Abstract
HMGA chromatin proteins, a family of gene regulatory factors found at only low concentrations in normal cells, are almost universally overexpressed in cancer cells. HMGA proteins are located in the nuclei of normal cells except during the late S/G2 phases of the cell cycle, when HMGA1, one of the members of the family, reversibly migrates to the mitochondria, where it binds to mitochondrial DNA (mtDNA). In many cancer cells, this controlled shuttling is lost and HMGA1 is found in mitochondria throughout the cell cycle. To investigate the effects of HMGA1 on mitochondria, we employed a genetically engineered line of human MCF-7 cells in which the levels of transgenic HMGA1 protein could be reversibly controlled. “Turn-ON” and “turn-OFF” time course experiments were performed with these cells to either increase or decrease intracellular HMGA1 levels, and various mitochondrial changes were monitored. Results demonstrated that changes in both mtDNA levels and mitochondrial mass inversely paralleled changes in HMGA1 concentrations, strongly implicating HMGA1 in the regulation of these parameters. Additionally, the level of cellular reactive oxygen species (ROS) increased and the efficiency of repair of oxidatively damaged mtDNA decreased as consequences of elevated HMGA1 expression. Increased ROS levels and reduced repair efficiency in HMGA1-overexpressing cells likely contribute to the increased occurrence of mutations in mtDNA frequently observed in cancer cells.
Aberrant alterations in mitochondrial function are observed in a wide range of human conditions and diseases (10, 109), including cancer (19, 22, 51). Because of their prevalence, mitochondrial abnormalities in cancer cells have been extensively investigated (17, 39, 84, 118). Even in the presence of an adequate oxygen supply, the mitochondria of cancer cells are almost universally deficient in their ability to generate ATP via oxidative phosphorylation and therefore rely heavily on glycolytic metabolism for energy production (the so-called Warburg effect) (119, 120). Likewise, cancer cells often exhibit electron transport chain deficiencies (90), generate smaller amounts of ATP and higher levels of reactive oxygen species (ROS) than normal cells (45, 65), display calcium signaling defects (101), and frequently exhibit alterations in their mitochondrial transmembrane potential (56, 57). Many cancers also exhibit reductions in both mitochondrial mass and mitochondrial DNA (mtDNA) levels (31, 80, 118, 125, 128), as well as significantly altered levels of mtDNA transcription (36, 50, 55, 66). It is also well documented that mtDNA in cancer cells is more susceptible to somatic mutation than nuclear DNA (nDNA) (8, 12, 22, 93, 104), partially as a consequence of the limited repair capabilities of mitochondria compared to the mechanisms responsible for the repair of nDNA (12, 13, 39, 76).
The underlying molecular causes for mitochondrial dysfunction in most cancers are unknown, although available evidence suggests that there are many possible contributing factors that interact with one another in complex ways. In a few instances, specific mutations in either mtDNA (17, 65, 91) or nDNA that codes for mitochondrial components (22, 83) have been found to result in mitochondrial dysfunction. But not all tumors have such identifiable mtDNA or nDNA mutations, and yet most still exhibit similar sets of abnormal mitochondrial characteristics. This aberrant phenotypic commonality suggests that additional, as yet unidentified factors likely contribute to mitochondrial dysfunction in cancer cells. In this regard, we recently demonstrated that human cells containing high levels of HMGA1 protein exhibit many of the same mitochondrial abnormalities found in naturally occurring cancers, which almost universally overexpress HMGA proteins (31).
The HMGA (formerly called HMG-I/Y) family of nonhistone proteins are architectural transcription factors that modulate chromatin structure and regulate the expression of well over 100 different vertebrate nuclear genes involved in normal biological processes (30, 54, 60, 82, 97, 99). For example, recent studies have implicated HMGA proteins in human height determination (103, 121), normal heart development in vertebrates (85), and the promotion of stem cell renewal in young mouse brains (88), as well as in the control of cellular aging and senescence of human cells (46, 129). HMGA proteins have also been implicated in a number of human pathological conditions. Members of the HMGA gene family (which includes HMGA1 and HMGA2) are the only genes coding for bona fide chromatin proteins so far demonstrated to be proto-oncogenes. Experimental overexpression of HMGA proteins induces cancerous transformation of normal cells and also promotes both neoplastic progression and metastasis (33, 35, 100, 111, 123, 124). In normal differentiated somatic cells, the concentration of HMGA proteins is very low (97, 124). In naturally occurring cancers, however, the levels of HMGA proteins are usually elevated, with increasing concentrations being correlated with increasing degrees of malignancy and poor patient prognosis (96, 96, 112, 122). This association is so consistent and widespread that elevated levels of HMGA proteins are being employed as clinical diagnostic markers for several different types of human cancers (1, 7, 25, 43, 89, 95).
Members of the HMGA1 protein subfamily (97) are almost exclusively localized in the nuclei of normal cells except during the late S and G2 phases of the cell cycle, when a minor protein fraction reversibly migrates out of the nucleus and into the mitochondria (32). In malignant cells that overexpress HMGA1, however, this highly regulated shuttling is frequently disrupted and the proteins are found in the mitochondria at all stages of the cell cycle (31, 32). Chromatin immunoprecipitation analyses have demonstrated that inside mitochondria, HMGA1 is bound to the regulatory D-loop of mtDNA (31). As noted above, the mitochondria of cancerous cells frequently exhibit many abnormal phenotypic and metabolic characteristics, but whether any of these features are causally associated with the constitutive presence of HMGA1 proteins inside these organelles remains to be determined.
Previously, employing genetically engineered human MCF-7 cells, we investigated observable differences between the mitochondria found in cells that express high levels of transgenic HMGA1 protein and the mitochondria found in nontransgenic parental MCF-7 cells that express only low levels of endogenous protein (31). Although the results from such all-or-none type experiments were very informative, by their nature they could not identify which of the mitochondrial changes observed in overexpressing cells were likely attributable to effects of the HMGA1 proteins themselves. In the present study, we have employed a transgenic line of MCF-7 cells in which the intracellular concentrations of HMGA1 protein can be reversibly controlled by altering the amount of tetracycline in the culture medium. In time course experiments, we analyzed changes in mitochondrial mass, mtDNA and ROS levels, and the efficiency of base excision repair (BER) of oxidatively damaged mtDNA in cells containing various amounts of transgenic HMGA1 protein. The results of these reversible HMGA1 protein “titration” experiments are discussed in terms of plausible molecular mechanisms by which high levels of HMGA1 protein may aberrantly influence mitochondrial structure/function in cancer cells in vivo.
MATERIALS AND METHODS
Cell culture and regulation of transgenic HMGA1 protein expression.
Details of the creation and use of the transgenic HA7C line of human MCF-7 breast adenocarcinoma cells containing a tetracycline-regulated hemagglutinin-tagged-HMGA1a cDNA expression cassette have been reported previously (2, 3, 81, 100, 116). When grown for several days in medium without tetracycline, HA7C cells express high levels (elevated ∼40-fold over basal levels) of the hemagglutinin-HMGA1a protein (hereinafter referred to as HMGA1) (117). For convenience, HA7C cells overexpressing HMGA1 are simply referred to as ON cells in this work. Following the growth of ON cells for several days in medium containing tetracycline (4 μg/ml), the concentration of the HMGA1 protein is reduced to nearly undetectable levels (the cells with these reduced levels of HMGA1 are referred to as OFF cells) (see Fig. 1). The MCF7-Tet cell line (also called MCF7 Tet-off or M/Tet-off; Clontech, Palo Alto, CA) is the line from which HA7C cells were derived, and MCF7-Tet cells, like HA7C cells, are transgenic with cDNA coding for the tetracycline transactivator protein. MCF7-Tet cells were originally derived from the parental, nontransgenic MCF-7 human breast adenocarcinoma cell line. The phenotypes and growth characteristics of the MCF7-Tet and MCF-7 cells are indistinguishable from each other, and both cell types contain only low endogenous levels of HMGA1 protein (32). MCF7-Tet cells, or in some cases parental MCF-7 cells, were therefore used as experimental controls for the HA7C cells. The basic growth medium used for all of these cell lines was Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin (100 μg/ml), and streptomycin (100 μg/ml). The growth medium for the MCF7-Tet cells and the ON and OFF cells also contained G418 (100 μg/ml) to maintain selection for the tetracycline transactivator protein gene. Additionally, the growth medium for both the ON and the OFF cells was supplemented with hygromycin (100 μg/ml) to maintain selection for the integrated HMGA1 transgene. The genetically matched set of human epithelial cell lines Hs578Bst (American Type Culture Collection [ATCC] catalog no. HTB-125) and Hs578T (ATCC catalog no. HTB-126) were obtained from the ATCC (Manassas, VA) and maintained as recommended by the supplier. All cell lines were culture at 37°C in an atmosphere containing 5% CO2.
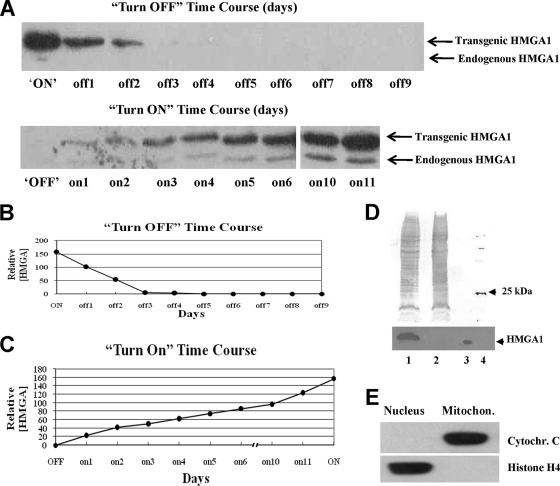
FIG. 1.
Time course turn-OFF and turn-ON experiments demonstrate that the concentrations of HMGA1 protein in transgenic cells can be reversibly controlled by the addition/removal of tetracycline to/from the culture medium. (A) WB analyses of the relative levels of HMGA1 protein present in ON cells as a function of time (days) following addition of tetracycline to the medium to inhibit transgene expression (top) and in OFF cells following removal of the drug from the medium to activate transgene expression (bottom). (B) Quantitative assessment of the results of the turn-OFF experiments shown in panel A, top. (C) Quantitative assessment of the results of the turn-ON experiments shown in panel A, bottom. All results shown are the averages of results from three or more independent experiments. (D, top) Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gel of total proteins from mitochondria (>95% pure) isolated from ON cells (lane 1) and OFF cells (lane 2). Lane 3 contains trace amounts of recombinant HMGA1 protein as a reference standard, and lane 4 contains molecular size marker proteins, with the 25-kDa protein labeled. (Bottom) WB analysis of a portion of the gel shown above, demonstrating that mitochondria isolated from ON cells (lane 1) contain readily detectable HMGA1 protein but that those isolated from OFF cells (lane 2) do not. The low abundance or apparent absence of detectable HMGA1 protein in the mitochondria from the OFF cells is consistent with the fact that in logarithmically growing populations of these cells the protein is found in the organelle only during a short, restricted phase of the cell cycle (32). Lane 3 contains recombinant HMGA1 included as a positive marker. No other anti-HMGA1 antibody-cross-reacting proteins were observed on the WB. (E) WB of total proteins found in mitochondria (mitochon.) isolated from ON cells using antibodies against either cytochrome C (cytochr. C; a mitochondrial marker) or histone H4 (a nuclear marker), demonstrating that there was very little nuclear contamination in these mitochondrial preparations.
Time course turn-OFF and turn-ON experiments.
Time course “turn-OFF” and “turn-ON” experiments involved the addition or removal of tetracycline to/from the culture medium of HA7C cells to either inhibit or allow expression of the transgenic HMGA1 protein. In turn-OFF experiments, tetracycline was added to the medium of HA7C ON cells to inhibit transgene expression and deplete cells of the HMGA1 protein. Changes in HMGA1 protein expression (or changes in various mitochondrial parameters) were then monitored as a function of the number of days following drug addition. Turn-ON experiments, on the other hand, involved the removal of tetracycline from the medium of HA7C OFF cells and monitoring of the accumulation of HMGA1 (or mitochondrial changes) in the cells as a function of the number of days following drug removal. Control MCF7-Tet cells were treated in an identical fashion in time course experiments except that, in this case, tetracycline addition to or removal from the medium did not alter the low endogenous levels of HMGA1 protein in the cells (see Fig. S3B in the supplemental material). Nevertheless, for the sake of nomenclature conformity with HA7C OFF and ON cells, control MCF7-Tet cells that had been grown in the presence of tetracycline for 2 weeks or more are referred to as MCF7-Tet (OFF) cells whereas control cells grown in medium lacking this drug for 2 weeks or more are referred to as MCF7-Tet (ON) cells.
WB analyses.
The cellular levels of both endogenous and transgenic HMGA1 proteins were routinely monitored by Western blot (WB) analysis prior to, during, and following all experiments by using the MR-19 anti-HMGA1 rabbit polyclonal antibody as described previously (100). Cellular levels of mitochondrial transcription factor A (TFAM) were also monitored using a commercially available anti-TFAM rabbit polyclonal antibody (mtTFA H-203; Santa Cruz Biotechnology). Peroxidase-conjugated goat anti-rabbit immunoglobulin G was employed as the secondary antibody in WB analyses, and bound proteins were detected using a SuperSignal West Pico chemiluminescent substrate kit according to the instructions of the manufacturer (Pierce Biotechnology, Rockford, IL).
Isolation of purified mitochondria.
Preparations of highly purified mitochondria were isolated from cells by using a mitochondrial isolation kit for mammalian cells according to protocols supplied by the manufacturer (Pierce Biotechnology, Inc., Rockford, IL). A number of control experiments demonstrated that these preparations were essentially free of cross-contamination with proteins or nucleic acids from other cell compartments. WB analyses using antibodies against either the mitochondrial marker protein cytochrome C (Abcam, Cambridge, United Kingdom) or the nuclear marker protein histone H4 (Abcam) demonstrated that there was very little nuclear contamination in these mitochondrial preparations (see Fig. 1E). Likewise, quantitative real-time PCR analyses using PCR primer sets based on published sequences and protocols for two nuclear genes, those encoding histone H4c (5) and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (25), demonstrated that less than 1% of the DNA in the mitochondrial preparations corresponded to nuclear contamination (data not shown). Additionally, WB analyses of the subcellular fractions of human colon cancer cells (which naturally overexpress HMGA1) using an anti-HMGA1 antibody revealed that both the nuclear and mitochondrial fractions (but not the cytoplasmic fraction) contained HMGA1 protein (unpublished data). Based on these controls, we conclude that the mitochondrial preparations used in our analyses were more than 95% pure.
Mitochondrial mass analyses.
Fluorescence-activated cell sorting (FACS) with an instrument from Becton Dickinson was used to monitor mitochondrial mass in cells at various time points during HMGA1 turn-ON and turn-OFF time course experiments. For these studies, cells were grown in Dulbecco's modified Eagle's medium lacking the pH indicator phenol red, which interferes with the cytometric detection method used. Mitochondrial mass was determined by staining cells with 100 nM nonylacridine orange (NAO; Molecular Probes [Invitrogen]), a metachromatic dye that binds to cardiolipin and accumulates in mitochondria regardless of organelle membrane potential or energetic state. At each experimental time point, cells at ∼80% confluence were incubated in the presence of NAO for 30 min, washed with phosphate-buffered saline (PBS), trypsinized, pelleted by gentle centrifugation, and then resuspended in 1 ml of PBS containing 100 nM NAO. Mitochondrial mass analyses of the stained cells were performed on a FACSCalibur instrument (Becton Dickinson; Immunocytometry Systems) with an emission wavelength of 488 nm and a 525-nm excitation detector. Data acquisition and analyses were performed as described previously (31) using Becton Dickinson CellQuest Pro software supplied with the instrument. In order to obtain consistent and statistically reliable results, a concentration of at least 106 cells/ml of counting solution was used for all FACS analyses.
mtDNA analyses.
Total cellular DNA (ratio of optical density at 260 nm to optical density at 280 nm, ∼1.9) was isolated by standard phenol-chloroform-isoamyl alcohol-ethanol (EtOH) extraction/precipitation methods (www.invitrogen.com/iprotocol) employing repeated digestions of the samples with DNase-free RNase A (Fermentas Life Sciences) and proteinase K (Fisher BioReagents, Germany). Treatment of the samples with RNase A during their isolation was important because any contaminating RNA in the preparations could anneal to PCR primers and interfere with the efficient amplification of target sequences from the DNA templates. mtDNA levels in the samples were determined by semiquantitative PCR (QPCR) analysis using mtDNA-specific primers that produced a 155-bp amplification product. The sequences of the mtDNA QPCR primers (based on the numbering system described on the Mitomap website [http://mitomap.org]) were as follows: sense (beginning at nucleotide [nt] 7773), 5′-CCG TCT GAA CTA TCC TGC CC-3′, and antisense (beginning at nt 7928), 5′-GCC GTA GTC GGT GTA CTC GT-3′. QPCR analysis was also used to establish the level of the nuclear gene coding for histone H4c in each DNA sample, and this value was used for normalization purposes in comparative mtDNA determinations. The sequences of the H4c QPCR primers, which amplified a 175-bp fragment of genomic DNA, were as follows: sense, 5′-GGG ATA ACA TCC AGG GCA TT-3′, and antisense, 5′-CCC TGA CGT TTT AGG GCA TA-3′. As an experimental control for the mtDNA PCRs, each sample was treated with DNase I (RNase free; Fermentas Life Sciences) for 30 min at 37°C and processed as described above prior to QPCR amplification of the 155-bp mtDNA fragment. As illustrated by the results presented below (see Fig. 4C), all samples treated with DNase I failed to yield amplified fragments corresponding to mtDNA, thus confirming the absence of significant PCR artifacts in the amplification reactions. All PCR analyses were performed in triplicate with 30 ng of template DNA using a Techne Genius thermal cycler. PCR parameters for amplifying mtDNA included an initial 5-min denaturation step at 94°C; 20 cycles of denaturation at 94°C for 30 s, annealing at 59°C for 30 s, and elongation at 72°C for 30 s; and a final extension period (10 min) at 72°C. QPCR conditions for H4c gene fragment amplification were the same except that the annealing temperature was 55°C and a total of 30 reaction cycles were used. The linearity of all of the PCR results was quantitatively confirmed by control reactions with 50% input template DNA. PCR products were separated by electrophoresis through 1% agarose gels, stained with ethidium bromide, captured in a digital image, and analyzed on a Gel Doc EQ system (Bio-Rad Labs Inc., Hercules, CA). The images were quantitatively assessed using Bio-Rad Quantity One one-dimensional analysis software supplied with the system.
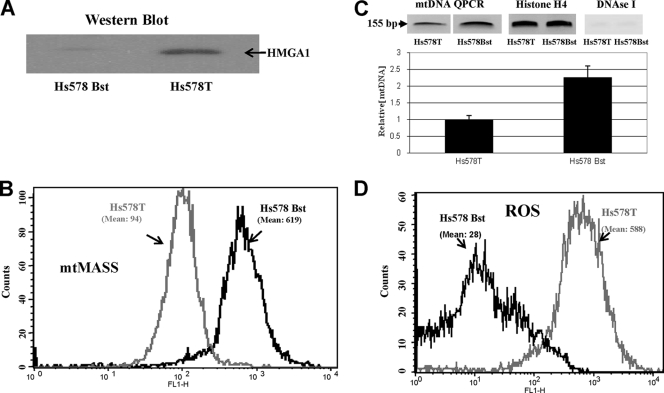
FIG. 4.
Nontransgenic human breast carcinoma cells (Hs578T) overexpress HMGA1 protein (A) and have reduced mitochondrial mass (B) and levels of mtDNA (C) and increased ROS levels (D) compared to normal breast epithelial cells (Hs578Bst). (A) WB with anti-HMGA1 antibody. (B) FACS analysis of mitochondrial mass (mtMASS) in Hs578Bst and Hs578T cells. Values are expressed as fluorescence intensity at 488 nm (see Materials and Methods). (C, bottom) Graph showing the results of QPCR analyses of mtDNA levels in Hs578Bst and Hs578T cells. (Top) The left two lanes show the ethidium bromide-stained 155-bp mtDNA QPCR products amplified from Hs578T and Hs578T cells. The middle two lanes show the 175-bp QPCR-amplified histone H4c nuclear gene fragments used for [mtDNA] normalization purposes (see Materials and Methods for discussion). The right lane shows the results of a negative control experiment demonstrating that no PCR amplification from DNA samples digested with DNase I occurs. (D) FACS analysis of ROS levels in Hs578Bst and Hs578T cells. Values are expressed as fluorescence intensity at 488 nm (see Materials and Methods).
FACS analysis of mitochondrial ROS levels.
Cells employed for the ROS time course studies were grown as described above for mitochondrial mass and mtDNA analyses. At each experimental time point, freshly prepared ROS indicator dye 2′,7′-dicholorodihydrofluorescein diacetate (H2DCFDA; Molecular Probes [Invitrogen]) was added to the cell growth medium at a final concentration of 10 μM. The cultures were incubated (37°C; 5% CO2) with the dye for 60 min, washed with PBS, and trypsinized, and the released cells were pelleted by centrifugation and then resuspended at a concentration of 106 cells/ml in PBS containing 10 μM H2DCFDA. The stained cells were immediately analyzed on a FACSCalibur machine utilizing excitation at 488 nm and fluorescein isothiocyanate filter detection parameters. CellQuest Pro software was used for both data acquisition and analysis to produce histogram plots and mean data values. OFF cells treated for 30 min with 200 μM H2O2 and then stained with H2DCFDA (as described above) served as positive ROS controls for the FACS analyses. Untreated OFF cells and OFF cells exposed to dilute 0.1% EtOH for 60 min both served as negative ROS FACS controls (data not shown).
mtDNA oxidative damage and BER analyses.
Twenty hours prior to treatment with either menadione sodium bisulfate (water soluble; Sigma-Aldrich, St. Louis, MO) or hydrogen peroxide (H2O2; Sigma-Aldrich), exponentially growing cells at ∼80% confluence were treated with hydroxyurea (4 mM final concentration) in order to lower the background level of replicative DNA synthesis and facilitate analysis of the repair of oxidatively damaged mtDNA. Hydroxyurea-treated cells were washed with PBS, and then warm serum-free medium was added to the culture dishes. In some experiments, menadione (40 to 400 μM) (31, 38) was added to the dishes and the cells were incubated for 30 to 60 min at 37°C and 5% CO2. In other experiments, H2O2 at a final concentration of 750 μM was added and the cells were incubated for 30 min at 37°C and 5% CO2. Following exposure to either agent, cells were washed with PBS, fresh complete medium was added, and the dishes were returned to the incubator for various lengths of time (0, 1, 2, and 4 h, etc.) to allow for the repair of oxidative lesions before the cells were harvested by trypsinization and their DNA was isolated by the phenol-chloroform-isoamyl alcohol-EtOH extraction method described above. DNA concentrations were determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Inc).
mtDNA repair time course experiments.
Time course analyses of the rate of repair of oxidative damage in nearly full-length (16.2-kb) mtDNA fragments were performed using modifications of the long-range QPCR methods developed by Santos and his coworkers (105, 106) as described previously (81). Briefly, prior to QPCR amplification, approximately 2 μg of purified total cellular DNA was digested to completion with the restriction enzyme XhoI (4 U; New England Biolabs) for 14 h at 37°C to linearize human mtDNA (GenBank accession no. J01415) in the sample. The XhoI-cut DNA was then digested for 4 h at 37°C with 2 U of the damage-specific enzyme FPG (formamidopyrimidine-DNA glycosylase, also known as 8-oxoguanine DNA glycosylase; New England Biolabs), which cleaves at sites of oxidized purine bases. The double-digested DNA was then repurified and accurately quantified (as described above) and used as a concentrated working stock solution.
The GeneAmp XL PCR kit (Applied Biosystems) was used for QPCR amplifications according to manufacturer-supplied protocols. Amplification reactions were performed with 25-μl reaction mixtures in 0.5-ml thin-wall PCR tubes (Phenix, Hayward, CA) on a Genius thermal cycler (Techne, Cambridge, United Kingdom). QPCR mixtures contained the following: 40 ng of total cellular DNA, 1× buffer XL II (Applied Biosystems), 100 ng/μl bovine serum albumin, a 200 μM concentration of each of the four deoxynucleoside triphosphates, 400 nM (each) forward and reverse mtDNA primers, 0.7 mM Mg2+, and 0.5 U of recombinant Tth DNA polymerase XL (Applied Biosystems). The human mtDNA primer set used for amplification was as follows: sense (beginning at nt 15149), 5′-TGA GGC CAA ATA TCA TTC TGA GGG GC-3′, and antisense (beginning at nt 14841), 5′-TTT CAT CAT GCG GAG ATG TTG GAT GG-3′ (105). The unique XhoI cut site employed to linearize mtDNA is situated between the two primers, at nt 14955, and hence the expected QPCR amplification fragment is 16.2 kb in length (rather than the full mtDNA length of 16.6 kb; GenBank RefSeq accession no. AC_000021.2). The amount of mtDNA in each DNA sample was quantitatively measured by QPCR amplification of a small (155-bp) mtDNA fragment as described above. In each mtDNA analysis, a fragment of the nuclear histone H4c gene present in each sample was QPCR amplified and used for normalization purposes. Controls for the QPCR amplifications followed the recommendations of Santos et al. (105). A half (50%)-template reaction was run for the first set of QPCR experiments to determine the linear amplification range for the template DNA. The 50% control mixtures contained the same concentrations of the long (16.2-kb) fragment and the short (∼155-bp) PCR primer set described above but only half the concentration of template DNA (i.e., 5 to 20 ng for total cell DNA). Amplification parameters included a PCR “hot start” in which samples were heated to 75°C for 5 min prior to the addition of recombinant Tth DNA polymerase. DNA samples were then denatured by heating at 94°C for 1 min and subjected to 20 cycles of denaturation at 94°C for 30 s and annealing and extension at 64°C for 12 min each. Samples were then heated to 72°C for 10 min for final elongation and stored at 4°C until electrophoretic separation and repair analyses of the amplified products could be completed.
mtDNA repair analyses.
Methods for the quantitative evaluation of DNA repair efficiency employing long-range QPCR amplification have been described in detail elsewhere (81, 105, 106). Briefly, 25 μl of each QPCR mixture was loaded onto a standard 0.7% agarose gel containing 2 μM ethidium bromide, and the amplified DNA products were separated by electrophoresis. Stained mtDNA bands resulting from QPCRs in the linear range of amplification (105) were visualized, captured in digital images, and quantified as described above. Band intensities were first corrected for gel background staining (i.e., the average above and below each band) and then normalized with respect to intensities from 100% control template reactions to measure variances observed between repair samples. The normalized value for each menadione- or H2O2-treated sample was then divided by the normalized band intensity for the untreated control sample to give the fraction of undamaged templates at a given dose of the oxidizing agent and/or at a given repair time point (105, 106). Assuming a Poisson distribution of oxidative lesions between amplified strands (e−S, where S is the average lesion frequency), the lesion frequency per strand was calculated using the following equation: S = −ln(AD/A0), where A0 is the intensity of the normalized full-length (undamaged) mtDNA band from untreated cells and AD is the intensity of the normalized full-length band from either menadione- or H2O2-treated samples at a given time after DNA damage. Repair efficiencies at each time point were determined from the difference in average lesion frequency between the sample at the given repair time point and the unrepaired DNA (0 h). In all analyses, the mean values and standard deviations for lesion incidence and repair efficiency were calculated from the results of at least three independent experiments. Best-fit curve plots of the data were derived by employing the PivotChart Wizard function of Microsoft Excel.
RESULTS
HMGA1 protein levels in transgenic cells can be reversibly controlled.
The WB results shown in Fig. 1A demonstrate that the concentrations of HMGA1 protein inside transgenic cells can be reversibly controlled by varying the amount of tetracycline in the culture medium. In turn-OFF experiments, the addition of tetracycline to the medium of ON cells, which initially contained high levels of HMGA1, completely inhibited the expression of this protein within about 3 days after drug exposure and, thereafter, the level remained very low for extended periods of time (Fig. 1A, top). The reverse occurred in turn-ON experiments: within 24 h of the removal of the drug from the medium of nonexpressing OFF cells, low levels of transgenic HMGA1 protein could be detected, and thereafter, the intracellular concentration continued to gradually increase with time until, after about 11 days, it nearly reached the maximum level found in fully ON cells (Fig. 1A, bottom). The graphs in Fig. 1B and C show plots of the relative concentrations of transgenic HMGA1 protein found inside ON and OFF cells during these time course experiments.
Two additional and significant observations are evident from the WB results in Fig. 1A to C. First, based on its rate of disappearance in turn-OFF experiments (Fig. 1B), the half-life of the HMGA1 protein is estimated to be about 30 to 35 h, a value consistent with data in previous reports (9, 63). Second, consistent with the half-life of the protein, it takes much (∼3.7 times) longer to accumulate nearly maximal levels of HMGA1 protein inside cells in turn-ON experiments (∼11 days) (Fig. 1C) than it does to deplete the protein in turn-OFF experiments (∼3 days). It is also interesting that in the turn-ON experiments (Fig. 1A), the levels of endogenous HMGA1 protein increased as transgenic HMGA1 accumulated. This increase in endogenous protein, which was transient and decreased after several weeks of turn-ON conditions, is likely the result of a feedback stimulatory loop in which the HMGA1 protein binds to an A/T-rich sequence in a promoter element of the HMGA1 gene and induces transcription (unpublished observations).
Mitochondria from overexpressing ON cells contain HMGA1.
The stained sodium dodecyl sulfate-polyacrylamide gel in the upper part of Fig. 1D shows the protein profiles of highly purified mitochondria isolated from ON (lane 1) and OFF (lane 2) cells, together with bands for marker proteins run as molecular size standards (lane 4; the 25-kDa marker is indicated). WB analyses of these mitochondrial preparations employing a specific antibody demonstrated that HMGA1 is present in mitochondria isolated from ON but not from OFF cells (lane 3 contains recombinant HMGA1 protein as a marker). The WB analyses of total mitochondrial proteins shown in Fig. 1E, employing antibodies to cytochrome c (a mitochondrial marker protein) and histone H4 (a nuclear marker protein), combined with the results of additional control analyses (see Materials and Methods for details), demonstrated that the mitochondrial preparations were more than 95% pure. It is, therefore, unlikely that the HMGA1 protein found in mitochondria isolated from ON cells is the result of spurious contamination. These findings are consistent with data in previous reports from both our own laboratory (31, 32) and others (24, 71) demonstrating the presence of HMGA1 in the mitochondria of a variety of mammalian cell types.
Mitochondria change in response to altered cellular HMGA1 levels.
Significant changes in mitochondrial structure, function, and metabolism in HMGA1-overexpressing ON cells compared to those in nontransgenic, parental MCF-7 cells have been demonstrated to occur (31). For example, compared to those in MCF-7 cells, mitochondrial mass, the mtDNA content, and cellular ATP levels are all substantially reduced in overexpressing ON cells, whereas ROS levels are elevated. Likewise, ON cells have a more glycolytic metabolism than MCF-7 cells and exhibit alterations in both mitochondrial membrane potential and mtDNA transcription rates. A limitation of earlier studies, however, is that the results were obtained by comparing genetically engineered cells containing high levels of transgenic HMGA1 protein with nontransgenic, parental MCF-7 cells that contained only low levels of the endogenous protein (31). Such a comparison leaves open the question of whether any of the changes observed in the mitochondria of ON cells are likely, or not, to be caused by the HMGA1 protein itself. The ability to reversibly alter intracellular HMGA1 protein levels in turn-OFF and turn-ON experiments (Fig. 1) provides an alternative strategy for addressing this important issue. The rationale behind the approach is that any mitochondrial change that clearly tracks with both increasing and decreasing intracellular concentrations of the HMGA1 protein is more likely to be causally related to some function of the HMGA1 protein itself than are changes that do not follow such a reciprocal pattern.
Changes in mitochondrial mass track with variations in HMGA1 protein levels.
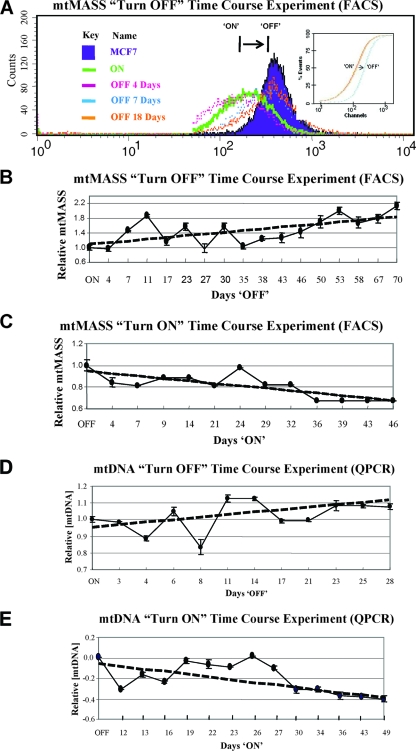
To experimentally test this idea, turn-OFF time course experiments were performed in which tetracycline was added to the culture medium of ON cells and FACS analysis was employed to monitor changes in mitochondrial mass as a function of time (days) following transgene turn-off. Previous work has shown that ON cells contain less than 70% of the mitochondrial mass of parental MCF-7 control cells (31). If mitochondrial mass is influenced by HMGA1 levels, then the predicted results of these turn-OFF experiments are that, over time, mitochondrial mass will increase in cells as a consequence of dropping HMGA1 protein levels. As shown in Fig. 2, this is indeed what was observed. Figure 2A shows the overlay of FACS histograms of the mitochondrial mass distributions found in cells at various times (0, 4, 7, and 18 days) following drug-induced transgene turn-off. The shift of the peak of mitochondrial mass intensity in the ON cells (at day 0) toward the peak intensity level found in nonexpressing MCF-7 control cells as a function of time (days) demonstrates that mitochondrial mass increases as the intracellular concentration of HMGA1 protein decreases. Additionally, Kolmogorov-Smirnov test analysis (Fig. 2A, inset) of the FACS scans from ON and parental MCF-7 cells statistically confirms that the mitochondrial mass in HMGA1-overexpressing cells is significantly reduced relative to that found in the OFF cells. The graph in Fig. 2B shows the results of turn-OFF experiments in which mitochondrial mass was monitored as a function of time following drug addition to the medium of ON cells and demonstrates that by day 70 the mitochondrial mass had risen to levels comparable to those found in parental MCF-7 cells. The graph in Fig. 2C shows the results of a reciprocal turn-ON time course experiment and demonstrates that following the removal of tetracycline from the culture medium of OFF cells, mitochondrial mass decreased by >30% over a period of about a month and a half.
FIG. 2.
Analysis of mitochondrial mass (mtMASS) and mtDNA levels found in cells during turn-OFF and turn-ON experiments as a function of time (days) following addition/removal of tetracycline to/from the culture medium. (A) Overlay of mitochondrial mass histograms for cells on various days (0, 4, 7, and 18) following drug addition to the medium in turn-OFF experiments. The fluorescence intensity (at 488 nm) of NAO-stained mitochondria (a measure of mitochondrial mass) is shown on the logarithmic x axis, and cell counts are shown on the y axis. (Inset) Kolmogorov-Smirnov statistical analysis of FACS results confirms that the mitochondrial mass in ON cells is significantly increased up to levels in control OFF cells following tetracycline treatment. (B) Quantitative plot of mitochondrial mass FACS data, such as that shown in panel A, as a function of time following the addition of the drug to the growth medium. (C) Quantitative plot of the results of turn-ON experiments in which mitochondrial mass was measured as a function of time following the removal of tetracycline from the medium. All results shown are the averages of those from three or more independent experiments. (D) Graph showing the results of QPCR analyses measuring mtDNA levels in cells following the addition of tetracycline to the medium of ON cells during turn-OFF experiments. (E) Graph showing the results of QPCR analyses measuring mtDNA levels in cells following the removal of tetracycline from the medium of OFF cells during turn-ON experiments.
An obvious concern with experiments involving tetracycline is that the observed changes in mitochondrial mass are due to effects of the drug on mitochondrial physiology and are not at all related to varying HMGA1 levels. To investigate this possibility, similar types of ON and OFF time course experiments were performed with control MCF7-Tet cells, the line from which HA7C was derived, and the results demonstrated that the growth of these cells in tetracycline had no effect on mitochondrial mass (see Fig. S1 in the supplemental material; see also Materials and Methods for details). The results of these control experiments, combined with the results shown in Fig. 2, clearly demonstrate that mitochondrial mass is influenced by the cellular concentration of HMGA1.
Changes in mtDNA track with variations in HMGA1 protein levels.
Overexpressing ON cells contain only about half the amount of mtDNA in parental MCF-7 cells (31). To determine whether changes in the levels of mtDNA also track with variations in cellular HMGA1 protein levels, similar types of turn-OFF and turn-ON experiments were performed and changes in mtDNA levels were assessed at various time points following addition/removal of tetracycline to/from the culture medium. In these experiments, mtDNA levels were determined by QPCR amplification of a 155-bp fragment of mtDNA from samples of total cellular DNA. Short fragments (175 bp) of a nuclear histone gene were amplified from the same DNA samples and used for normalization purposes (see Materials and Methods). The graphs in Fig. 2D and E plot the results of these turn-OFF and turn-ON experiments. As shown in Fig. 2D, mtDNA levels increased in cells as a function of time following the addition of tetracycline to the medium of ON cells. Reciprocally, mtDNA levels decreased with time after the removal of the drug from the medium of OFF cells (Fig. 2E). Results from similar types of ON and OFF time course experiments performed with MCF7-Tet cells demonstrated that tetracycline had no observable effect on mtDNA levels in these control cells (see Fig. S2 in the supplemental material). Thus, both mitochondrial mass and mtDNA vary in predictable ways with changes in the intracellular HMGA1 protein concentration.
ROS concentrations track with HMGA1 protein levels.
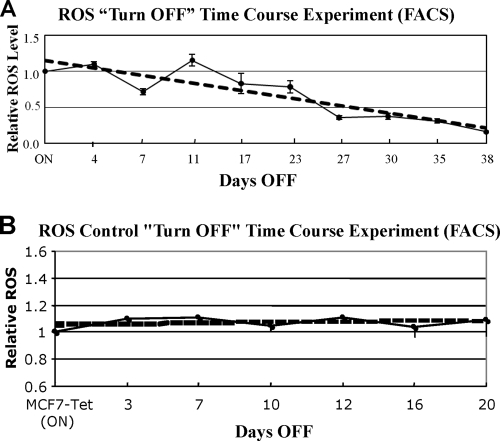
Consistent with their overtly cancerous phenotype (100), HMGA1-overexpressing ON cells contain elevated levels of ROS (31). FACS analyses were employed to monitor ROS levels in cells during turn-OFF experiments to determine whether ROS levels would drop as HMGA1 levels decreased, as would be expected if these two parameters were somehow linked. The results of these time course experiments are shown in Fig. 3A and demonstrate that ROS levels did indeed decline as predicted. Furthermore, the results of control time course turn-OFF experiments in which tetracycline was added to the culture medium of MCF7-Tet (ON) cells (see Materials and Methods) demonstrated that ROS levels in these control cells did not change over a period of several weeks (Fig. 3B). Together, these results suggest that ROS levels are, either directly or indirectly, linked to intracellular concentrations of the HMGA1 protein.
FIG. 3.
ROS levels in cells decline as HMGA1 protein levels decrease in turn-OFF experiments. (A) Graph showing the results of FACS analyses of ROS levels in cells following the addition of tetracycline to the culture medium of ON cells during turn-OFF experiments. (B) Graph showing the results of FACS analyses of ROS levels in control MCF7-Tet cells following the addition of tetracycline to the culture medium during turn-OFF experiments. In these experiments, the MCF7-Tet cells had been grown in medium lacking tetracycline for 2 weeks prior to drug addition and are therefore referred to as ON cells (see Materials and Methods for details).
Mitochondria in nontransgenic human cells are similar to those in transgenic ON and OFF cells.
Although the level of HMGA1 protein overexpression in the ON cells is well within the range of overexpression of this protein found in naturally occurring human cancers (117), a concern with experimental results obtained with transgenic cells is that they may not faithfully reflect the responses of naturally occurring, nontransgenic cells. If the results shown in Fig. 1 to 3 are generally applicable to nontransgenic cells, a number of biologically relevant and testable predictions can be made. Among these is the expectation that HMGA1 protein concentrations in naturally occurring cancers will be significantly higher than those in normal cells. Likewise, both mitochondrial mass and mtDNA levels in naturally occurring cancers are predicted to be noticeably lower than those in normal cells, whereas ROS levels are expected to be higher. To test these predictions, we determined the HMGA1 protein concentrations, mitochondrial masses, and mtDNA and ROS levels in a matched set of human mammary cells (Hs578Bst and Hs578T cells) that were derived from tissue biopsy samples from the same individual but exhibit markedly different tumorigenic phenotypes (53). The aneuploid Hs578T cell line originated from an epithelial carcinoma and causes aggressive, highly metastatic tumors when injected into nude mice. In contrast, the Hs578Bst myoepithelial cell line is diploid and is both phenotypically and biochemically normal in all respects examined (53, 77), including susceptibility to natural senescence (67). From the results shown in Fig. 4, it is evident that all of the predictions were confirmed: the malignant Hs578T cancer cells have much higher levels of HMGA1 protein (Fig. 4A), lower mitochondrial masses and mtDNA levels (Fig. 4B and C), and larger amounts of ROS (Fig. 4D) than the normal Hs578Bst cells. The upper part of Fig. 4C shows the results of control experiments demonstrating both the specificity and sensitivity of the QPCR analyses employed for determining mtDNA levels (see Materials and Methods for discussion). Interestingly, the differences in mitochondrial mass and mtDNA and ROS levels between the malignant Hs578Bst and normal Hs578T cells are either equal to, or greater than, the differences in these parameters between the transgenic OFF and ON cells. This disparity may be attributed to genetic differences between these cell types or may even be caused by factors other than the differences in HMGA1 protein levels found in the cells. An equally plausible explanation for such a difference, however, is that whereas the Hs578Bst cells are indeed normal by all criteria examined, both the HA7C OFF and ON cells are derivatives of MCF-7, a line of adenocarcinoma cells. It is reasonable to suspect that the mitochondrial differences observed between truly normal cells and cancerous cells (e.g., Hs578Bst versus Hs578T cells) are likely to be greater than the mitochondrial differences found between two established cell lines (i.e., ON versus OFF cells), neither of which is entirely normal. In any event, the results obtained with these normal and cancerous cells are consistent with those obtained with the transgenic ON and OFF cells and, therefore, support the biological relevance of the findings with the latter cells.
HMGA1 overexpression inhibits BER of oxidatively damaged mtDNA.
Nucleotide excision repair (NER) of DNA lesions induced by UV light is severely compromised in cancer cells that overexpress HMGA1 proteins (2, 81). At least two molecular mechanisms contribute to this NER deficiency: (i) inhibition by HMGA1 of the transcription of DNA repair genes (e.g., in xeroderma pigmentosum A) whose encoded protein products are required for NER to occur (2) and (ii) direct binding of HMGA1 to UV-induced DNA lesions [e.g., cyclobutane pyrimidine dimers and (6-4) photoproducts], which prevents access of repair enzymes to the lesions (2).
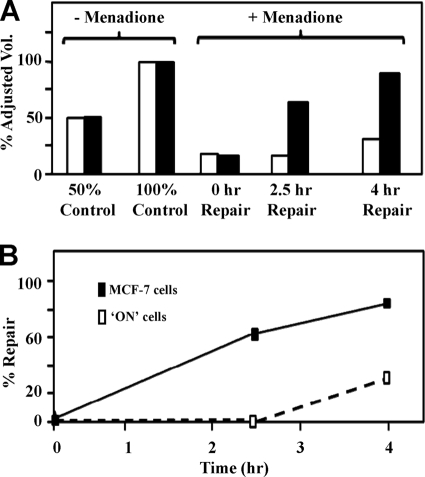
Unlike the nucleus, mitochondria lack the enzymes required for NER (14, 15, 27, 44). The mechanisms for the repair of lesions in mtDNA are complex and highly dependent on both the type and extent of damage involved (44, 72), with BER playing a primary role in removing oxidatively damaged bases (34, 78, 86). Preliminary cell survival studies using the base alkylating agent dimethyl sulfate demonstrated that BER of lesions in nDNA is, like NER, reduced in cells that overexpress HMGA1 (unpublished data). We therefore examined whether BER of oxidatively damaged mtDNA is also compromised in ON cells. For these repair experiments, both ON and MCF-7 control cells were briefly treated with the vitamin K3 analog menadione, a redox cycler that selectively induces excess superoxide radicals in mitochondria. Following menadione treatment, the removal of oxidative lesions from mtDNA was monitored as a function of time by employing long-range QPCR to monitor the extent of repair in nearly full-length (16.2-kb) mtDNA. Oxidatively damaged bases interfere with polymerase elongation during PCR amplification and hence reduce the yield of the 16.2-kb product. Thus, in these assays, the amount of amplified product is a quantitative measure of the relative number of lesions in the mtDNA template. Figure 5A shows experimental results in which the percentage of the adjusted volume of amplified mtDNA products (i.e., a direct measure of the extent of lesion removal) is plotted as a function of time posttreatment. It is evident from this graph that HMGA1 overexpression significantly reduces the efficiency of BER, as demonstrated by the fact that ∼60% of the lesions in mtDNA in the MCF-7 control cells had been removed by 2.5 h postdamage but that little, if any, repair had occurred in the mtDNA from overexpressing ON cells. By 4 h postdamage, >80% of the mtDNA damage in MCF-7 control cells had been repaired, in contrast to only around 30% in the ON cells. Figure 5A also shows the results of control experiments using DNA isolated from untreated cells (minus menadione; 100% and 50% input), demonstrating that the QPCR results for mtDNA were in the required linear range of amplification (106). Figure 5B graphically depicts these results as the percentage of repair and clearly shows the dramatic difference observed between ON cells (open boxes) and MCF-7 control cells (closed boxes) during the first 2 h following menadione treatment.
FIG. 5.
Time course of repair of oxidative damage in mtDNA following treatment of HMGA1-overexpressing ON and parental MCF-7 control cells with menadione. (A) Bar graph of the percentages of adjusted volumes (vol.) of quantified QPCR bands for mtDNA from ON cells (open bars) and parental MCF-7 control cells (closed bars) following drug treatment. −, without; +, with. (B) Results from panel A plotted as the percentages of repair in ON cells (open boxes) and parental MCF-7 control cells (closed boxes) following drug treatment. Data averages are based on the results of three independent experiments.
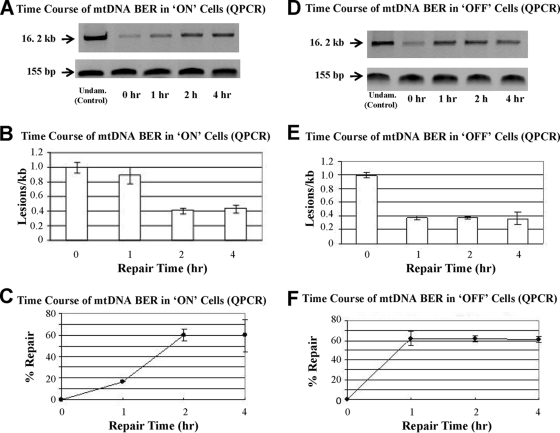
Hydrogen peroxide (H2O2) is frequently used as an alternative/complement to menadione as an oxidative damaging agent in studies of mtDNA damage and repair in mammalian cells (23, 52, 87). Optimization experiments in which ON and OFF cells were exposed to various concentrations of H2O2 (50 μM to 1 mM) for different lengths of time (15 to 60 min) demonstrated that exposure to 750 μM H2O2 for 30 min induced, on average, approximately 1 oxidative lesion/kb of mtDNA in these cells (data not shown). The time courses of mtDNA repair after the exposure of both ON and OFF cells to this dosage of H2O2 are shown in Fig. 6A and D, respectively. From these results, it is apparent that immediately following H2O2 treatment (i.e., at 0 h, before any repair has occurred), the amounts of the amplified 16.2-kb mtDNA product from both the ON and OFF cells are significantly reduced (>75%) compared to that from the undamaged mtDNA controls. Following H2O2 treatment (i.e., at 1, 2, and 4 h), the amount of the amplified 16.2-kb product from both the ON and OFF cells increases with time, indicating that oxidative lesions are being progressively removed from the damaged mtDNA by in vivo repair processes. Importantly, however, it is obvious that at 1 h postdamage, significantly less mtDNA repair has occurred in the ON cells (Fig. 6A) than in the OFF cells (Fig. 6D). Quantitative assessments of these QPCR results in terms of both the relative number of lesions per kilobase removed and the percentage of repair that occurs with time are shown, respectively, in Fig. 6B and C for ON cells and Fig. 6E and F for OFF cells. From these graphs, it is seen that by 1 h postdamage, OFF cells have repaired more than 60% of the oxidative lesions in mtDNA but that only around 15% of the lesions in the ON cells have been repaired. The results of control QPCRs in which a small, 155-bp sub-mtDNA fragment was amplified at each point during the repair time course experiments are shown in the bottom rows in Fig. 6A and D and demonstrate that the reaction mixtures contained approximately the same amounts of mtDNA. We therefore conclude that the observed variations in the intensity of the amplified 16.2-kb product were due to variations in the amount of repair that had occurred at each time point and were not the result of differences in the amount of mtDNA present in the reaction mixtures themselves (105).
FIG. 6.
Time course of mtDNA repair following treatment of ON and MCF-7 control cells with H2O2 under conditions that produce approximately 1 oxidative lesion/kb of mtDNA. (A and D) Results of long-range QPCR analyses employed to monitor the repair of mtDNA in both ON cells (A, top) and OFF cells (D, top) as a function of time (hours) post-H2O2 treatment. The bottom rows in panels A and D show the results of control QPCR amplifications of a 155-bp sub-mtDNA fragment and demonstrate the loading of equal amounts of input for all of the samples. Undam., undamaged. (B, C, E, and F) Quantitative assessments of the QPCR results shown in panels A and D in terms of both the relative number of lesions per kilobase remaining in mtDNA and the percentage of repair that has occurred as a function of time are shown, respectively, in panels B and C for ON cells and panels E and F for the parental MCF-7 cells.
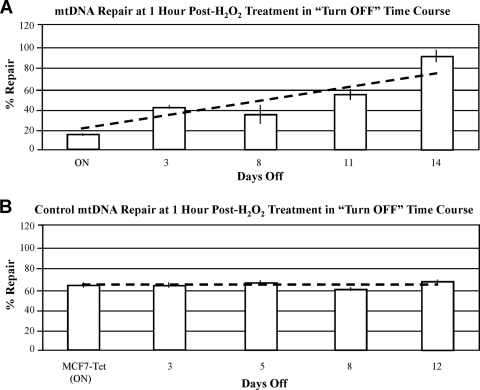
Having established that the greatest observed difference in mtDNA repair between the ON and OFF cells occurs at 1 h following H2O2 treatment, we conducted turn-OFF time course experiments to determine whether HMGA1 protein levels influence the efficiency of mitochondrial BER. In these experiments, tetracycline was added to the medium of ON cells to inhibit HMGA1 protein production and, on subsequent days, aliquots of cells were exposed to H2O2 and allowed to recover for 1 h and the amount of mtDNA repair that had occurred during this recovery period was quantitatively assessed by long-range QPCR. Figure 7A shows the results of these experiments and clearly demonstrates that as the levels of HMGA1 protein decrease in cells following transgene turn-off, the efficiency of repair of oxidative damage in mtDNA increases as a function of time. The results of experiments in which tetracycline was added to the medium of MCF7-Tet cells [yielding MCF7-Tet (ON) cells; see Materials and Methods] demonstrated that the drug had no effect on repair in these control cells (Fig. 7B). Thus, in addition to influencing both mitochondrial mass and mtDNA levels, the overexpression of HMGA1 decreases the efficiency of BER of oxidatively damaged mtDNA, and this inhibition is relieved once the intracellular levels of HMGA1 drop. This, to our knowledge, is the first reported evidence suggesting that the overexpression of HMGA proteins influences the efficiency of BER of either mtDNA or nDNA.
FIG. 7.
Results of turn-OFF time course experiments in which the efficiency of repair of mtDNA damage (at 1 h post-H2O2 treatment) was measured as a function of time (days) following the addition of tetracycline to the cell culture medium. (A) Results of time course experiments in which tetracycline was added to the medium of HA7C ON cells. (B) Results of time course experiments in which tetracycline was added to the culture medium of control MCF7-Tet cells. In these experiments, the MCF7-Tet cells are, for convenience, referred to as ON because they had been grown in medium lacking tetracycline for 2 weeks prior to drug addition (see Materials and Methods for details).
DISCUSSION
Previous studies demonstrated that, in contrast to OFF cells, the ON cells used in the present experiments acquire abilities for anchorage-independent growth in soft agarose and for the formation of both primary and metastatic tumors when injected into nude mice (100). Immunohistochemical analyses of these metastatic tumors indicated that the ON cells had undergone an epithelial-mesenchymal transition, demonstrating that HMGA1 overexpression promotes both neoplastic progression and metastatic potential of cancer cells. Furthermore, and of considerable importance for the present study, these cancerous phenotypic changes were reversible following turn-off of HMGA1 transgene expression (100). Additionally, stable transfection of nontransgenic human cancer cells (e.g., Hs578T and HeLa cells) with either antisense or dominant-negative HMGA1 constructs has been demonstrated to inhibit their ability to grow in soft agar (100). The reversible nature of many of the cancerous phenotypic changes induced in cells by HMGA overexpression has been confirmed by researchers in numerous laboratories (33, 37, 64, 123, 124) and serves as the basis for the suppression of HMGA gene expression as a promising new therapeutic approach to the treatment of cancer (11, 92, 107, 114).
A comprehensive picture of the complex and varied molecular mechanisms by which overexpression of the HMGA1 gene is initially induced in cancer cells, as well as the way in which this aberrant expression promotes tumor progression, is beginning to emerge (reviewed in references 30, 42, 47, 49, 99, and 123). For example, among the most significant biological changes induced by HMGA1 overexpression are (i) alterations in the transcription of a subset of cancer-related target genes whose promoters are directly regulated by the HMGA1 protein (3, 28, 40, 59, 113, 116) and (ii) the activation of signal transduction pathways that can lead to anomalous downstream gene activation/repression and cancer (29, 100, 115, 117, 124).
The results reported here demonstrate that several mitochondrial characteristics found to be altered in naturally occurring cancers (e.g., mitochondrial mass and mtDNA and ROS levels) (Fig. 4) change in response to varying cellular levels of the HMGA1 protein (Fig. 1 to 3). These findings strongly imply that HMGA1 is somehow causally involved with these changes, but the underlying molecular mechanisms possibly associated with such linkage are unknown. In connection with this issue, it is important to point out the marked difference in the time scale between the changes in HMGA1 protein expression levels and the changes in mitochondrial parameters observed in the turn-OFF experiments. While the overexpression of HMGA1 quickly ceased (within ∼72 h) after tetracycline was added to the culture medium of ON cells (Fig. 1), changes in the mitochondria (e.g., changes in mitochondrial mass and mtDNA levels [Fig. 2], ROS levels [Fig. 3], and mtDNA repair efficiency [Fig. 7A]) occurred on a much longer and more gradual time scale, i.e., in weeks rather than days. This marked temporal difference suggests that the molecular events connecting HMGA1 levels and alterations in mitochondrial structure/function are undoubtedly complex, with some parameters likely to be more closely linked to changes in HMGA1 concentration than others.
That causal relationships between variations in HMGA1 levels and changes in both mtDNA levels and mitochondrial mass potentially exist is supported by the results shown in Fig. 2 and 4. Of these relationships, a causal link between the cellular concentration of HMGA1 and the level of mtDNA (Fig. 2D and E) is perhaps the easier to rationalize in mechanistic terms. Two competing models for how animal mtDNA is replicated in vivo have been advanced. One, the strand asymmetric model, proposes that mtDNA replication initiates in the D-loop through the synthesis of RNA primers from the light-strand promoter (PL) and that both strands are synthesized continuously from physically and temporally distinct sites termed OH and OL (18, 20, 26, 110). The other model proposes that mtDNA replicates in a bidirectional manner from an initiation zone including portions of the D-loop and adjacent gene coding regions and involves conventional double-stranded replication intermediates, with coupled leading- and lagging-strand syntheses (16, 62, 126). Elevated levels of mitochondrial HMGA1 protein may well interfere with either of these replication mechanisms and result in the reduced levels of mtDNA observed. For example, chromatin immunoprecipitation analyses have demonstrated that in ON cells HMGA1 protein binds to regions of the D-loop referred to as conserved sequence blocks (CSBs) (32), regulatory units involved in controlling mtDNA synthesis. Given such localized and specific D-loop binding, we previously suggested that HMGA1 proteins in overexpressing cells likely inhibit mitochondrial replication processes in vivo (32), and findings from the present study are consistent with this proposal. One possible mechanistic explanation for how this inhibition may occur is that HMGA1 is acting at the level of transcript cleavage during the formation of RNA primers from the PL (18, 110). These short RNA primers are required for the transition from RNA to DNA synthesis and for the initiation of mtDNA replication at the heavy-strand origin (OH) by mtDNA polymerase γ (79). During asymmetric mtDNA replication, transcripts from the PL are cleaved in the vicinity of CSB sites, the first of which (CSB I) has a very high A/T content (20, 21) and is a specific binding site for HMGA1 both in living cells and in vitro (32). Since HMGA1 binding induces bends and other structural distortions in DNA substrates (99), any alterations to the CSB region resulting from HMGA1 binding may potentially hinder the transition process, interfere with replication, and result in decreased amounts of mtDNA in overexpressing cells.
The processes of transcription and replication in mitochondria are so intimately coupled (21, 110) that any protein involved in regulating transcription will also likely influence mtDNA replication, as has been demonstrated for the TFAM protein. TFAM is a DNA-binding protein that interacts with mtDNA polymerase γ and another transcription factor (either TFB1M or TFB2M) and stimulates mtDNA transcription through binding to specific D-loop sequences called upstream enhancers (6, 48, 70). In addition to exhibiting sequence-specific promoter recognition, TFAM binds nonspecifically to DNA and, due to its high abundance, plays an architectural role in the stabilization and maintenance of the entire mtDNA molecule and its compaction into a nucleoid structure (4, 68). Substantial evidence, including results from experiments demonstrating that the amount of mtDNA closely parallels (both upward and downward) changes in TFAM levels (41, 69), indicates that TFAM is required for mtDNA replication (reviewed in references 4, 68, and 69).
TFAM is an HMG box-containing protein that, like HMGA1, can bend and unwind DNA—properties linked to the biological functions of both proteins. We have previously demonstrated (58) that HMGA1 has the ability to outcompete HMG box-containing proteins for binding to both B-form DNA and distorted DNA structures similar to those thought to exist in the mtDNA D-loop in vivo (58, 94, 110). More recently, competitive in vitro electrophoretic mobility shift assay experiments (K. J. Wertzler, Z. Wang, K. Ohgaki, D. Kang, and R. Reeves, submitted for publication) have demonstrated that HMGA1 can outcompete TFAM for binding to (and also displace it from) fragments of DNA corresponding to both the D-loop region and the initiation zone proposed in the bidirectional replication model (16, 127). Therefore, a second possible mechanistic explanation for the decreased levels of mtDNA found in ON cells is that HMGA1 binding to these regulatory regions displaces TFAM and interferes with bidirectional mtDNA replication. A caveat to such an explanation, however, is that the same phenotypic result could be produced if TFAM protein production was inhibited by HMGA1 overexpression. To investigate this possibility, the levels of TFAM were monitored in turn-ON and turn-OFF experiments, and as shown by the WBs in Fig. S3A in the supplemental material, the total amount of TFAM in the HA7C cells did not change under these conditions. Nonetheless, in contrast to the total cellular TFAM levels, the amounts of TFAM protein present inside mitochondria did differ between ON and OFF cells. WB analyses demonstrated that mitochondria isolated from HMGA1 ON cells contained significantly less TFAM than mitochondria isolated from OFF cells (Wertzler et al., submitted). Since the mtDNA copy number is directly proportional to TFAM protein levels (41), these electrophoretic mobility shift assay and WB results are consistent with the suggestion that the reduced amounts of mtDNA found in ON cells may be partially attributed to HMGA1 displacement of TFAM from regions of the mitochondrial genome that control replication.
The results of the ON and OFF time course experiments shown in Fig. 2A to C demonstrate that, like mtDNA levels, mitochondrial mass is closely linked to the intracellular levels of HMGA1. In this case, however, a plausible mechanistic explanation that could account for the observed changes in mitochondrial mass is not easily discernible, and these changes may well be the result of complex and more indirect effects of the HMGA1 protein. Mitochondrial biogenesis is controlled through highly coordinated regulatory circuits involving the transcription of both the nuclear and mitochondrial genomes (70, 108). More than 98% of the protein content of mitochondria (i.e., most of the mitochondrial mass) is, however, coded for by nuclear genes (102) whose transcription is controlled by a plethora of external and internal factors (70). HMGA1 proteins are well-characterized accessory transcription factors involved in both positive and negative nuclear gene regulation (99). Hence, the reduction of mitochondrial mass seen in overexpressing cells (Fig. 2C and 4B) may be the result of alterations in nuclear gene expression induced by the HMGA1 protein. Indeed, gene expression microarray analyses have shown that HMGA1 overexpression modulates the transcription of a number of nuclear genes that code for mitochondrial proteins whose altered expression may contribute to reduction in mitochondrial mass (115, 117). Additional research is, therefore, needed in order to elucidate the molecular mechanisms linking mitochondrial mass and HMGA1 levels.
Like the basis for the relationship between HMGA1 levels and mitochondrial mass, possible mechanistic explanations for why cellular ROS levels are influenced by HMGA1 concentration (Fig. 3A and 4D) are not readily apparent and may well involve complex effects of the HMGA1 protein on the transcription of nuclear genes coding for proteins involved in mitochondrial oxidative processes (115). On the other hand, HMGA1 overexpression influences transcription from both the heavy and light strands of mtDNA (31), causing alterations that may also affect ROS levels since the mitochondrial genome codes for integral membrane proteins of the ATP-generating oxidative phosphorylation system. Further investigations are required in order to determine the relationship of HMGA1 to nuclear and mitochondrial transcriptional changes and how these changes relate to cellular ROS levels.
mtDNA is more susceptible to mutation than genomic DNA, and somatic mutations in the mtDNA of several types of cancers have been observed previously (22). Although some of these mtDNA mutations may eventually turn out to be experimental artifacts, a considerable number are likely to be authentic and have a direct relationship with cancer development (65, 104). The present work suggests that HMGA1 overexpression may contribute to increased mutation rates in the mtDNA of cancer cells in at least two ways: first, by inducing increased levels of ROS (Fig. 3 and 4) and, second, by decreasing the efficiency of BER of oxidative lesions (Fig. 5 to 7). The mechanism(s) by which HMGA1 overexpression inhibits BER in mitochondria is unknown but, by analogy to the way HMGA1 proteins inhibit NER of lesions in nDNA (2, 3), is likely to be a combination of (i) binding to lesions in mtDNA and preventing access of repair enzymes and (ii) repressing the transcription of nuclear BER genes that are essential for repair to occur. Although the former possibility has not yet been explored, there is experimental evidence supporting the latter. Gene expression array analyses have shown that transcription of the nuclear gene coding for DNA ligase III is significantly downregulated in HMGA1-overexpressing cells (98). DNA ligase III, in a complex with XRCC1, is the main nick-sealing enzyme involved in nuclear short-patch BER (44, 61). The human DNA ligase III gene encodes a transcript containing two in-frame ATG translation initiation start sites, one used to produce an isoform of the protein with an amino-terminal mitochondrial targeting sequence and the other used to produce a shorter form of the protein involved in BER in the nucleus (73). Inside the mitochondria, DNA ligase III functions independently of XRCC1 (74) and forms a complex with DNA polymerase γ (14) that is involved in both BER of oxidative lesions (75) and the maintenance of mtDNA integrity and stability (74). Given that cells deficient in DNA ligase III are also defective in BER (74), perhaps it is not surprising that, as demonstrated here (Fig. 5 to 7), the efficiency of repair of oxidative lesions in mtDNA is reduced in ON cells. Regardless of the cause, however, the decrease in efficiency of BER in HMGA1-overexpressing cells likely contributes to the increased levels of somatic mutations frequently observed in the mtDNA of tumors.
Results from this study suggest a novel and unexpected biological role for the HMGA1 protein in the complex communication network that exists in cells between the nucleus and the mitochondria (102). In normal cells, a subpopulation of HMGA1 proteins reversibly migrates between the nucleus and mitochondria in a cell cycle-dependent manner (32). The effects of the HMGA1 protein on both mtDNA levels and mitochondrial mass demonstrated here raise the possibility that the tightly controlled shuttling of this protein between the nucleus and mitochondria in normal cells serves as a previously unrecognized means for coordinating the metabolic and replication activities of these two subcellular compartments.
Supplementary Material
Acknowledgments
This work was supported by NIH grant no. GM071760 (to R.R.).
Footnotes
Published ahead of print on 17 August 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abe, N., T. Watanabe, M. Sugiyama, H. Uchimura, G. Chiappetta, A. Fusco, and Y. Atomi. 1999. Determination of high mobility group I(Y) expression level in colorectal neoplasias: a potential diagnostic marker. Cancer Res. 59:1169-1174. [PubMed] [Google Scholar]
- 2.Adair, J. E., Y. Kwon, G. A. Dement, M. J. Smerdon, and R. Reeves. 2005. Inhibition of nucleotide excision repair by high mobility group protein HMGA1. J. Biol. Chem. 280:32184-32192. [DOI] [PubMed] [Google Scholar]
- 3.Adair, J. E., S. C. Maloney, G. A. Dement, K. J. Wertzler, M. J. Smerdon, and R. Reeves. 2007. High-mobility group A1 proteins inhibit expression of nucleotide excision repair factor xeroderma pigmentosum group A. Cancer Res. 67:6044-6052. [DOI] [PubMed] [Google Scholar]
- 4.Alam, T. I., T. Kanki, T. Muta, K. Ukaji, Y. Abe, H. Nakayama, K. Takio, N. Hamasaki, and D. Kang. 2003. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 31:1640-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alvarez, D., C. J. Chou, L. Latella, S. G. Zeitlin, S. Ku, P. L. Puri, P. B. Dervan, and J. M. Gottesfeld. 2006. A two-hit mechanism for pre-mitotic arrest of cancer cell proliferation by a polyamide-alkylator conjugate. Cell Cycle 5:1537-1548. [DOI] [PubMed] [Google Scholar]
- 6.Asin-Cayuela, J., and C. M. Gustafsson. 2007. Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem. Sci. 32:111-117. [DOI] [PubMed] [Google Scholar]
- 7.Bandiera, A., D. Bonifacio, G. Manfioletti, F. Mantovani, A. Rustighi, F. Zanconati, A. Fusco, L. Di Bonito, and V. Giancotti. 1998. Expression of HMGI(Y) proteins in squamous intraepithelial and invasive lesions of the uterine cervix. Cancer Res. 58:426-431. [PubMed] [Google Scholar]
- 8.Basso, D., F. Navaglia, P. Fogar, C. F. Zambon, E. Greco, S. Schiavon, M. Fasolo, A. Stranges, A. Falda, A. Padoan, E. Fadi, S. Pedrazzoli, and M. Plebani. 2007. DNA repair pathways and mitochondrial DNA mutations in gastrointestinal carcinogenesis. Clin. Chim. Acta 381:50-55. [DOI] [PubMed] [Google Scholar]
- 9.Begum, N., J. M. Pash, and J. S. Bhorjee. 1990. Expression and synthesis of high mobility group chromosomal proteins in different rat skeletal cell lines during myogenesis. J. Biol. Chem. 265:11936-11941. [PubMed] [Google Scholar]
- 10.Berdanier, C. D. 2005. Mitochondria in health and disease. Taylor & Francis, New York, NY.
- 11.Berlingieri, M. T., G. M. Pierantoni, V. Giancotti, M. Santoro, and A. Fusco. 2002. Thyroid cell transformation requires the expression of the HMGA1 proteins. Oncogene 21:2971-2980. [DOI] [PubMed] [Google Scholar]
- 12.Berneburg, M., Y. Kamenisch, and J. Krutmann. 2006. Repair of mitochondrial DNA in aging and carcinogenesis. Photochem. Photobiol. Sci. 5:190-198. [DOI] [PubMed] [Google Scholar]
- 13.Berneburg, M., Y. Kamenisch, J. Krutmann, and M. Rocken. 2006. “To repair or not to repair—no longer a question”: repair of mitochondrial DNA shielding against age and cancer. Exp. Dermatol. 15:1005-1015. [DOI] [PubMed] [Google Scholar]
- 14.Bogenhagen, D. F., K. G. Pinz, and R. M. Perez-Jannotti. 2001. Enzymology of mitochondrial base excision repair. Prog. Nucleic Acid Res. Mol. Biol. 68:257-271. [DOI] [PubMed] [Google Scholar]
- 15.Bohr, V. A., and G. L. Dianov. 1999. Oxidative DNA damage processing in nuclear and mitochondrial DNA. Biochimie 81:155-160. [DOI] [PubMed] [Google Scholar]
- 16.Bowmaker, M., M. Y. Yang, T. Yasukawa, A. Reyes, H. T. Jacobs, J. A. Huberman, and I. J. Holt. 2003. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J. Biol. Chem. 278:50961-50969. [DOI] [PubMed] [Google Scholar]
- 17.Brandon, M., P. Baldi, and D. C. Wallace. 2006. Mitochondrial mutations in cancer. Oncogene 25:4647-4662. [DOI] [PubMed] [Google Scholar]
- 18.Brown, T. A., C. Cecconi, A. N. Tkachuk, C. Bustamante, and D. A. Clayton. 2005. Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes Dev. 19:2466-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carew, J. S., and P. Huang. 2002. Mitochondrial defects in cancer. Mol. Cancer 1:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang, D. D., and D. A. Clayton. 1985. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc. Natl. Acad. Sci. USA 82:351-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang, D. D., W. W. Hauswirth, and D. A. Clayton. 1985. Replication priming and transcription initiate from precisely the same site in mouse mitochondrial DNA. EMBO J. 4:1559-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatterjee, A., E. Mambo, and D. Sidransky. 2006. Mitochondrial DNA mutations in human cancer. Oncogene 25:4663-4674. [DOI] [PubMed] [Google Scholar]
- 23.Chatterjee, A., E. Mambo, Y. Zhang, T. Deweese, and D. Sidransky. 2006. Targeting of mutant hogg1 in mammalian mitochondria and nucleus: effect on cellular survival upon oxidative stress. BMC Cancer 6:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi, Y. S., B. K. Ryu, H. K. Min, S. W. Lee, and Y. K. Pak. 2005. Analysis of proteome bound to D-loop region of mitochondrial DNA by DNA-linked affinity chromatography and reverse-phase liquid chromatography/tandem mass spectrometry. Ann. N. Y. Acad. Sci. 1042:88-100. [DOI] [PubMed] [Google Scholar]
- 25.Chuma, M., N. Saeki, Y. Yamamoto, T. Ohta, M. Asaka, S. Hirohashi, and M. Sakamoto. 2004. Expression profiling in hepatocellular carcinoma with intrahepatic metastasis: identification of high-mobility group I(Y) protein as a molecular marker of hepatocellular carcinoma metastasis. Keio J. Med. 53:90-97. [DOI] [PubMed] [Google Scholar]
- 26.Clayton, D. A. 1982. Replication of animal mitochondrial DNA. Cell 28:693-705. [DOI] [PubMed] [Google Scholar]
- 27.Clayton, D. A., J. N. Doda, and E. C. Friedberg. 1974. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc. Natl. Acad. Sci. USA 71:2777-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cleynen, I., J. R. Brants, K. Peeters, R. Deckers, M. Debiec-Rychter, R. Sciot, W. J. Van de Ven, and M. M. Petit. 2007. HMGA2 regulates transcription of the Imp2 gene via an intronic regulatory element in cooperation with nuclear factor-κB. Mol. Cancer Res. 5:363-372. [DOI] [PubMed] [Google Scholar]
- 29.Cleynen, I., C. Huysmans, T. Sasazuki, S. Shirasawa, W. Van de Ven, and K. Peeters. 2007. Transcriptional control of the human high mobility group A1 gene: basal and oncogenic Ras-regulated expression. Cancer Res. 67:4620-4629. [DOI] [PubMed] [Google Scholar]
- 30.Cleynen, I., and W. J. Van de Ven. 2008. The HMGA proteins: a myriad of functions (review). Int. J. Oncol. 32:289-305. [PubMed] [Google Scholar]
- 31.Dement, G. A., S. C. Maloney, and R. Reeves. 2007. Nuclear HMGA1 nonhistone chromatin proteins directly influence mitochondrial transcription, maintenance, and function. Exp. Cell Res. 313:77-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dement, G. A., N. R. Treff, N. S. Magnuson, V. Franceschi, and R. Reeves. 2005. Dynamic mitochondrial localization of nuclear transcription factor HMGA1. Exp. Cell Res. 307:388-401. [DOI] [PubMed] [Google Scholar]
- 33.Dhar, A., J. Hu, R. Reeves, L. M. Resar, and N. H. Colburn. 2004. Dominant-negative c-Jun (TAM67) target genes: HMGA1 is required for tumor promoter-induced transformation. Oncogene 23:4466-4476. [DOI] [PubMed] [Google Scholar]
- 34.Dianov, G. L., N. Souza-Pinto, S. G. Nyaga, T. Thybo, T. Stevnsner, and V. A. Bohr. 2001. Base excision repair in nuclear and mitochondrial DNA. Prog. Nucleic Acid Res. Mol. Biol. 68:285-297. [DOI] [PubMed] [Google Scholar]
- 35.Di Cello, F., J. Hillion, A. Hristov, L. J. Wood, M. Mukherjee, A. Schuldenfrei, J. Kowalski, R. Bhattacharya, R. Ashfaq, and L. M. Resar. 2008. HMGA2 participates in transformation in human lung cancer. Mol. Cancer Res. 6:743-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dmitrenko, V., K. Shostak, O. Boyko, O. Khomenko, V. Rozumenko, T. Malisheva, M. Shamayev, Y. Zozulya, and V. Kavsan. 2005. Reduction of the transcription level of the mitochondrial genome in human glioblastoma. Cancer Lett. 218:99-107. [DOI] [PubMed] [Google Scholar]
- 37.Dolde, C. E., M. Mukherjee, C. Cho, and L. M. Resar. 2002. HMG-I/Y in human breast cancer cell lines. Breast Cancer Res. Treat. 71:181-191. [DOI] [PubMed] [Google Scholar]
- 38.Druzhyna, N. M., S. B. Hollensworth, M. R. Kelley, G. L. Wilson, and S. P. Ledoux. 2003. Targeting human 8-oxoguanine glycosylase to mitochondria of oligodendrocytes protects against menadione-induced oxidative stress. Glia 42:370-378. [DOI] [PubMed] [Google Scholar]
- 39.Druzhyna, N. M., G. L. Wilson, and S. P. Ledoux. 2008. Mitochondrial DNA repair in aging and disease. Mech. Ageing Dev. 129:383-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duncan, B., and K. Zhao. 2007. HMGA1 mediates the activation of the CRYAB promoter by BRG1. DNA Cell Biol. 26:745-752. [DOI] [PubMed] [Google Scholar]
- 41.Ekstrand, M. I., M. Falkenberg, A. Rantanen, C. B. Park, M. Gaspari, K. Hultenby, P. Rustin, C. M. Gustafsson, and N. G. Larsson. 2004. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 13:935-944. [DOI] [PubMed] [Google Scholar]
- 42.Fedele, M., G. M. Pierantoni, R. Visone, and A. Fusco. 2006. Critical role of the HMGA2 gene in pituitary adenomas. Cell Cycle 5:2045-2048. [DOI] [PubMed] [Google Scholar]
- 43.Franco, R., F. Esposito, M. Fedele, G. Liguori, G. M. Pierantoni, G. Botti, D. Tramontano, A. Fusco, and P. Chieffi. 2008. Detection of high-mobility group proteins A1 and A2 represents a valid diagnostic marker in post-pubertal testicular germ cell tumours. J. Pathol. 214:58-64. [DOI] [PubMed] [Google Scholar]
- 44.Friedberg, E. C., G. C. Walker, W. Siede, R. D. Wood, R. A. Schultz, and T. Ellenberger. 2006. DNA repair and mutagenesis. ASM Press, Washington, DC.
- 45.Frisard, M., and E. Ravussin. 2006. Energy metabolism and oxidative stress: impact on the metabolic syndrome and the aging process. Endocrine 29:27-32. [DOI] [PubMed] [Google Scholar]
- 46.Funayama, R., M. Saito, H. Tanobe, and F. Ishikawa. 2006. Loss of linker histone H1 in cellular senescence. J. Cell Biol. 175:869-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fusco, A., and M. Fedele. 2007. Roles of HMGA proteins in cancer. Nat. Rev. Cancer 7:899-910. [DOI] [PubMed] [Google Scholar]
- 48.Gaspari, M., N. G. Larsson, and C. M. Gustafsson. 2004. The transcription machinery in mammalian mitochondria. Biochim. Biophys. Acta 1659:148-152. [DOI] [PubMed] [Google Scholar]
- 49.Giannini, G., F. Cerignoli, M. Mellone, I. Massimi, C. Ambrosi, C. Rinaldi, and A. Gulino. 2005. Molecular mechanism of HMGA1 deregulation in human neuroblastoma. Cancer Lett. 228:97-104. [DOI] [PubMed] [Google Scholar]
- 50.Glaichennhaus, N., and P. Leopold. 1986. Increased levels of mitochondrial gene expression in rat fibroblast cells immortalized or transformed by viral and cellular oncogenes. EMBO J. 5:1261-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gogvadze, V., S. Orrenius, and B. Zhivotovsky. 2008. Mitochondria in cancer cells: what is so special about them? Trends Cell Biol. 18:165-173. [DOI] [PubMed] [Google Scholar]
- 52.Grishko, V. I., L. I. Rachek, D. R. Spitz, G. L. Wilson, and S. P. Ledoux. 2005. Contribution of mitochondrial DNA repair to cell resistance from oxidative stress. J. Biol. Chem. 280:8901-8905. [DOI] [PubMed] [Google Scholar]
- 53.Hackett, A. J., H. S. Smith, E. L. Springer, R. B. Owens, W. A. Nelson-Rees, J. L. Riggs, and M. B. Gardner. 1977. Two syngeneic cell lines from human breast tissue: the aneuploid mammary epithelial (Hs578T) and the diploid myoepithelial (Hs578Bst) cell lines. J. Natl. Cancer Inst. 58:1795-1806. [DOI] [PubMed] [Google Scholar]
- 54.Hammond, S. M., and N. E. Sharpless. 2008. HMGA2, microRNAs, and stem cell aging. Cell 135:1013-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haugen, D. R., O. Fluge, L. J. Reigstad, J. E. Varhaug, and J. R. Lillehaug. 2003. Increased expression of genes encoding mitochondrial proteins in papillary thyroid carcinomas. Thyroid 13:613-620. [DOI] [PubMed] [Google Scholar]
- 56.Heerdt, B. G., M. A. Houston, and L. H. Augenlicht. 2005. The intrinsic mitochondrial membrane potential of colonic carcinoma cells is linked to the probability of tumor progression. Cancer Res. 65:9861-9867. [DOI] [PubMed] [Google Scholar]
- 57.Heerdt, B. G., M. A. Houston, and L. H. Augenlicht. 2006. Growth properties of colonic tumor cells are a function of the intrinsic mitochondrial membrane potential. Cancer Res. 66:1591-1596. [DOI] [PubMed] [Google Scholar]
- 58.Hill, D. A., and R. Reeves. 1997. Competition between HMG-I(Y), HMG-1 and histone H1 on four-way junction DNA. Nucleic Acids Res. 25:3523-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hillion, J., S. Dhara, T. F. Sumter, M. Mukherjee, F. Di Cello, A. Belton, J. Turkson, S. Jaganathan, L. Cheng, Z. Ye, R. Jove, P. Aplan, Y. W. Lin, K. Wertzler, R. Reeves, O. Elbahlouh, J. Kowalski, R. Bhattacharya, and L. M. Resar. 2008. The high-mobility group A1a/signal transducer and activator of transcription-3 axis: an Achilles heel for hematopoietic malignancies? Cancer Res. 68:10121-10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hock, R., T. Furusawa, T. Ueda, and M. Bustin. 2007. HMG chromosomal proteins in development and disease. Trends Cell Biol. 17:72-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366-374. [DOI] [PubMed] [Google Scholar]
- 62.Holt, I. J., and H. T. Jacobs. 2003. Response: the mitochondrial DNA replication bubble has not burst. Trends Biochem. Sci. 28:355-356. [DOI] [PubMed] [Google Scholar]
- 63.Holth, L. T., A. E. Thorlacius, and R. Reeves. 1997. Effects of epidermal growth factor and estrogen on the regulation of the HMG-I/Y gene in human mammary epithelial cell lines. DNA Cell Biol. 16:1299-1309. [DOI] [PubMed] [Google Scholar]
- 64.Hommura, F., M. Katabami, V. D. Leaner, H. Donninger, T. F. Sumter, L. M. Resar, and M. J. Birrer. 2004. HMG-I/Y is a c-Jun/activator protein-1 target gene and is necessary for c-Jun-induced anchorage-independent growth in Rat1a cells. Mol. Cancer Res. 2:305-314. [PubMed] [Google Scholar]
- 65.Ishikawa, K., K. Takenaga, M. Akimoto, N. Koshikawa, A. Yamaguchi, H. Imanishi, K. Nakada, Y. Honma, and J. Hayashi. 2008. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science 320:661-664. [DOI] [PubMed] [Google Scholar]
- 66.Jakupciak, J. P., W. Wang, M. E. Markowitz, D. Ally, M. Coble, S. Srivastava, A. Maitra, P. E. Barker, D. Sidransky, and C. D. O'Connell. 2005. Mitochondrial DNA as a cancer biomarker. J. Mol. Diagn. 7:258-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang, Y., D. Tong, G. Lou, Y. Zhang, and J. Geng. 2008. Expression of RUNX3 gene, methylation status and clinicopathological significance in breast cancer and breast cancer cell lines. Pathobiology 75:244-251. [DOI] [PubMed] [Google Scholar]
- 68.Kanki, T., H. Nakayama, N. Sasaki, K. Takio, T. I. Alam, N. Hamasaki, and D. Kang. 2004. Mitochondrial nucleoid and transcription factor A. Ann. N. Y. Acad. Sci. 1011:61-68. [DOI] [PubMed] [Google Scholar]
- 69.Kanki, T., K. Ohgaki, M. Gaspari, C. M. Gustafsson, A. Fukuoh, N. Sasaki, N. Hamasaki, and D. Kang. 2004. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol. Cell. Biol. 24:9823-9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kelly, D. P., and R. C. Scarpulla. 2004. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 18:357-368. [DOI] [PubMed] [Google Scholar]
- 71.Kislinger, T., B. Cox, A. Kannan, C. Chung, P. Hu, A. Ignatchenko, M. S. Scott, A. O. Gramolini, Q. Morris, M. T. Hallett, J. Rossant, T. R. Hughes, B. Frey, and A. Emili. 2006. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell 125:173-186. [DOI] [PubMed] [Google Scholar]
- 72.Kraytsberg, Y., M. Schwartz, T. A. Brown, K. Ebralidse, W. S. Kunz, D. A. Clayton, J. Vissing, and K. Khrapko. 2004. Recombination of human mitochondrial DNA. Science 304:981. [DOI] [PubMed] [Google Scholar]
- 73.Lakshmipathy, U., and C. Campbell. 1999. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol. Cell. Biol. 19:3869-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lakshmipathy, U., and C. Campbell. 2000. Mitochondrial DNA ligase III function is independent of Xrcc1. Nucleic Acids Res. 28:3880-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lakshmipathy, U., and C. Campbell. 2001. Antisense-mediated decrease in DNA ligase III expression results in reduced mitochondrial DNA integrity. Nucleic Acids Res. 29:668-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Larsen, N. B., M. Rasmussen, and L. J. Rasmussen. 2005. Nuclear and mitochondrial DNA repair: similar pathways? Mitochondrion 5:89-108. [DOI] [PubMed] [Google Scholar]
- 77.Laury-Kleintop, L. D., E. C. Coronel, M. K. Lange, T. Tachovsky, S. Longo, S. Tucker, and J. A. Alhadeff. 1995. Western blotting and isoform analysis of cathepsin D from normal and malignant human breast cell lines. Breast Cancer Res. Treat. 35:211-220. [DOI] [PubMed] [Google Scholar]
- 78.Ledoux, S. P., and G. L. Wilson. 2001. Base excision repair of mitochondrial DNA damage in mammalian cells. Prog. Nucleic Acid Res. Mol. Biol. 68:273-284. [DOI] [PubMed] [Google Scholar]
- 79.Lee, D. Y., and D. A. Clayton. 1998. Initiation of mitochondrial DNA replication by transcription and R-loop processing. J. Biol. Chem. 273:30614-30621. [DOI] [PubMed] [Google Scholar]
- 80.Lee, H. C., S. H. Li, J. C. Lin, C. C. Wu, D. C. Yeh, and Y. H. Wei. 2004. Somatic mutations in the D-loop and decrease in the copy number of mitochondrial DNA in human hepatocellular carcinoma. Mutat. Res. 547:71-78. [DOI] [PubMed] [Google Scholar]
- 81.Maloney, S. C., J. E. Adair, M. J. Smerdon, and R. Reeves. 2007. Gene-specific nucleotide excision repair is impaired in human cells expressing elevated levels of high mobility group A1 nonhistone proteins. DNA Repair (Amsterdam) 6:1371-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martinez, H. J., M. Fedele, S. Battista, F. Pentimalli, M. Kruhoffer, C. Arra, T. F. Orntoft, C. M. Croce, and A. Fusco. 2004. Identification of the genes up- and down-regulated by the high mobility group A1 (HMGA1) proteins: tissue specificity of the HMGA1-dependent gene regulation. Cancer Res. 64:5728-5735. [DOI] [PubMed] [Google Scholar]
- 83.Maximo, V., T. Botelho, J. Capela, P. Soares, J. Lima, A. Taveira, T. Amaro, A. P. Barbosa, A. Preto, H. R. Harach, D. Williams, and M. Sobrinho-Simoes. 2005. Somatic and germline mutation in GRIM-19, a dual function gene involved in mitochondrial metabolism and cell death, is linked to mitochondrion-rich (Hurthle cell) tumours of the thyroid. Br. J. Cancer 92:1892-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Modica-Napolitano, J. S., M. Kulawiec, and K. K. Singh. 2007. Mitochondria and human cancer. Curr. Mol. Med. 7:121-131. [DOI] [PubMed] [Google Scholar]
- 85.Monzen, K., Y. Ito, A. T. Naito, H. Kasai, Y. Hiroi, D. Hayashi, I. Shiojima, T. Yamazaki, K. Miyazono, M. Asashima, R. Nagai, and I. Komuro. 2008. A crucial role of a high mobility group protein HMGA2 in cardiogenesis. Nat. Cell Biol. 10:567-574. [DOI] [PubMed] [Google Scholar]
- 86.Myers, K. A., R. Saffhill, and P. J. O'Connor. 1988. Repair of alkylated purines in the hepatic DNA of mitochondria and nuclei in the rat. Carcinogenesis 9:285-292. [DOI] [PubMed] [Google Scholar]
- 87.Nakamura, J., E. R. Purvis, and J. A. Swenberg. 2003. Micromolar concentrations of hydrogen peroxide induce oxidative DNA lesions more efficiently than millimolar concentrations in mammalian cells. Nucleic Acids Res. 31:1790-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nishino, J., I. Kim, K. Chada, and S. J. Morrison. 2008. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf expression. Cell 135:227-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nucci, M. R., D. H. Castrillon, H. Bai, B. J. Quade, T. A. Ince, D. R. Genest, K. R. Lee, G. L. Mutter, and C. P. Crum. 2003. Biomarkers in diagnostic obstetric and gynecologic pathology: a review. Adv. Anat. Pathol. 10:55-68. [DOI] [PubMed] [Google Scholar]
- 90.Pedersen, P. L. 1978. Tumor mitochondria and the bioenergetics of cancer cells. Prog. Exp. Tumor Res. 22:190-274. [DOI] [PubMed] [Google Scholar]
- 91.Penta, J. S., F. M. Johnson, J. T. Wachsman, and W. C. Copeland. 2001. Mitochondrial DNA in human malignancy. Mutat. Res. 488:119-133. [DOI] [PubMed] [Google Scholar]
- 92.Pentimalli, F., M. Dentice, M. Fedele, G. M. Pierantoni, L. Cito, P. Pallante, M. Santoro, G. Viglietto, P. Dal Cin, and A. Fusco. 2003. Suppression of HMGA2 protein synthesis could be a tool for the therapy of well differentiated liposarcomas overexpressing HMGA2. Cancer Res. 63:7423-7427. [PubMed] [Google Scholar]
- 93.Petros, J. A., A. K. Baumann, E. Ruiz-Pesini, M. B. Amin, C. Q. Sun, J. Hall, S. Lim, M. M. Issa, W. D. Flanders, S. H. Hosseini, F. F. Marshall, and D. C. Wallace. 2005. mtDNA mutations increase tumorigenicity in prostate cancer. Proc. Natl. Acad. Sci. USA 102:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pohjoismaki, J. L., S. Goffart, H. Tyynismaa, S. Willcox, T. Ide, D. Kang, A. Suomalainen, P. J. Karhunen, J. D. Griffith, I. J. Holt, and H. Jacobs. 2009. Human heart mitochondrial DNA is organized in complex catenated networks containing abundant four-way junctions and replication forks. J. Biol. Chem. 284:21446-21457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rajeswari, M. R., D. Singh, A. Jain, and R. Ray. 2001. Elevated levels of high-mobility-group chromosomal proteins, HMGA1, in murine skin carcinoma. Cancer Lett. 173:93-99. [DOI] [PubMed] [Google Scholar]
- 96.Reeves, R. 2000. Structure and function of the HMGI(Y) family of architectural transcription factors. Environ. Health Perspect. 108(Suppl. 5):803-809. [DOI] [PubMed] [Google Scholar]
- 97.Reeves, R. 2001. Molecular biology of HMGA proteins: hubs of nuclear function. Gene 277:63-81. [DOI] [PubMed] [Google Scholar]
- 98.Reeves, R., and J. E. Adair. 2005. Role of high mobility group (HMG) chromatin proteins in DNA repair. DNA Repair (Amsterdam) 4:926-938. [DOI] [PubMed] [Google Scholar]
- 99.Reeves, R., and L. Beckerbauer. 2001. HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim. Biophys. Acta 1519:13-29. [DOI] [PubMed] [Google Scholar]
- 100.Reeves, R., D. D. Edberg, and Y. Li. 2001. Architectural transcription factor HMGI(Y) promotes tumor progression and mesenchymal transition of human epithelial cells. Mol. Cell. Biol. 21:575-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Richter, C. 1993. Pro-oxidants and mitochondrial Ca2+: their relationship to apoptosis and oncogenesis. FEBS Lett. 325:104-107. [DOI] [PubMed] [Google Scholar]
- 102.Ryan, M. T., and N. J. Hoogenraad. 2007. Mitochondrial-nuclear communications. Annu. Rev. Biochem. 76:701-722. [DOI] [PubMed] [Google Scholar]
- 103.Sanna, S., A. U. Jackson, R. Nagaraja, C. J. Willer, W. M. Chen, L. L. Bonnycastle, H. Shen, N. Timpson, G. Lettre, G. Usala, P. S. Chines, H. M. Stringham, L. J. Scott, M. Dei, S. Lai, G. Albai, L. Crisponi, S. Naitza, K. F. Doheny, E. W. Pugh, Y. Ben Shlomo, S. Ebrahim, D. A. Lawlor, R. N. Bergman, R. M. Watanabe, M. Uda, J. Tuomilehto, J. Coresh, J. N. Hirschhorn, A. R. Shuldiner, D. Schlessinger, F. S. Collins, S. G. Davey, E. Boerwinkle, A. Cao, M. Boehnke, G. R. Abecasis, and K. L. Mohlke. 2008. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat. Genet. 40:198-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Santos, C., M. Martinez, M. Lima, Y. J. Hao, N. Simoes, and R. Montiel. 2008. Mitochondrial DNA mutations in cancer: a review. Curr. Top. Med. Chem. 8:1351-1366. [DOI] [PubMed] [Google Scholar]
- 105.Santos, J. H., B. S. Mandavilli, and B. Van Houten. 2002. Measuring oxidative mtDNA damage and repair using quantitative PCR, p. 159-176. In W. C. Copeland (ed.), Mitochondrial DNA: methods and protocols, vol. 197. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 106.Santos, J. H., J. N. Meyer, B. S. Mandavilli, and B. Van Houten. 2006. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol. Biol. 314:183-199. [DOI] [PubMed] [Google Scholar]
- 107.Scala, S., G. Portella, M. Fedele, G. Chiappetta, and A. Fusco. 2000. Adenovirus-mediated suppression of HMGI(Y) protein synthesis as potential therapy of human malignant neoplasias. Proc. Natl. Acad. Sci. USA 97:4256-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Scarpulla, R. C. 2008. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 88:611-638. [DOI] [PubMed] [Google Scholar]
- 109.Schaffer, S. W., and M. S. Suleiman. 2007. Mitochondria: the dynamic organelle. Springer, Berlin, Germany.
- 110.Shadel, G. S., and D. A. Clayton. 1997. Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 66:409-435. [DOI] [PubMed] [Google Scholar]
- 111.Takaha, N., L. M. Resar, D. Vindivich, and D. S. Coffey. 2004. High mobility group protein HMGI(Y) enhances tumor cell growth, invasion, and matrix metalloproteinase-2 expression in prostate cancer cells. Prostate 60:160-167. [DOI] [PubMed] [Google Scholar]
- 112.Tallini, G., and P. Dal Cin. 1999. HMGI(Y) and HMGI-C dysregulation: a common occurrence in human tumors. Adv. Anat. Pathol. 6:237-246. [PubMed] [Google Scholar]
- 113.Tesfaye, A., F. Di Cello, J. Hillion, B. M. Ronnett, O. Elbahloul, R. Ashfaq, S. Dhara, E. Prochownik, K. Tworkoski, R. Reeves, R. Roden, L. H. Ellenson, D. L. Huso, and L. M. Resar. 2007. The high-mobility group A1 gene up-regulates cyclooxygenase 2 expression in uterine tumorigenesis. Cancer Res. 67:3998-4004. [DOI] [PubMed] [Google Scholar]
- 114.Trapasso, F., M. Sarti, R. Cesari, S. Yendamuri, K. R. Dumon, R. I. Aqeilan, F. Pentimalli, L. Infante, H. Alder, N. Abe, T. Watanabe, G. Viglietto, C. M. Croce, and A. Fusco. 2004. Therapy of human pancreatic carcinoma based on suppression of HMGA1 protein synthesis in preclinical models. Cancer Gene Ther. 11:633-641. [DOI] [PubMed] [Google Scholar]
- 115.Treff, N. R. 2003. Molecular genetics of HMGA1-mediated malignant transformation of breast cancer cells. Ph.D. dissertation. Washington State University, Pullman.
- 116.Treff, N. R., G. A. Dement, J. E. Adair, R. L. Britt, R. Nie, J. E. Shima, W. E. Taylor, and R. Reeves. 2004. Human KIT ligand promoter is positively regulated by HMGA1 in breast and ovarian cancer cells. Oncogene 23:8557-8562. [DOI] [PubMed] [Google Scholar]
- 117.Treff, N. R., D. Pouchnik, G. A. Dement, R. L. Britt, and R. Reeves. 2004. High-mobility group A1a protein regulates Ras/ERK signaling in MCF-7 human breast cancer cells. Oncogene 23:777-785. [DOI] [PubMed] [Google Scholar]
- 118.Tseng, L. M., P. H. Yin, C. W. Chi, C. Y. Hsu, C. W. Wu, L. M. Lee, Y. H. Wei, and H. C. Lee. 2006. Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosomes Cancer 45:629-638. [DOI] [PubMed] [Google Scholar]
- 119.Warburg, O. 1956. On respiratory impairment in cancer cells. Science 124:269-270. [PubMed] [Google Scholar]
- 120.Warburg, O. 1956. On the origin of cancer cells. Science 123:309-314. [DOI] [PubMed] [Google Scholar]
- 121.Weedon, M. N., H. Lango, C. M. Lindgren, C. Wallace, D. M. Evans, M. Mangino, R. M. Freathy, J. R. Perry, S. Stevens, A. S. Hall, N. J. Samani, B. Shields, I. Prokopenko, M. Farrall, A. Dominiczak, T. Johnson, S. Bergmann, J. S. Beckmann, P. Vollenweider, D. M. Waterworth, V. Mooser, C. N. Palmer, A. D. Morris, W. H. Ouwehand, J. H. Zhao, S. Li, R. J. Loos, I. Barroso, P. Deloukas, M. S. Sandhu, E. Wheeler, N. Soranzo, M. Inouye, N. J. Wareham, M. Caulfield, P. B. Munroe, A. T. Hattersley, M. I. McCarthy, and T. M. Frayling. 2008. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 40:575-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wisniewski, J. R., and R. Schwanbeck. 2000. High mobility group I/Y: multifunctional chromosomal proteins causally involved in tumor progression and malignant transformation (review). Int. J. Mol. Med. 6:409-419. [DOI] [PubMed] [Google Scholar]
- 123.Wood, L. J., J. F. Maher, T. E. Bunton, and L. M. Resar. 2000. The oncogenic properties of the HMG-I gene family. Cancer Res. 60:4256-4261. [PubMed] [Google Scholar]
- 124.Wood, L. J., M. Mukherjee, C. E. Dolde, Y. Xu, J. F. Maher, T. E. Bunton, J. B. Williams, and L. M. Resar. 2000. HMG-I/Y, a new c-Myc target gene and potential oncogene. Mol. Cell. Biol. 20:5490-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yamada, S., S. Nomoto, T. Fujii, et al. 2006. Correlation between copy number of mitochondrial DNA and clinico-pathologic parameters of hepatocellular carcinoma. Eur. J. Surg. Oncol. 32:303-307. [DOI] [PubMed] [Google Scholar]
- 126.Yasukawa, T., A. Reyes, T. J. Cluett, M. Y. Yang, M. Bowmaker, H. T. Jacobs, and I. J. Holt. 2006. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 25:5358-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yasukawa, T., M. Y. Yang, H. T. Jacobs, and I. J. Holt. 2005. A bidirectional origin of replication maps to the major noncoding region of human mitochondrial DNA. Mol. Cell 18:651-662. [DOI] [PubMed] [Google Scholar]
- 128.Yip, P. H., H. C. Lee, G. Y. Chau, Y. T. Wu, S. H. Li, W. Y. Lui, Y. H. Wei, T. Y. Liu, and C. W. Chi. 2004. Alteration of the copy number and deletion of mitochondrial DNA in human hepatocellular carcinoma. Br. J. Cancer 90:2390-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang, R., W. Chen, and P. D. Adams. 2007. Molecular dissection of formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 27:2343-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.