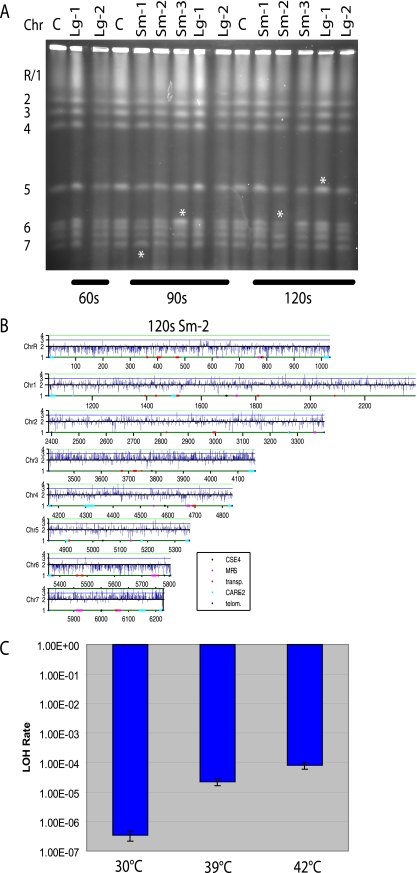
Transformation of C. albicans is generally performed by exposure of cells to LiOAc followed by a heat shock at 42°C for 1 h. In early studies of C. albicans tetraploids, it was reported that chromosome loss was induced by a heat shock of 50°C for 1 to 2 min (47). This conclusion was based on the loss of multiple markers that mapped to the same chromosome and allowed detection of chromosome loss but not chromosome gain events. Since most aneuploidies detected are trisomies (chromosome gain) rather than monosomies (chromosome loss) (78, 81), we used molecular methods to determine whether exposure to 50°C for 60, 90, or 120 s induces chromosome gain as well as chromosome loss in the C. albicans strain SC5314. We used CHEF gel analysis to identify colonies that had undergone obvious changes in chromosome mobility or alterations of chromosome band intensity suggestive of changes in chromosome copy number (Fig. 5A). We then performed aCGH to specifically analyze individuals suspected of having altered chromosome copy numbers (Fig. 5B).
FIG. 5.
Elevated temperature causes genetic instability. (A) CHEF gel of large and small colonies isolated after SC5314 was subjected to heat shock at 50°C for 60, 90, or 120 s, as indicated. Asterisks are located below (Sm-1 at 90 s) or above the chromosomes with suspected alterations in band intensity. C, control (SC5314 with no heat shock); Lg, large colony isolate; Sm, small colony isolate. (B) aCGH analysis of small colony isolate 2 after heat shock at 50°C for 120 s (from panel A) indicates multiple changes in chromosome copy number relative to SC5314 without heat shock: ChrR and -6 have become monosomic, while Chr3 and -7 have become trisomic. Some of these changes are detected by CHEF gel analysis (A). (C) Strain YJB9834 (heterozygous for URA3 on Chr5) was analyzed by fluctuation analysis at 30°C, 39°C, and 42°C. Bars indicate the LOH rate at the URA3 marker for each temperature tested.
As reported previously (47), heat shock affected strain viability as a function of time of exposure to 50°C. Following exposure to the heat shock, strain SC5314 was spread on rich medium and individual colonies were analyzed. Interestingly, all cultures exposed to 50°C for 90 or 120 s gave rise to a mixture of large and small colonies. Small colonies reflect slow growth and are often indicative of strains with monosomies or other chromosome rearrangements (53; reviewed in references 18 and 78). In general, the proportion of small colonies increased as a function of the time of exposure to 50°C.
CHEF analysis of 12 to 14 heat shock-treated colonies revealed that about twice as many small colonies had a predicted karyotype change (32%) as large colonies (15%). aCGH analysis of several of these colonies confirmed chromosome copy number changes in all small colony isolates and often revealed additional aneuploidies that were not evident by CHEF karyotype analysis. The number of chromosome changes detected by CHEF and aCGH increased with increasing time of exposure to heat shock, and in many cases multiple chromosomes were aneuploid within individual isolates (Fig. 5B). Finally, the aneuploidies that appeared included some increases, as well as many decreases, in chromosome copy number. Thus, heat shock most likely leads to an increase in chromosome nondisjunction, resulting in the gain and loss of chromosomes.
Exposure to 42°C results in increased LOH.
The experiments described above indicate that very high temperature affects chromosome stability in C. albicans. However, DNA transformation is performed with a milder heat treatment over a longer period (42°C for 1 h). C. albicans strains are known to survive at 42°C, and thus, these conditions are likely to have a much less dramatic effect on genome rearrangements. Thus, we asked if exposure to 42°C affects the genome integrity of C. albicans cells. Chromosome loss from a diploid cell results in LOH at multiple genetic markers along that chromosome. We used strain YJB9834, which is heterozygous for a URA3 marker on Chr5, to determine whether exposure to 42°C for 16 h affected rates of LOH as determined by fluctuation analysis (59). Importantly, the rate of LOH at 42°C was 200-fold higher than the LOH rate at 30°C and >3-fold higher than the LOH rate at 39°C (Fig. 5C). This indicates that exposure to elevated temperatures can result in high levels of LOH and/or changes in chromosome copy number.
Aneuploids are also unstable when transformed by electroporation.
All transformation experiments described above utilized the LiOAc-heat shock protocol (100). Electroporation is an alternative method of DNA transformation that does not involve a heat treatment (88). Rather, it requires use of an electric shock coupled with LiOAc incubation. Because heat treatments increased the levels of recombination and/or aneuploidy, we asked if transformation by electroporation would have a similar or different effect on transformation compared to the LiOAc-heat shock protocol. For this experiment we transformed CAI4-F3 with the URA3 designed to delete one allele of BUD7 that was used in the LiOAc-heat shock transformation experiment described above. Interestingly, the heat shock and electroporation DNA transformation protocols gave rise to similar levels of chromosome copy number changes (Fig. 4D). Thus, while exposure to elevated temperature can cause alterations in chromosome number, transformation by electroporation also results in the instability of chromosome number in aneuploid strains.
DISCUSSION
Taken together, our retrospective and prospective analysis of C. albicans transformants detected a relatively high degree of aneuploidy, especially in strains resulting from transformation of an aneuploid parent. In general, the chromosome that was targeted by transforming DNA did not become aneuploid. Additionally, transformation of aneuploid strains resulted in ∼3-fold more changes in chromosome copy number than that of nonaneuploid strains, although the change usually involved loss of the aneuploidy. While these levels of aneuploidy and chromosome change are high and clearly should be of concern to those working with transformed C. albicans strains, it is important to note that strains maintained in the lab do not generally become aneuploid during conventional growth in rich medium. For example, aCGH analysis of strain SC5314 relative to itself has never detected the acquisition of aneuploidy (data not shown). Similarly, Rustchenko and coworkers did not see changes when strain CAF4-2 was propagated nonselectively (1). Thus, there must be something about the process of transformation with DNA that results in increased genome instability.
The effect of aneuploidy on genome stability in C. albicans is reminiscent of the situation in cancer cells, where cells become polyploid or aneuploid and then continue to undergo karyotype changes at a high frequency (38, 92). Furthermore, it has been proposed that aneuploidy itself can initiate tumor formation, although some aneuploid cells grow less rapidly than normal cells (14, 24-27, 92), and cells that become aneuploid may also acquire point mutations that promote tumor growth (64). Here, retrospective and prospective experiments suggest that aneuploid cells are ∼3-fold more likely to undergo changes in chromosome copy number than diploid cells. Thus, like cancer cells, C. albicans is prone to become aneuploid, and once it is aneuploid, changes in chromosome copy number become even more frequent. Studies of cancer and yeast cells indicate that not all aneuploidies promote cell proliferation (64) but that some aneuploidies are well tolerated and may provide selective advantages under specific stress conditions (81, 82).
Exploiting aneuploidy to generate specialized strains.
Altered ploidy can be useful for generating some types of specialized strains. A classic example is the use of sorbose treatment to induce loss of one copy of Chr5, resulting in strains homozygous for the mating type-like locus MTL (MTLhom strains), which are useful for mating studies. Our work suggests that treatment with UV/sorbose induces changes in chromosome copy number that are not limited to loss of Chr5 (Fig. 3B), and thus, strains subjected to sorbose treatment should be tested for chromosome copy number prior to their use in other experiments. An alternative is to incubate cells for a short period at 50°C (47). In this case, inclusion of a counterselectable marker on Chr5 would permit identification of the rare cells that undergo LOH of the counterselectable marker, and screening of these selected cells by PCR would quickly identify those that are MTLhom (63).
Suggested methods for detecting aneuploid strains.
Ultimately, strains that are constructed by transformation and are to be used in additional experiments should be screened for aneuploidy using one of the approaches discussed below.
If transformants exhibit a mixture of colony phenotypes, aneuploidy should be suspected, and it is especially important to test the chromosome copy number (78). We note that selection of larger colonies may reduce the likelihood that a strain is monosomic, yet it is not sufficient to rule out aneuploidy. Indeed, in S. cerevisiae, sometimes the desired mutant colonies are small, and those that have acquired suppressor mutations, such as segmental aneuploidies that suppress the original mutation, form larger colonies (for example, see reference 49). We also suggest that more than one independent transformant be analyzed and, preferably, that any mutation of interest be complemented by reintroduction of a wild-type copy of the gene of interest.
Aneuploidy can be detected in a number of ways. aCGH is the most comprehensive (and should be performed with a reference control strain such as SC5314, which is known to be a stable diploid). An alternative is to analyze transcription profile data as a function of chromosome position (Fig. 1) (49). Aneuploidy detected in this manner generally correlates well with aCGH data (Fig. 1) (49). Of course, analysis of expression profiles that reveal aneuploidy must take into account the altered copy number (51), which can overemphasize relative changes in gene expression on the aneuploid chromosome(s). For labs that do not have access to microarray technology, quantitative real-time PCR of multiple markers (Fig. 5) can be used to detect aneuploidies in chromosomes of interest and ideally should be performed for markers on all eight chromosomes.
Our detection of aneuploidy in a reference strain raises two important points. First, expression data obtained using relative hybridization from two strains can exhibit biased gene expression along a whole chromosome or large chromosomal segment because of aneuploidy in either one of the two strains; therefore, the choice of reference strain is critical. We suggest that a known diploid strain (e.g., SC5314) always be used. Second, when the same isolate (2-79 [isolate 2], described above) was obtained directly, rather than via the lab that performed the microarray analysis, the trisomy was not detected by aCGH. This suggests that an increase in the copy number of Chr3 occurred during transfer of the strain or its subsequent propagation prior to transcription profile analysis. One possibility is that exposure to heat during transfer of strains between laboratories, especially during summer months or in warm climates, may cause the strains to incur alterations in chromosome copy number and/or increased recombination. We recommend that exposure of strains to high temperatures be avoided whenever possible.
Aneuploidy is present in some but certainly not all C. albicans strains; it rarely arises during propagation in the laboratory. Furthermore, the major change we detected in this study was loss of one or more aneuploid chromosomes (Fig. 5). Indeed, while the CAI-4 strains all carry aneuploidies, laboratory strains derived from them (e.g., RM1000) do not. Furthermore, important conclusions have been reached, even from array analysis of a set of strains, many of which were aneuploid (94, 102). Importantly, C. albicans is not unique: aneuploidy is seen in many other organisms, including the model yeast S. cerevisiae, where an extra copy of a single chromosome affects not only the expression of genes on the aneuploid chromosomes but also genes that sense altered levels of DNA or altered stoichiometry of protein complexes (91). We suggest that C. albicans transformants may tolerate aneuploidy better because they are diploid: aneuploidy is more detrimental to haploid strains (92), and accordingly, it arises more frequently in diploid than haploid S. cerevisiae strains (45). Thus, it is important to be aware that aneuploidy may occur in transformed strains of C. albicans, as well as in other organisms. Fortunately, tools are now available to detect it.
Acknowledgments
We thank Merima Helic, Mary Ann Weinzierl, and Aaron Christensen-Quick for technical support. We are also grateful to Anne McBride, Darren Abbey, and Amnon Koren for critical reading of the manuscript. We thank Ted White and P. David Rogers for sending strains and all of the researchers whose work provided the microarray data summarized in Table 1.
This work was supported by a Minnesota Supercomputing Institute Fellowship to K.E.S.T., a Microbial and Plant Genomics Institute Integrative fellowship to A.S., a 3M graduate fellowship award and an NIH T32 Biotechnology Training fellowship to K.B., and NIH/NIAID grant R01 AI0624273 to J.B.
Footnotes
Published ahead of print on 21 August 2009.
REFERENCES
- 1.Ahmad, A., M. A. Kabir, A. Kravets, E. Andaluz, G. Larriba, and E. Rustchenko. 2008. Chromosome instability and unusual features of some widely used strains of Candida albicans. Yeast 25:433-448. [DOI] [PubMed] [Google Scholar]
- 2.Andaluz, E., J. Gomez-Raja, B. Hermosa, T. Ciudad, E. Rustchenko, R. Calderone, and G. Larriba. 2007. Loss and fragmentation of chromosome 5 are major events linked to the adaptation of rad52-DeltaDelta strains of Candida albicans to sorbose. Fungal Genet. Biol. 44:789-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andes, D., A. Lepak, J. Nett, L. Lincoln, and K. Marchillo. 2006. In vivo fluconazole pharmacodynamics and resistance development in a previously susceptible Candida albicans population examined by microbiologic and transcriptional profiling. Antimicrob. Agents Chemother. 50:2384-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andes, D., A. Lepak, A. Pitula, K. Marchillo, and J. Clark. 2005. A simple approach for estimating gene expression in Candida albicans directly from a systemic infection site. J. Infect. Dis. 192:893-900. [DOI] [PubMed] [Google Scholar]
- 5.Bachewich, C., A. Nantel, and M. Whiteway. 2005. Cell cycle arrest during S or M phase generates polarized growth via distinct signals in Candida albicans. Mol. Microbiol. 57:942-959. [DOI] [PubMed] [Google Scholar]
- 6.Bennett, R. J., and A. D. Johnson. 2003. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 22:2505-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett, R. J., and A. D. Johnson. 2006. The role of nutrient regulation and the Gpa2 protein in the mating pheromone response of C. albicans. Mol. Microbiol. 62:100-119. [DOI] [PubMed] [Google Scholar]
- 8.Bensen, E. S., S. J. Martin, M. Li, J. Berman, and D. A. Davis. 2004. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol. Microbiol. 54:1335-1351. [DOI] [PubMed] [Google Scholar]
- 9.Bolstad, B. M., R. A. Irizarry, M. Astrand, and T. P. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185-193. [DOI] [PubMed] [Google Scholar]
- 10.Bruno, V. M., S. Kalachikov, R. Subaran, C. J. Nobile, C. Kyrarsous, and A. P. Mitchell. 2006. Control of the C. albicans cell wall damage response by transcriptional regulator Cas5. PLoS Pathogens 2:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruno, V. M., and A. P. Mitchell. 2005. Regulation of azole drug susceptibility by Candida albicans protein kinase CK2. Mol. Microbiol. 56:559-573. [DOI] [PubMed] [Google Scholar]
- 12.Cao, F., S. Lane, P. P. Raniga, Y. Lu, Z. Zhou, K. Ramon, J. Chen, and H. Lui. 2006. The Flo8 transcription factor is essential for hyphal formation and virulence in Candida albicans. Mol. Biol. Cell 17:295-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castillo, L., A. L. Martinez, A. Garcera, J. Garcia-Martinez, J. Ruiz-Herrera, E. Valentin, and R. Sentandreu. 2006. Genomic response programs of Candida albicans following protoplasting and regeneration. Fungal Genet. Biol. 43:124-134. [DOI] [PubMed] [Google Scholar]
- 14.Chandhok, N. S., and D. Pellman. 2009. A little CIN may cost a lot: revisiting aneuploidy and cancer. Curr. Opin. Genet. Dev. 19:74-81. [DOI] [PubMed] [Google Scholar]
- 15.Chen, H. Y., M. Fujita, Q. Feng, J. Clardy, and G. Fink. 2004. Tyrosol is a quorum-sensing molecule in Candida albicans. Proc. Natl. Acad. Sci. USA 101:5048-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, X., B. B. Magee, D. Dawson, P. T. Magee, and C. A. Kumamoto. 2004. Chromosome 1 trisomy compromises the virulence of Candida albicans. Mol. Microbiol. 51:551-565. [DOI] [PubMed] [Google Scholar]
- 17.Cheng, G., K. M. Yeater, and L. L. Hoyer. 2006. Cellular and molecular biology of Candida albicans estrogen response. Eukaryot. Cell 5:180-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chibana, H., S. Iwaguchi, M. Homma, A. Chindamporn, Y. Nakagawa, and K. Tanaka. 1994. Diversity of tandemly repetitive sequences due to short periodic repetitions in the chromosomes of Candida albicans. J. Bacteriol. 176:3851-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Copping, V. M., C. J. Barelle, B. Hube, N. A. Gow, A. J. Brown, and F. C. Odds. 2005. Exposure of Candida albicans to antifungal agents affects expression of SAP2 and SAP9 secreted proteinase genes. J. Antimicrob. Chemother. 55:645-654. [DOI] [PubMed] [Google Scholar]
- 20.Coste, A. T., M. Karababa, F. Ischer, J. Bille, and D. Sanglard. 2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell 3:1639-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cowen, L. E., A. Nantel, M. S. Whiteway, D. Y. Thomas, D. C. Tessier, L. M. Kohn, and J. B. Anderson. 2002. Population genomics of drug resistance in Candida albicans. Proc. Natl. Acad. Sci. USA 99:9284-9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dignard, D., and M. Whiteway. 2006. SST2, a regulator of G-protein signaling for the Candida albicans mating response pathway. Eukaryot. Cell 5:192-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doedt, T., S. Krishnamurthy, D. P. Bockmuhl, B. Tebarth, C. Stempel, C. L. Russell, A. J. Brown, and J. F. Ernst. 2004. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell 15:3167-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duesberg, P. 2007. Chromosomal chaos and cancer. Sci. Am. 296:52-59. [DOI] [PubMed] [Google Scholar]
- 25.Duesberg, P., and R. Li. 2003. Multistep carcinogenesis: a chain reaction of aneuploidizations. Cell Cycle. 2:202-210. [PubMed] [Google Scholar]
- 26.Duesberg, P., R. Li, A. Fabarius, and R. Hehlmann. 2005. The chromosomal basis of cancer. Cell Oncol. 27:293-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duesberg, P., D. Rasnick, R. Li, L. Winters, C. Rausch, and R. Hehlmann. 1999. How aneuploidy may cause cancer and genetic instability. Anticancer Res. 16:4887-4906. [PubMed] [Google Scholar]
- 28.Enjalbert, B., A. Nantel, and M. Whiteway. 2003. Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol. Biol. Cell 14:1460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enjalbert, B., D. A. Smith, M. J. Cornell, I. Alam, S. Nicholls, A. J. Brown, and J. Quinn. 2006. Role of the Hog1 stress-activated protein kinase in the global transcription response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell 17:1018-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enjalbert, B., and M. Whiteway. 2005. Release from quorum-sensing molecules triggers hyphal formation during Candida albicans resumption of growth. Eukaryot. Cell 4:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forche, A., K. Alby, D. Schaefer, A. D. Johnson, J. Berman, and R. J. Bennett. 2008. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 6:e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forche, A., P. T. Magee, B. B. Magee, and G. May. 2004. Genome-wide Single-nucleotide polymorphism map for Candida albicans. Eukaryot. Cell 3:705-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forche, A., P. T. Magee, A. Selmecki, J. Berman, and G. May. 2009. Evolution in Candida albicans populations during a single passage through a mouse host. Genetics 182:799-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forche, A., G. May, and P. T. Magee. 2005. Demonstration of loss of heterozygosity by single-nucleotide polymorphism microarray analysis and alterations in strain morphology in Candida albicans during infection. Eukaryot. Cell 4:156-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forche, A., M. Steinbach, and J. Berman. 22 June 2009. Efficient and rapid identification of Candida albicans allelic status using SNP-RFLP. FEMS Yeast Res. [Epub ahead of print.] doi: 10.1111/j.1567-1364.2009.00542.x. [DOI] [PMC free article] [PubMed]
- 37.Fradin, C., P. De Groot, D. MacMallum, M. Schaller, F. Klis, and F. Odds. 2005. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol. Microbiol. 56:397-415. [DOI] [PubMed] [Google Scholar]
- 38.Ganem, N. J., Z. Storchova, and D. Pellman. 2007. Tetraploidy, aneuploidy and cancer. Curr. Opin. Genet. Dev. 17:157-162. [DOI] [PubMed] [Google Scholar]
- 39.Garcera, A., L. Castillo, A. L. Martinez, M. V. Elorza, E. Valentin, and R. Sentandreu. 2005. Anchorage of Candida albicans Ssr1 to the cell wall, and transcript profiling of the null mutant. Res. Microbiol. 156:911-920. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Sanchez, S., A. L. Mavor, C. L. Russell, S. Argimon, P. Dennison, B. Enjalbert, and M. Whiteway. 2005. Global roles of Ssn6 in Tup1-and Nrg1-dependent gene regulation in the fungal pathogen Candida albicans. Mol. Biol. Cell 16:2913-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerami-Nejad, M., J. Berman, and C. A. Gale. 2001. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 18:859-864. [DOI] [PubMed] [Google Scholar]
- 42.Gerami-Nejad, M., J. Berman, D. L. Hausauer, and C. A. Gale. 2004. Cassettes for the PCR-mediated construction of regulatable alleles in Candida albicans. Yeast 21:429-436. [DOI] [PubMed] [Google Scholar]
- 43.Gerami-Nejad, M., K. Dulmage, M. McClellan, and J. Berman. 2009. Cassettes for epitope tagging genes in Candida albicans. Yeast 26:399-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greenberg, J. R., N. P. Price, R. P. Oliver, F. Sherman, and E. Rustchenko. 2005. Candida albicans SOU1 encodes a sorbose reductase required for l-sorbose utilization. Yeast 22:957-969. [DOI] [PubMed] [Google Scholar]
- 45.Gresham, D., M. M. Desai, C. M. Tucker, H. T. Jenq, D. A. Pai, A. Ward, C. G. DeSevo, D. Botstein, and M. J. Dunham. 2008. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 4:e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harcus, D., A. Nantel, T. Rigby, and M. Whiteway. 2004. Transcription profiling of cyclic AMP signaling in Candida albicans. Mol. Biol. Cell 15:4490-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hilton, C., D. Markie, B. Corner, E. Rikkerink, and R. Poulter. 1985. Heat shock induces chromosome loss in the yeast Candida albicans. Mol. Gen. Genet. 200:162-168. [DOI] [PubMed] [Google Scholar]
- 48.Hromatka, B. S., S. M. Noble, and A. D. Johnson. 2005. Transcriptional response of Candida albicans to nitric oxide and the role of the YHB1 gene in nitrosative stress and virulence. Mol. Biol. Cell 16:4814-4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hughes, T. R., C. J. Roberts, H. Dai, A. R. Jones, M. R. Meyer, D. Slade, J. Burchard, S. Dow, T. R. Ward, M. J. Kidd, S. H. Friend, and M. J. Marton. 2000. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat. Genet. 25:333-337. [DOI] [PubMed] [Google Scholar]
- 50.Hull, C. M., and A. D. Johnson. 1999. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science 285:1271-1275. [DOI] [PubMed] [Google Scholar]
- 51.Huttenhower, C., M. Schroeder, M. D. Chikina, and O. G. Troyanskaya. 2008. The Sleipnir library for computational functional genomics. Bioinformatics 24:1559-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ihmels, J., S. Bergmann, M. Gerami-Nejad, I. Yanai, M. McClellan, J. Berman, and N. Barkai. 2005. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science 309:938-940. [DOI] [PubMed] [Google Scholar]
- 53.Janbon, G., F. Sherman, and E. Rustchenko. 1998. Monosomy of a specific chromosome determines L-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc. Natl. Acad. Sci. USA 95:5150-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kabir, M. A., A. Ahmad, J. R. Greenberg, Y. K. Wang, and E. Rustchenko. 2005. Loss and gain of chromosome 5 controls growth of Candida albicans on sorbose due to dispersed redundant negative regulators. Proc. Natl. Acad. Sci. USA 102:12147-12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kadosh, D., and A. D. Johnson. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16:2903-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karababa, M., A. T. Coste, B. Rognon, J. Bille, and D. Sanglard. 2004. Comparison of gene expression profiles of Candida albicans azole-resistant clinical isolates and laboratory strains exposed to drugs inducing multidrug transporters. Antimicrob. Agents Chemother. 48:3064-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karababa, M., E. Valentino, G. Pardini, A. Coste, J. Bille, and D. Sanglard. 2006. CRZ1, a target of the calcineurin pathway in Candida albicans. Mol. Microbiol. 59:1429-1451. [DOI] [PubMed] [Google Scholar]
- 58.Kohler, G. A., T. C. White, and N. Agabian. 1997. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J. Bacteriol. 179:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lea, D. E., and C. A. Coulson. 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49:264-285. [DOI] [PubMed] [Google Scholar]
- 60.Lee, C. M., A. Nantel, L. Jiang, M. Whiteway, and S. H. Shen. 2004. The serine/threonine protein phosphatase SIT4 modulates yeast-to-hypha morphogenesis and virulence in Candida albicans. Mol. Microbiol. 51:691-709. [DOI] [PubMed] [Google Scholar]
- 61.Lee, R. E., T. T. Liu, K. S. Barker, R. E. Lee, and P. D. Rogers. 2005. Genome-wide expression profiling of the response to ciclopirox olamine in Candida albicans. J. Antimicrob. Chemother. 55:655-662. [DOI] [PubMed] [Google Scholar]
- 62.Legrand, M., A. Forche, A. Selmecki, C. Chan, D. T. Kirkpatrick, and J. Berman. 2008. Haplotype mapping of a diploid non-meiotic organism using existing and induced aneuploidies. PLoS Genet. 4:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Legrand, M., P. Lepart, A. Forche, F.-M. C. Mueller, T. J. Walsh, P. T. Magee, and B. B. Magee. 2004. Homozygosity at the MTL locus in clinical strains of Candida albicans: karyotypic rearrangements and tetraploid formation. Mol. Microbiol. 52:1451-1462. [DOI] [PubMed] [Google Scholar]
- 64.Li, X. C., J. C. Schimenti, and B. K. Tye. 2009. Aneuploidy and improved growth are coincident but not causal in a yeast cancer model. PLoS Biol. 7:e1000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lorenz, M., J. A. Bender, and G. R. Fink. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 3:1076-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Magee, B. B., and P. T. Magee. 2000. Induction of mating in Candida albicans by construction of MTLa and MTLα strains. Science 289:310-313. [DOI] [PubMed] [Google Scholar]
- 67.Nantel, A., D. Dignard, C. Bachewich, D. Harcus, A. Marcil, A. P. Bouin, C. W. Sensen, H. Hogues, M. van het Hoog, P. Gordon, T. Rigby, F. Benoit, D. C. Tessier, D. Y. Thomas, and M. Whiteway. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13:3452-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Navarro-Garcia, F., R. M. Perez-Diaz, B. B. Magee, J. Pla, C. Nombela, and P. Magee. 1995. Chromosome reorganization in Candida albicans 1001 strain. J. Med. Vet. Mycol. 33:361-366. [PubMed] [Google Scholar]
- 69.Nicholls, S., M. Straffon, B. Enjalbert, A. Nantel, S. Macaskill, M. Whiteway, and A. J. Brown. 2004. Msn2- and Msn4-like transcription factors play no obvious roles in the stress responses of the fungal pathogen Candida albicans. Eukaryot. Cell 3:1111-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nobile, C. J., and A. P. Mitchell. 2005. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1. Curr. Biol. 15:1150-1155. [DOI] [PubMed] [Google Scholar]
- 71.Oberholzer, U., A. Nantel, J. Berman, and M. Whiteway. 2006. Transcript profiles of Candida albicans cortical actin patch mutants reflect their cellular defects: contribution of the Hog1p and Mkc1p signaling pathways. Eukaryot. Cell 5:1252-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perepnikhatka, V., F. J. Fischer, M. Niimi, R. A. Baker, R. D. Cannon, Y. K. Wang, F. Sherman, and E. Rustchenko. 1999. Specific chromosome alterations in fluconazole-resistant mutants of Candida albicans. J. Bacteriol. 181:4041-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rancati, G., N. Pavelka, B. Fleharty, A. Noll, R. Trimble, K. Walton, A. Perera, K. Staehling-Hampton, C. W. Seidel, and R. Li. 2008. Aneuploidy underlies rapid evolution of yeast cells deprived of a conserved cytokinesis motor. Cell 135:879-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Redding, S., J. Smith, G. Farinacci, M. Rinaldi, A. Fothergill, J. Rhine-Chalberg, and M. Pfaller. 1994. Resistance of Candida albicans to fluconazole during treatment of oropharyngeal candidiasis in a patient with AIDS: documentation by in vitro susceptibility testing and DNA subtype analysis. Clin. Infect. Dis. 18:240-242. [DOI] [PubMed] [Google Scholar]
- 75.Ritchie, M. E., J. Silver, A. Oshlack, M. Holmes, D. Diyagama, A. Holloway, and G. K. Smyth. 2007. A comparison of background correction methods for two-colour microarrays. Bioinformatics 23:2700-2707. [DOI] [PubMed] [Google Scholar]
- 76.Rogers, P. D., and K. S. Barker. 2003. Genome-wide expression profile analysis reveals coordinately regulated genes associated with stepwise acquisition of azole resistance in Candida albicans clinical isolates. Antimicrob. Agents Chemother. 47:1220-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rubin-Bejerano, I., I. Fraser, P. Grisafi, and J. R. Fink. 2003. Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc. Natl. Acad. Sci. USA 100:11007-11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rustchenko, E. 2007. Chromosome instability in Candida albicans. FEMS Yeast Res. 7:2-11. [DOI] [PubMed] [Google Scholar]
- 79.Scherer, S., and P. T. Magee. 1990. Genetics of Candida albicans. Microbiol. Rev. 54:226-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Selmecki, A., S. Bergmann, and J. Berman. 2005. Comparative genome hybridization reveals widespread aneuploidy in Candida albicans laboratory strains. Mol. Microbiol. 55:1553-1565. [DOI] [PubMed] [Google Scholar]
- 81.Selmecki, A., A. Forche, and J. Berman. 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313:367-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Selmecki, A., M. Gerami-Nejad, C. Paulson, A. Forche, and J. Berman. 2008. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol. Microbiol. 68:624-641. [DOI] [PubMed] [Google Scholar]
- 83.Setiadi, E. R., T. Doedt, F. Cottier, C. Noffz, and J. F. Ernst. 2006. Transcriptional response of Candida albicans to hypoxia: linkage of oxygen sensing and Efg1-regulatory networks. J. Mol. Biol. 361:399-411. [DOI] [PubMed] [Google Scholar]
- 84.Sigle, H. C., S. Thewes, M. Niwerth, H. C. Korting, M. Schaefer-Korting, and B. Hube. 2005. Oxygen accessibility and iron levels are critical factors for the antifungal action of ciclopirox against Candida albicans. J. Antimicrob. Chemother. 55:663-673. [DOI] [PubMed] [Google Scholar]
- 85.Singh, V., I. Sinha, and P. P. Sadhale. 2005. Global analysis of altered gene expression during morphogenesis of Candida albicans in vitro. Biochem. Biophys. Res. Commun. 334:1149-1158. [DOI] [PubMed] [Google Scholar]
- 86.Smyth, G. K., and T. Speed. 2003. Normalization of cDNA microarray data. Methods 31:265-273. [DOI] [PubMed] [Google Scholar]
- 87.Spell, R. M., and S. Jinks-Robertson. 2004. Determination of mitotic recombination rates by fluctuation analysis in Saccharomyces cerevisiae, p. 3-12. In A. S. Waldman (ed.), Genetic recombination: review and protocols, vol. 262. Humana Press Inc., Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 88.Staib, P., S. Michel, G. Kohler, and J. Morschhauser. 2000. A molecular genetic system for the pathogenic yeast Candida dubliniensis. Gene 242:393-398. [DOI] [PubMed] [Google Scholar]
- 89.Thewes, S., M. Kretschmar, H. Park, M. Schaller, S. G. Filler, and B. Hube. 2007. In vivo and ex vivo comparative profiling of invasive and non-invasive Candida albicans isolates identifies genes associated with tissue invasion. Mol. Microbiol. 63:1606-1628. [DOI] [PubMed] [Google Scholar]
- 90.Thrash-Bingham, C., and J. A. Gorman. 1992. DNA translocations contribute to chromosome length polymorphisms in Candida albicans. Curr. Genet. 22:93-100. [DOI] [PubMed] [Google Scholar]
- 91.Torres, E. M., T. Sokolsky, C. M. Tucker, L. Y. Chan, M. Boselli, M. J. Dunham, and A. Amon. 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317:916-924. [DOI] [PubMed] [Google Scholar]
- 92.Torres, E. M., B. R. Williams, and A. Amon. 2008. Aneuploidy: cells losing their balance. Genetics 179:737-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tournu, H., G. Tripathi, G. Bertram, S. Macaskill, A. Mavor, L. Walker, F. Odds, N. A. Gow, and A. J. Brown. 2005. Global role of the protein kinase Gcn2 in the human pathogen Candida albicans. Eukaryot. Cell 4:1687-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tsong, A. E., M. G. Miller, R. M. Raisner, and A. D. Johnson. 2003. Evolution of a combinatorial transcript circuit: a case study in yeasts. Cell 115:389-399. [DOI] [PubMed] [Google Scholar]
- 95.Tsong, A. E., B. B. Tuch, H. Li, and A. D. Johnson. 2006. Evolution of alternative transcriptional circuits with identical logic. Nature 443:415-420. [DOI] [PubMed] [Google Scholar]
- 96.van het Hoog, M., T. J. Rast, M. Martchenko, S. Grindle, D. Dignard, H. Hogues, C. Cuomo, M. Berriman, S. Scherer, B. B. Magee, M. Whiteway, H. Chibana, A. Nantel, and P. T. Magee. 2007. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biol. 8:R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang, Y., Y. Y. Cao, X. M. Jia, Y. B. Cao, P. H. Gao, X. P. Fu, K. Ying, W. S. Chen, and Y. Y. Jiang. 2006. Cap1p is involved in multiple pathways of oxidative stress response in Candida albicans. Free Radic. Biol. Med. 40:1201-1209. [DOI] [PubMed] [Google Scholar]
- 98.Whelan, W. L., R. M. Partridge, and P. T. Magee. 1980. Heterozygosity and segregation in Candida albicans. Mol. Gen. Genet. 180:107-113. [DOI] [PubMed] [Google Scholar]
- 99.Whelan, W. L., and D. R. Soll. 1982. Mitotic recombination in Candida albicans: recessive lethal alleles linked to a gene required for methionine biosynthesis. Mol. Gen. Genet. 187:477-485. [DOI] [PubMed] [Google Scholar]
- 100.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhao, R., K. J. Daniels, D. J. Lockhart, K. M. Yeater, L. L. Hoyer, and D. R. Soll. 2005. Unique aspects of gene expression during Candida albicans mating and possible G(1) dependency. Eukaryot. Cell 4:1175-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zordan, R. E., D. J. Galgoczy, and A. D. Johnson. 2006. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc. Natl. Acad. Sci. USA 103:12807-12812. [DOI] [PMC free article] [PubMed] [Google Scholar]